Abstract
HIV-1 mutations which reduce or abolish cytotoxic T lymphocyte responses against virus-infected cells are frequently selected in acute and chronic HIV-infection. Among population HIV-1 sequences, immune selection is evident as HLA allele-associated substitutions of amino acids within or near CD8 T cell epitopes. In these cases, the non-adapted epitope is susceptible to immune recognition until an escape mutation renders the epitope less immunogenic. However, several population-based studies have independently identified HLA-associated viral changes which lead to formation of a new T cell epitope, suggesting that the immune responses which these variants or “neo-epitopes” elicit provide an evolutionary advantage to the virus rather than the host. Here, we examined functional characteristics of eight CD8 T cell responses that result from viral adaptation in 125 HLA-genotyped individuals with chronic HIV-1 infection. Neo-epitopes included well-characterised immunodominant epitopes restricted by common HLA alleles and in most cases, the T cell responses against the neo-epitope exhibited significantly greater functional avidity and higher IFNγ production than T cells for non-adapted epitopes but were not more cytotoxic. Neo-epitope formation and emergence of the cognate T cell response co-incident with a rise in viral load was then observed in-vivo in an acutely infected individual. These findings demonstrate that HIV-1 adaptation not only abrogates immune recognition of early targeted epitopes, but may also increase immune recognition to other epitopes, which elicit immunodominant but non-protective T cell responses. These data have implications for immunodominance associated with polyvalent vaccines based on the diversity of chronic HIV-1 sequences.
Keywords: HIV, Immune escape, T cell response
Introduction
The cytotoxic T lymphocyte (CTL) response is associated with marked reduction of viremia during acute HIV-1 infection in most individuals,1 but ongoing replicative infection in the presence of partially suppressive responses rapidly selects for mutations that allow viral escape from these responses.2 The emergence of HIV-specific CD8 T cell responses commensurate with falling plasma HIV-1 RNA concentration and positive selection of mutations within the targeted T cell epitopes have been directly observed 14 days after HIV p24 antigen detection and preceding HIV-1 antibody seroconversion.3 Studies of HIV-1 seropositive individuals have found evidence of CTL escape mutations in viral sequences as early as 4 weeks after the peak viremia of acute infection4–6 as well as at later time points in chronic infection.7,8 Loss or reduction of HLA-viral epitope binding,8 reduction in T cell receptor (TCR) recognition of the HLA/epitope complex9 or disruption of intracellular processing of viral epitopes10–12 are all well documented mechanisms by which viral mutation leads to evasion from T cell recognition and cytotoxic clearance of virus-infected cells.
Once the virus escapes from initial immunodominant CTL responses, new responses to alternative subdominant epitopes may be generated and select further mutation networks in the viral genome, such that there is often a hierarchy of immunodominant and subdominant CTL responses with successively selected viral adaptations and broadening of the overall T cell response.13,14 At the population level, this intra-host viral evolution is manifest as inter-host viral diversity that correlates with HLA class I alleles.15–25 Population-based studies of HLA-HIV polymorphism associations have revealed a greater diversity of HLA alleles and viral epitope targets putatively associated with immune selection in vivo. Several examples of experimentally characterised HIV-1 mutations known to cause CTL escape have been consistently apparent as HLA allele-associated HIV-1 polymorphisms at the population level.
For example, a mutational network involving Gag residues 210, 242, 248 and others associated with escape from HLA-B57-restricted T cell responses targeting the Gag TW10 (240–249) epitope has been detected consistently as HLA-B57 associated polymorphisms in a number of population-based studies.9,20,25 Somewhat unexpectedly however, this genetics-based approach has also revealed instances where a viral epitope which is more susceptible to CTL cell recognition in T cell assays, is enriched in individuals with carriage of the restricting HLA allele.16,18,25 For example, HLA-A*0301 was associated with an arginine (R) to lysine (K) substitution at position 3 of a known HLA-A*0301-restricted CD8 T cell p7 epitope LR9, however the −A*0301-adapted form of the epitope (LAKNCRAP) elicited higher magnitude and more functionally avid responses than the non-adapted variant in some tested individuals.16 Similarly, a well characterised HLA-B*0702-restricted Gag epitope GL9 which elicited interferon-gamma (IFNγ) responses was relatively enriched in those with carriage of HLA-B*0702 in a subtype C population-based association study.19 In both these cases, the epitope which was “CTL-adapted” in genetic analyses was also immunogenic by standard immunological measures, in contradistinction to the classical description of viral adaptation causing loss of immunogenicity. Whilst de novo CD8 T cell responses to epitope variants26 and cross reactive T cell responses,27 have been described in chronic HIV-1 infection, these have not been explored as a product of viral adaptation and therefore offering strategic advantages for the virus, rather than the host. A population-wide association between enrichment of a particular viral epitope sequence and an HLA allele suggests that the maintenance of the epitope and the cognate epitope-specific T cell response is adaptive for the virus in some way. Furthermore, according to an evolutionary argument, the immune responses that are evaded by HIV-1 may be qualitatively different from those immune responses which are tolerated by the virus and do not select further changes. Understanding the differences between these responses may therefore reveal key aspects of TCR-epitope/HLA interactions and functional T cell immunity which should be either harnessed or avoided by HIV vaccines.
A study of associations between HLA alleles and HIV-1 polymorphisms in a large population of predominantly HIV-1 subtype B infected individuals in the United States of America and Australia detected over 800 HLA class I allele-specific polymorphisms across the HIV-1 genome.25 Scanning for known or predicted CD8 T cell epitopes around these polymorphic sites in the study cohort sequences identified 97 HLA-viral polymorphism associations in which the amino acid substitution led to the creation rather than abolition of an epitope with the same HLA restriction overlapping or adjacent to the original epitope. There were nine HLA-epitope combinations in particular in which the HLA-adapted epitope sequence corresponded to a published epitope for which CD8 T cell recognition had been well established in the scientific literature (see supplementary data Table).13, 28–38 For example, HLA-B*0702 was associated with a serine (S) to glycine (G) change at position 357 of Gag, creating the previously mentioned HLA-B*0702-restricted Gag epitope GL9 (GPGHKARVL). Rather than the restricting HLA allele driving a departure from this sequence in keeping with loss of the epitope, HLA-B*0702 was associated with creation of the epitope, making the immune-susceptible epitope also HLA-adapted, rather than non-adapted or “wild-type”. Other such adapted “neo-epitopes” included the extensively studied immunodominant HLA-A*0201 restricted Gag 77–85 SL9 (SLYNTVATL) epitope and Vpr 59–67 AL9 (AIIRILQQL), −A*0301-restricted Pol 424–432 QR9 (QIYPGIKVR), −B*0702 and −B*4201 restricted Nef 128–137 TL10 (TPGPGVRYPL), −B*1503 Nef 183–191 WF9 (WRFDSRLAF) and −B*4402-restricted Pol 724–734 QW11 (QEEHEKYHSNW) and HLA-C*0702 restricted Nef 105–115 KY11 (KRQEILDLWVY) epitope, all of which have been detected as one of few commonly detected responses in chronically infected individuals39 (see supplementary data Table). Independent HLA association studies and observational studies of HIV sequence evolution conducted in geographically distinct populations have also shown the same associations and direction of substitutions for four of these cases: HLA-A*03 Pol QR9,17,19,24 HLA−B*0702 Gag GL9,13,18,19 HLA-A*0201 Vpr AL917,19 and HLAB*1503 WF9.13 We chose to focus on these nine HLA-epitope combinations in detail as they were all well characterised published optimal epitopes restricted by prevalent HLA alleles and demonstrated by others to elicit measureable and usually immunodominant CD8 T cell responses in chronically infected subjects (http://www.hiv.lanl.gov/content/immunology/tables/optimal_ctl_summary.html).39 The concordance in significance level and direction of the specific amino acid substitution in four of these cases from independently conducted studies reduces the likelihood that the non-adapted and adapted epitope sequences had been falsely assigned in this study.25 Functional characteristics of the memory CD8 T cell response against the non-adapted and adapted epitopes in 125 individuals in the Western Australian HIV Cohort Study (WAHCS) with chronic HIV infection, the relevant HLA genotypes and sufficient cryopreserved peripheral blood mononuclear cells (PBMC) were examined. In addition, we serially tracked and characterised viral adaptation and neo-epitope formation over time in an individual in whom primary HIV-1 infection was diagnosed five days after an epidemiologically proven transmission event and preceding seroconversion. This individual expressed two of the HLA alleles associated with viral adaptation to neoepitopes and one HLA allele associated with more classically described CTL escape.
Results
We performed assays of HIV-1 epitope-specific responses on 216 peripheral blood mononuclear cell (PBMC) samples from 125 individuals. The medians and ranges for CD4 and CD8 T cell counts were 540 cells/μL (16–1591) cells/μL and 932 (110–3519) cells/μL respectively. 87% of the individuals tested had some anti-retroviral therapy experience with a median HIV RNA viral load of 50 copies/mL (range <50 to >1 million copies/mL). Fifty seven percent of individuals had undetectable viral load (<50 copies/ml) and 69% had viral loads less than 1000 copies/mL.
Memory CD8 T cell responses against neo-epitopes
The ELISpot assay was initially used to screen for memory IFNγ responses after ex vivo stimulation with peptides representing both non-adapted and adapted neo-epitopes in individuals carrying HLA-A*0201 (n=36), HLA-A*0301 (n=19), HLA-B*0702 (n=56), HLA-B*1503 (n=16), HLA-B*4402 (n=32) and HLA-C*0702 (n=57). Positive IFNγ responses were detected to both non-adapted and adapted epitopes in 21% (n=46) of samples on screening. The median and [range] of values for non-adapted and adapted responses were 450 [53–2225] and 575 [55–3700] SFU respectively (n=46, p=0.09, mixed models). These 46 IFNγ responses included six against the HLA-A*0201-restricted Gag SL9 epitope, two against the HLA-A*0201 Vpr AL9 epitope, six against the HLA-B*0702-restricted Gag GL9 epitope, 15 against the HLA-B*0702-restricted Nef TL10 epitope, one against the HLA-B*1503-restricted Nef WF9 epitope, three against the HLA-B*4402 restricted Pol QW11 epitope and 13 against the HLA-C*0702-restricted Nef KY11 epitope. In the subset of samples with detectable viral load and detectable IFNγ responses to both non-adapted and adapted epitopes (n=22), responses to the adapted epitopes were marginally greater than responses to non-adapted epitopes (792 [55–3700] vs. 462 [58–1982] SFU for adapted and non adapted responses respectively), although this was not statistically significant in this sample set (Figure 1). No responses were detected to the A*0301 restricted Pol QR9 peptides in the 19 samples with carriage of HLA-A*0301. Of note 63% (12 of 19) of HLA-A*0301 samples had undetectable viral loads.
Figure 1.
Comparison of IFNγ responses elicited by non-adapted and adapted peptides from individuals with detectable HIV viremia in the study. Each data point represents the average SFU of triplicate wells after subtraction of background.
Forty-six of the 216 samples tested were from individuals who had previously been severely immunodeficient with a nadir CD4 T cell count <50 cells/μL. There was no significant overall correlation between the nadir CD4 T-cell counts and the magnitude of IFNγ responses, however individuals with nadir CD4 T cell counts less than 50 cells/μL had lower rates of detectable IFNγ responses against non-adapted and adapted epitopes (p<0.007, for non-adapted, p=0.014 for adapted epitopes, n=215, mixed models logistic regression) compared with individuals with nadir CD4 T-cell counts >50 cells/μL.
HLA-adapted neo-epitopes stimulate higher avidity memory CD8 T cell responses in contrast to epitopes associated with `classical' CTL escape
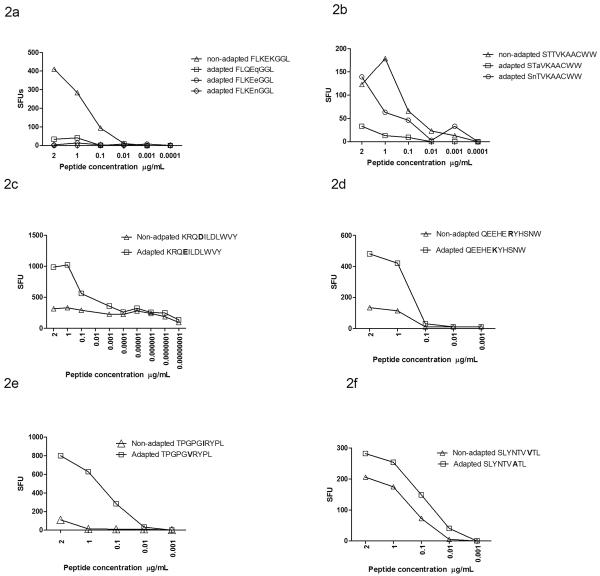
The tendency to equivalent or in some cases even increased IFNγ responses to the HLA-adapted forms of these epitopes on screening assays with excess peptide concentrations suggested that the avidity of these responses may be higher than anti-non-adapted responses. This contrasts with previous demonstrations of intra-patient mutational escape from CTLs, in which immune recognition of variant epitopes is reduced or completely abrogated.40 We therefore formally compared the functional avidity of responses against non-adapted and adapted neo-epitopes in those individuals with positive responses to both peptides on screening using serial dilutions of peptides and IFNγ production as the functional readout (Figure 2). This included seven epitopes after exclusion of HLA-A*0301 Pol QR9. To indicate the contrasting patterns with“classically” described CTL escape mutations, we also tested in parallel the comparative avidity of two well described CTL escape variants. These were variants of HLA-B*0801-restricted Nef 90–97 FL8 (FLKEKGGL) in which lysine (K) to glutamine (Q), glutamic acid (E) or asparagine (N) substitutions at position five mediates functional viral CTL escape2,4 (Figure 2a) as well as variants of the HLA-B*5701-restricted Integrase 128–132 SW10 (STTVKAACWW) epitope, in which mutations at amino acid positions two and three abrogate CTL responses38 (Figure 2b). Combined data from these HLA-B*0801 FL8 and −B*57 SW10 responses (n=14 samples from 11 individuals) measured at 2μg/ml peptide concentration showed the expected pattern of significantly reduced IFNγ responses to the adapted epitopes, accounting for repeated measures in individuals (raw median 203 SFU for non-adapted versus 65 SFU for adapted, p=0.03).
Figure 2.
Comparative functional avidity of IFNγ responses against non-adapted and adapted epitopes associated with “classical escape” from (a) HLA-B*0801-restricted responses against FLKEKGGL and (b) HLA-B57-restricted response against STTVKAACWW. In both examples, responses are abrogated by the HLA-associated substitution. In contrast, CTL responses against neo-epitopes have higher functional avidity than their non-adapted epitopes (c–f). Representative plots of peptide stimulated IFNγ responses to HLA- C*0702-restricted KRQ(D/E)ILDLWVY (c), −B*4402-restricted QEEHE(R/K)THSNW (d), −B*0702-restricted TPGPG(I/V)RYPL (e), and −A*0201-restricted SLYNTV(V/A)TL. Peptide concentrations (X-axis) from 2μg/ml followed by 1:2 dilution, and thereafter 1:10 dilutions dependant on cell availability. Non-adapted epitopes are shown as triangles and neo-epitopes (adapted epitopes) are squares in all plots.
In contrast responses to the seven adapted neo-epitopes and their non-adapted epitope pair frequently showed the opposite pattern of avidity differences with adapted neo-epitopes inducing greater IFNγ production at lower peptide concentrations compared with the non-adapted epitopes (Figures 2c–f). The pattern of equivalent or increased IFNγ production induced by the adapted epitope was observed across multiple individuals for most of the epitopes studied (8 subjects for Nef KY11, 6 subjects for Nef TL10 epitope, 5 subjects for Gag GL9, 4 subjects Gag SL9 and 2 subjects for Pol QW11) and in one subject each for Nef WF9 and Vpr AL9. In the subset of 46 individuals with measureable responses to both non-adapted and adapted epitopes on screening, the combined data of all HLA-epitope pairs measured at 2μg/ml peptide concentration showed no significant decline in IFNγ to the epitope variants (449 SFU for non-adapted epitopes compared with 572 SFU for adapted epitopes, n=46 p=0.6, mixed model sign test, p=0.09 based on the square roots of absolute differences), (Figure 3), though this subset would tend to select out the more equivalent responses. In the larger dataset of 216 screening assays where low level or absent responses to either peptide were included, there was a trend towards increased IFNγ responses to the adapted peptide relative to the non adapted peptide (median 14.4 SFU versus 12.5 SFU p=0.06 for adapted and non-adapted peptide responses). Peptide binding scores using SFYPEITHI, IMAS or IEDB for epitope pairs that could be compared across the three software programs were not consistent. Three of the five peptide pairs demonstrated similar binding scores whilst the remaining two epitope pairs showed differences and thus peptide binding scores did not completely explain the observed patterns of avidity (see supplementary data). Differences in avidity may therefore be attributed to peptide HLA interaction with the T cell receptor. Only three of 15 individuals with HLA-C*0702 expressed HLA-B*18, −B*1302 or HLA-B*44, which have overlapping restriction to KY11 and two of the 21 HLA-B*0702 cases expressed HLA-B*5301 which could potentially bind to the B*0702 restricted TL10 epitope, indicating that for the majority of cases tested the avidity differences could not be explained by alternative HLA alleles with overlapping peptide restriction.
Figure 3.
Patterns of immunoreactivity associated with neo-epitopes in which IFNγ responses display equivalent or marginally enhanced IFN-γ responses to the HLA-adapted variant epitope, in contrast to “classical escape” in which there is typically abrogation of IFNγ responses to escape variants.
GL9, TL10 and KY11 neo-epitope-stimulated CD8 T cell responses are not associated with differences in production of IL-2, dual IFNγ/IL-2 or cytotoxicity
HLA-B*0702-expressing PBMCs from two individuals were cultured with the HLA-B*0702 restricted Gag (GL9) and Nef (TL10) non-adapted and adapted epitopes. In addition HLA-C*0702- expressing PBMCs from seven individuals were cultured with HLA-C*0702-restricted Nef (KY11) adapted and non adapted epitopes to generate epitope-specific CD8 T cell lines. PBMC were cultured for a median of 12 days (range 9–17 days) after which the frequency of single IL-2 and IFNγ-producing cells and dual IL-2/IFNγ producing cells in effector memory and central memory populations were assessed by flow cytometry after overnight peptide re-stimulation. Central memory and effector memory CD8 T-cells were determined by expression levels of CCR7 and CD45RA. Across all three epitope pairs, there were overall greater proportions of effector memory CD8 T cells compared with central memory CD8 T cells among all epitope-specific cell lines, with no significant difference in these proportions between T cells cultured with either non-adapted or adapted epitopes (20[3–35] vs 14[3–29] % median[range] of CCR7+CD45RA− central memory CD8 T cells and 58[25–78] vs 61 [21–82] %CCR7−CD45RA− effector memory CD8 T cells for adapted and non adapted stimulated cultures respectively n=11, data not shown). This was also observed in overnight stimulations of T cell lines with media alone, or PHA/SEB (data not shown), suggesting that there was peptide driven differentiation of central memory cells to effector cells, in addition to the proliferation of effector memory cells during the 12 day culture.
For all three epitope-pairs associated with HLA-B*0702 and −C*0702, frequencies of IL-2 and dual IL-2/IFNγ–producing central memory and effector memory T cells were not significantly different whether directed against non-adapted or adapted epitopes (Figure 4), though central memory populations in general were associated with increased numbers of IL-2 producing cells compared with effector memory populations (1.4 [0.4–2.8] vs 0.2 [0.1–0.6] % of IL2 producing central memory and effector memory cells for non-adapted cells (p=0.001) and (2.1 [0.2–6.5] vs 0.1 [0.05–2.8] % of IL2 producing central memory and effector memory cells for adapted cells (p=0.002) in keeping with a central memory functional phenotype.41 In the case of single IFNγ producing cells, adapted (peptide-stimulated) cultures were associated with higher frequencies of single IFNγ producing central memory and effector memory cells compared with non-adapted peptide-stimulated cultures (30[6–67] and 37[10–66] %IFNγ producing central memory and effector memory CD8 T cells respectively from adapted cultures compared with 7(1–55) vs 10(1–58) %IFNγ central memory and effector memory CD8 T cells for non-adapted cultures), in keeping with the high IFNγ levels observed in the ELISpot screening assays and functional avidity profiles of adapted peptide -stimulated responses observed in PBMCs, although values did not reach significance for central memory (p=0.11) or for effector memory (p=0.09) cultures.
Figure 4.
Single IFNγ producing cells are marginally increased in adapted cultures compared with non-adapted cultures while single IL-2 producing cells are significantly increased in central memory cells from both adapted and non-adapted peptide-stimulated T cell cultures (p<0.05 mixed models analysis). The frequency of cytokine producing cells is plotted for central memory (CM) and effector memory (EM) populations from adapted and non-adapted cultures. Single IFNγ producing cells from non-adapted, and adapted cultures, dual IFNγ/IL-2 producing cells from non-adapted and adapted cultures and single IL-2 producing cells from non-adapted and adapted cultures are shown.
To assess differences in functional cytotoxicity, chromium release assays were performed on CD8 T cell lines cultured from one individual expressing HLA-C*0702. Co-expressed HLA class I alleles were HLA-A*03, A*23, B*0702, B*4001 and C*0304 in this case. As expected, B cell targets pulsed with non-adapted Nef KY11 (KRQDILDLWVY) were killed more readily by non-adapted KY11 specific T cells compared with targets pulsed with adapted Nef KY11 (KRQEILDLWVY) (18% vs 12% lysis). However the reverse was observed for adapted KY11-specific T cells, which killed fewer numbers of adapted KY11 pulsed targets compared with non-adapted KY11 pulsed target cells (33% vs 43% lysis n=1). That is, though adapted neo-epitope specific T cells exhibited demonstrable cytotoxicity, this was more evident for non-adapted peptide pulsed targets (33 vs 12% than for adapted peptide pulsed target and 42 vs 18% for non-adapted peptide pulsed targets n=1), which would predict outgrowth of the adapted virus over the non-adapted species in the presence of adapted CTLs.
An alternative target cell line (expressing HLA-A*01, A*02, B*0702, B*4001, C*0304, and C*04) was used to determine cytotoxicity for the HLA-B*0702 restricted CTLs derived from two individuals with the HLA-B*0702 allele common to the HLA class I genotypes of the patients and the target cell line. Non-adapted and adapted HLA-B*0702- restricted CTLs did not show differential killing of the target cells (n=4 assays).
TCR variable beta gene family expression levels were similar between adapted and nonadapted CTLs
T cell receptor Vβ chain family diversity was assessed in HLA-C*0702 and −B*0702-restricted adapted and non-adapted peptide-specific CTLs. We found no differences in TCR Vβ family repertoire between the adapted and non-adapted CTLs examined in the three samples tested (data not shown). Vβ 13.1 was the most common Vβ family for the HLAC*0702 restricted CTLs with more than 50% of the CD8 IFNγ producing cells expressing Vβ 13.1. The predominant Vβ family for the HLA-B*0702 Gag specific CTLs was Vβ 12 contributing to more than 30% of CD8 IFNγ producing cells. Greater than 55% of the HLA-B*0702 Nef restricted CTLs expressed Vβ 23. As expected CTLs directed against Gag and Nef epitopes from the same patient expressing HLA-B*0702 displayed different Vβ family hierarchies with Vβ 12 predominating in the Gag CTLs and Vβ 23 more commonly expressed by the Nef CTLs thus reflecting distinct clonotypes for different HIV epitopes.
Directly observed neo-epitope formation during acute HIV infection
We were able to examine neo-epitope selection on a longitudinal basis in an individual who was diagnosed with acute subtype A HIV-1 infection days after an epidemiologically proven sexual transmission event. At first testing HIV-1 p24 antigen was detected in the absence of p24 antibody indicating the patient was Fiebig stage II.42 Repeat testing on day 15 post-transmission indicated detectable p24 antigen with reactive EIA screen and indeterminate group 4 Western Blot with antibodies detected to p24, gp160 and equivocally against gp120 (Fiebig stage IV). At this time the HIV RNA concentration (viral load) was >106 copies/mL. By day 98 the Western blot was fully reactive and the viral load had declined without antiretroviral medication to 125,893 HIV RNA copies/mL (Fiebig VI). The HLA class I genotype of the individual was HLA-A*23, −A*3004, −B*08, −B*15, −C*04 and −C*07, which included two HLA class I alleles, HLA-B*15 and −C*0702 associated with neoepitopes studied here; HLA-B*1503-restricted Nef (183–191) WRFDSRLAF and −C*0702 restricted Nef (105–115) KRQEILDLWVY, as well as the HLA-B*08 allele associated with the “classical” escape in Nef FL8 as described previously. Nine consecutive PBMC samples were collected and cryopreserved from days 37 to 658 post transmission. IFNγ responses to 28 HIV epitopes were evaluated in the ELISpot assay. These epitopes included the HLA-B*15 and −C*07 adapted and non adapted epitopes, the HLA-B*08 restricted Nef FL8 epitope with three of its adapted variants, in addition to 23 other well characterised CD8 T cell epitopes restricted to the individual's HLA class I alleles.39 Eight plasma samples were also collected from day 15 to day 469 post-transmission, seven of which were collected prior to commencement of antiretroviral therapy.
IFNγ responses to HLA-B*0801-restricted Nef FLKEKGGL (3,735 SFU) and its variant FLKENGGL (1298 SFU) were among the three responses detected in the first available time point for immunological testing 37 days post transmission, the other being HLA-B*1503-restricted Pol RKAKIIRDY (230 SFU). The FL8 epitope was in the non-adapted form in the patient's viral sequence from the earliest plasma samples (day 15 post transmission) and remained unchanged despite detection of high magnitude IFNγ responses. Ex vivo responses to the FLKEQGGL adapted variant were reduced at all nine time points tested. The HLA-C*0702-restricted KY11 epitope remained invariant at all timepoints, however the position associated with HLA-C*0702 selection in the subtype B population analysis within this epitope is known to be a site of subtype-specific variation, with E (KRQEILDLWVY) present in subtype A reference sequences43 and therefore likely to be present in the transmitted founder sequence. Indeed, the E was also present in donor sequence suggesting this was the case. Low level responses to the KY11 epitope were detected at days 98 (62 SFU) and 385 (60 SFU) post transmission. In Nef WF9, the non-adapted epitope WKFDSRLAF was present in the individual's sequence on day 15. A mixture of the adapted WRFDSRLAF and the non-adapted WKFDSRLAF sequence was detected in the patient sample by day 266, and the adapted sequence was detected in subsequent patient samples (days 385 and 469) in keeping with the specific substitution predicted by population-based HLA association studies.25 IFNγ responses to the non-adapted WF9 epitope were detected on day 98 (216 SFU) and day 266 (178 SFU). Responses then decreased to 88 SFU by day 385 after the autologous viral adaptation was detected. Responses further declined to 50 SFU on day 469 and were not detected thereafter. Low level responses to the adapted WF9 neoepitope reached 70 SFU by day 385, having been below the positive cut off previously (36 SFU on day 98 and 38 SFU on day 266), (Figure 5). The patient commenced therapy thereafter and no further responses were detected to the adapted peptide. The change in WF9 following strong WF9 T cell responses coincided with a rise in viral load over 3 months from an apparent setpoint of 70,795 copies/mL to 245,471 HIV RNA copies/mL (Figure 5) coincident with a drop in CD4 T cell count from 572 to 357 cells/μL over the same time points. There was a broadening of CD8 T cell responses overall, peaking at 20 epitope-specific responses against HIV epitopes in Gag, Pol, Env and Nef at this time. The patient subsequently commenced anti-retroviral therapy. Overall, T cell responses were detected to 21 of the 28 peptides evaluated, median 7 (inclusive of responses to the 5 variants) ranging from 3 to 20 responses over 9 time points (figures 6a∓b). All Nef variants examined in the study were included in the analysis and are shown separately.
Figure 5.
Evolution of the HLA-B*1503-restricted CD8 T cell response against WKFDSRLAF and following adaptation, against WRFDSRLAF in a single individual following sexual transmission of HIV-1. K to R substitution within the epitope occurs by day 266 in the autologous viral sequence. Antiretroviral therapy commenced 393 post-transmission and all HIV-1 specific CD8 T-cell responses were not detected (<50 SFU) thereafter.
Figure 6.
Breadth (a) and Magnitude (b) of all tested HIV protein-specific responses in a single individual following sexual transmission of HIV-1. (a) The number of IFNγ responses detected at each time point against epitopes in Gag, Pol, Env, Nef and Nef variants are shown. HIV RNA levels at each time point in relation to number of responses are also shown. (b) The Magnitude of IFNγ responses (SFU) against Gag, Pol, Env, Nef and Nef variants.
Discussion
Studies of HIV-1 in natural infection, post-vaccination, transmission between hosts and on exposure to antiretroviral drugs have shown that patterns of HIV-1 genetic variation in all these scenarios are strongly shaped by the virus' evolutionary drive to preserve and optimise replicative fitness.2–12, 44–46 HIV mutation which reduces or abolishes CTL recognition of single epitopes is a pre-eminent example of this. Mutation which leads to persistent and immunodominant CD8 T cell responses against epitopes which then remain invariant imply that these responses either do not hinder, or possibly even enhance, viral fitness. HIV exploiting immunodominance by actively promoting particular non-suppressive responses would effectively block alternative epitope-specific T cells from which the virus would have to accommodate further mutation networks with potential compromises in replicative capacity. Several population based studies have independently generated HLA-HIV polymorphism associations which match established cases of `classical' T cell escape15–25 and these cases have in part been used to provide experimental validation of the statistical associations and the association approach in general. In the same studies, including the population-based study used here, associations generated by the same methods and stratified by the same statistical criteria show that HIV-1 adaptation serves to create epitopes as well. If the former examples of “classical” CTL escape are accepted as validating observations, then these associations showing creation of epitopes cannot be rejected a priori. Many neoepitopes are commonly presented, and are among the relatively narrow repertoire of immunodominant responses documented in several studies of chronic HIV-1 infection.
We have systematically studied a group of these epitopes selected on the basis of genetic signals and pre-existing immunological observations. We show that the HLA allele-specific selection pressures on single codons which create these epitopes in vivo cause significant enhancement of the functional avidity of memory responses to them, compared to the nonadapted epitopes. These responses did not display a more “polyfunctional” cytokine profile as determined by either increased IL-2 secretion or dual IL-2/IFNγ secretion. Instead neoepitope-specific responses had a high IFNγ producing effector memory and central memory phenotype and an inconsistent profile of cytotoxicity. In one of the five cases tested, the cytotoxicity of the adapted epitope-specific T cell response was lower against the adapted epitope than the non-adapted epitope, predicting the persistence and selection of viruses bearing the adapted epitope in-vivo. We did not prove that T cell populations reacting to adapted and non-adapted sequences were distinct T cell clonotypes using Vβ chain family usage, though a clear signal of selection away from one response and towards the other was evident in the viral sequences (supplementary data Table). Finally, in an acutely infected individual we directly observed the early presence of the HLA-B*1503- non-adapted Nef WF9 epitope sequence and cognate CD8 T cell IFNγ response, followed by viral adaptation within the epitope and emergence of the neo-epitope-specific CD8 T cell response, confirming that the genetic analyses had correctly identified the direction of amino acid substitution typically associated with HLA-B*1503-restricted selection pressure and that the functional characteristics of the neo-epitope CD8 T cell response could not be explained by “original antigenic sin” of the transmitted sequence in this case.47 The emergence of HLA-B*1503-driven selection coincided temporally with the first increase in the viral load after a decreasing viral load associated with the other early CD8 T cell responses. Taken together, these data may provide evidence of HIV-1 adaptation having a broader influence on the immunodominance patterns of chronic infection beyond abrogating the viral inhibition of individual epitope-specific CTLs. In particular, the induction of high avidity CD8 T cell responses in this context appears the result, rather than the cause of, immune selection, with the effect of facilitating the immunodominance of high IFNγ producing responses (even within the central memory compartment) rather than improving cytotoxic viral control.
High functional avidity or antigen sensitivity, measured as recognition of peptide-MHC at low densities on the target cell surface, has been generally considered a favourable qualitative property of antiviral CTLs, associated with greater elimination of target cells and viral clearance.48–52 In the setting of HIV and SIV infection, high avidity CD8 T cells are associated with the intense selection pressure of acute infection53 and greater anti-viral control in established chronic infection.54,55 Furthermore, the functional profile of HIV-specific CD8 T cell responses is strongly determined by levels of antigen concentration and TCR engagement.56,57 For example, highly avid HLA B*2705-restricted T cell clones and PBMC-derived T cells against HIV-1 p24 Gag KK10, displayed greater IFNγ, TNFα and CD107a mobilisation per cell compared to less avid T cells.58 However, the rapid lymphoproliferation triggered by T cell activation at low antigen concentrations, represents a potential “double edged sword” in which high avidity responses can preferentially expand and dominate the antigen-specific response,59 but also progress to a state of replicative and functional senescence more rapidly in the presence of ongoing antigen stimulation.60 While the multiplicity of general (non-epitope specific) immune defects in HIV infection affecting the circulating naïve T cell pool, CD4 T cell help, thymic output and haematopoiesis61 all contribute to the failure of clonotype renewal in the initial wave of early immunodominant CTL responses, our data suggests that there can be replacement by high avidity variant-specific clonotypes but this is not necessarily useful for immune containment if the targeted variants are HLA-adapted.
These data have further implications for understanding complex interactions between T cell avidity, viral sequence evolution and cell function. Data from the influenza field suggests that high T cell avidity may in fact be inversely correlated with cytotoxicity. Among influenza A virus-specific CTLs recovered from the infected lung of B6 mice, T cells with lower epitope avidity associated with short-lived “kiss and run” engagement with the HLA-peptide complex lysed more target cells than high avidity, high IFNγ producing T cells.62 In a study of CD8 T cell responses against HIV-1, cytomegalovirus (CMV), EBV and influenza, greater IL-2 production and antigen-specific proliferation of more protective HLA-B-restricted T cell responses were associated with lower-avidity T cell receptors compared with less favourable IFNγ secreting, effector-only responses expressing the T cell exhaustion marker PD-1 and high avidity receptors.63 HLA-restricted T cell responses to mutated epitopes in Gag restricted by HLA A*11 and B*57 and in Pol, Nef and Env restricted by HLA-B*35 have also demonstrated decreased proliferative capacity associated with variant-specific responses.26,64,65 A number of studies in chronic HIV-1 infection have also documented high avidity CD8 T cells associated with high viremia66 and lack of a simple correlation between magnitude or breadth of IFNγ responses alone and viral load.67 While this reflects the likely importance of qualitative markers aside from IFNγ, our data suggests that epitope specificity is also important. Despite the degeneracy of TCR recognition, single viral polymorphisms can facilitate particular epitope-specific responses over others once infection is established. We have not obtained clear evidence of a superior functional profile such as IL-2 production associated with the non-adapted epitope responses here, however such profiles are presumably rare when considering memory responses in chronically infected individuals. Our observations support a study by Karlsson et al13 who found that HIV-1 escapes temporarily effective, acute immune responses but then broadens to target epitope variants which match consensus subtype B sequence. Such broadening was associated with higher viral load during the first year of infection. The authors argue that T cell breadth in this context was not a favourable host response to HIV variation, but the “immunological footprint” of viral adaptation, which helps to maintain some elements of consensus viral sequence in a population. Here we show that this phenomenon is evident at the population level. By escaping from early, strongly cytotoxic CD8 T cell responses and then creating neo-epitopes which elicit highly avid but exhausted effector phenotype T cell responses, HIV is effectively exploiting immunodominance to shift to non-inhibitory responses.
Aside from the particular epitopes examined here, there were a large number of overlapping putative epitopes/neo-epitopes associated with HLA associated polymorphism in population-based studies,25 suggesting that this could be applied to many more of the measurable responses in HIV-infected individuals. This model would reconcile conflicting arguments about the utility of CD8 T cell immunity in HIV infection. The findings that high-frequency HIV-specific CTLs are detectable in individuals with AIDS and the adverse interference between Env- and accessory protein-responses with Gag-specific responses68,69 would all be expected if HIV can itself promote certain non-suppressive responses over suppressive ones as an adaptive strategy. Several mechanisms of viral enhancement are possible - `decoying' from other better epitopes, direct effects of epitope variation on T cells to IFNγ-driven exhaustion or activatory recruitment of CD4 T cell targets. Regardless of the mechanism, the possibility that some epitopes serve the virus and not the host has implications for vaccine design. Polyvalent vaccines designed to combat HIV strain diversity by incorporating as much of HIV-1 variation as possible 70,71 may increase the breadth of CD8 T cell reactivity but this may not be desirable if this recapitulates the immunodominance hierarchies that such variation facilitates in nature. Diversity coverage strategies based on very early acute or founder sequences may circumvent this issue, however such sequences may also include adaptive changes made in the donor host that have not yet reverted or are slow to revert in the recipient host because of minimal or enhancing effects on viral fitness. We were limited in this study to assaying for memory CD8 T cell responses in a cohort of chronically infected individuals, many of which with highly suppressed HIV viral load on antiretroviral medication. This probably accounted for the high rate of CD8 T cell non-responders generally in the study, as previous studies have shown reduced HIV-specific CTL responses in patients on antiretroviral therapy,72,73 including absence of responses against the HLA-A*0301 restricted responses against Pol QR9.74 Furthermore, we have not taken account of other factors which do modulate T cell avidity in vivo such as dendritic cell function75 and it is also possible that the functional avidity apparent in the ELISpot assay does not reflect in vivo reactivity with natural antigen presentation mechanisms. Nevertheless, we have in each case compared the relative functional avidity profiles and immunological characteristics of two epitope variants- non-adapted and adapted- under equivalent assay conditions, and the fact that the differences between the profiles are consistent across seven different epitope pairs and 125 individuals tested, and the consistent direction of amino acid substitution associated with creation of neo-epitopes in independent genetic studies, suggests this is a robust observation. In the absence of detailed immunological characterisation of the large number of prevalent or possible HLA-restricted CD8 T cell responses in naturally infected or vaccinated individuals, population-based genetic signals as used here can serve to delineate empirically which HLA-restricted responses are likely to mediate relative suppression, non-suppression or enhancement of viral replication based on the evolutionary solutions of HIV to evade or exploit them.
Materials and Methods
Study cohort
The 125 subjects used for all cellular studies were enrolled into the WAHCS which is a prospective, observational study of HIV-infected individuals established in 1983 and approved by the Royal Perth Hospital Ethics Committee and conforms to the provisions of the Declaration of Helsinki. All subjects gave informed consent to these studies. Longitudinal CD4, CD8 T cell counts, viral load measurements and antiretroviral medication history were available on all subjects.
HIV peptides
Eight HIV-1 9–11 mer peptide pairs were synthesized by Invitrogen (Melbourne, Australia); HLA-A*0201-restricted Vpr (59–67) AIIRILQQL and ALIRILQQL, HLA-B*0702 restricted Gag (355–363) GPGHKARVL and GPSHKARVL, HLA-B*0702 and HLA-B*4201-restricted Nef (128–137) TPGPGVRYPL and TPGPGIRYPL, HLA-B*1503 restricted Nef (183–91) WRFDSRLAF and WKFDSRLAF, HLA-C*0702 restricted Nef (105–115) KRQEILDLWVY and KRQDILDLWVY, HLA-A*0301 restricted Pol (424–432) QIYPGIKVR and QIYPGIKVK, HLA-A*0201 restricted Gag (77–85) SLYNTVATL and SLYNTVVTL epitope and HLA-B*4402 restricted Pol 724–734 QEEHEKYHSNW and QEEHERYHSNW. Peptides were reconstituted in dimethyl sulphoxide to 10mg/mL and stored at minus 80°C prior to dilution for use in assays.
IFNγ ELISpot assay
Cryopreserved PBMC were thawed and left to settle over night at 37°C. Viable lymphocytes were enumerated by trypan blue exclusion using a Neubauer haemacytometer. 100,000 cells resuspended in culture medium (10%FCS/RPMI-1640) were dispensed per well using a modified version of the previously described IFNγ ELISpot assay.76 Sterile 96 well nitrocellulose backed plates (MAIP S4510, Millipore, Bedford, USA) were coated with 2μg/mL IFNγ coating antibody (Mabtech, Naka Strand, Sweden) overnight 4°C in sterile Phosphate buffered Saline (PBS). Plates were washed with sterile PBS, blocked with culture medium (CM, 10% FCS/RPMI-1640 30minutes, room temperature [RT]), after which cells and stimulants (synthetic HIV peptides dispensed in triplicate at limiting dilution [2μg/mL, 1μg/mL then 10 fold dilutions, dependant on cell numbers], with positive [anti-CD3 antibody Mabtech, Victoria, Australia] and negative [culture media alone] controls)- were dispensed for overnight incubation CO2, 37°C. Plates were then washed with sterile PBS. Biotinylated IFNγ (Mabtech, Victoria, Australia) was added (2 hrs, RT), after which plates were washed and streptavidin horseradish peroxidise (Mabtech, Victoria, Australia) added (1 hr, RT). Plates were washed and developed with 100μL/well tetramethylbenzidine substrate (Mabtech, Victoria, Australia) for 10minutes at RT. Plates were washed extensively with MilliQ H20 and left to dry prior to analysis on the AID iSpot reader (AID, Strassburg, Germany) with AID software (5.0 B7337). Responses were determined by subtracting the mean of the negative control wells from the mean of triplicate stimulated wells. Results are presented as spot forming units/106cells (SFU). Responses were considered positive if they were > 50 SFU based on the distribution of the negative controls and test data (see statistical analysis).
EBV transformed B cells
EBV transformed B-lymphoblastoid cell lines were generated in-house in the Department of Clinical Immunology (DCI), Royal Perth Hospital, Western Australia. The HLA homozygous B cell line (workshop HHKB) R86 12350C, expressing HLA-A*0301, B*0702 and C*0702 was used to present the HLA-C*0702 restricted Nef KY11 peptide and non adapted variant to CTLs from the HLA-C*0702 expressing patient. The R04 0244215 (HLA- A*01, −A*02, −B*0702, −B*4001, −C*0304, −C*04) cell line was used to present HLA-B*0702 restricted Gag GL9 and Nef TL10 peptides and variants. All cells were maintained in culture media for a minimum period of two weeks prior to use in cytotoxicity assays.
CD8 T cell lines
HLA restricted peptide specific CD8 T cell lines were generated from cryopreserved PBMC. PBMCs were thawed, left to settle overnight in culture medium and counted. Two to five ×106 cells were incubated with 20μg of HIV specific peptide to give a final concentration of 20μg peptide in a total volume of 1 mL and incubated for 2 hours at 37°C in a CO2 incubator. Cells were then washed and resuspended in 1mL of T-stim media (15mL -T cell growth media harvested from a T lymphoblastoid cell line [kindly donated by Professor James McCluskey, University of Melbourne, Australia], 25mL - culture media, 7.5mL - FCS) supplemented with 50 IU IL-2 (Chiron, USA) in a single well of a 24 well plate and left for 3 days at 37°C in a CO2 incubator. On day 4, 1mL of T-stim media, supplemented with IL-2 at final concentration of 100 IU/mL, was added to the culture. Cultures were fed or split as required every 2–3 days up to day 14. On day 14 to16, peptide specific CTLs were confirmed by flow cytometry. The cytotoxicity of peptide positive CTLs were determined using the standard 51Cr release assay.77
T cell immunophenotyping
CD3+/CD8+ T cell effector and central memory cell phenotypes were differentiated by surface expression of CCR7 and CD45RA78 by flow cytometry using the BD Biosciences FACSCanto instrument. Results were analysed using FlowJo Software (TreeStar, Ashland, OR). 0.5 × 106 CTLs/mL were re-stimulated with specific HLA-restricted peptides and appropriate positive (Staphylococcus Enterotoxin B / Phytohaemmagglutinin) or negative (culture media alone) controls in overnight cultures. 10uL each (from 1:10 stock dilutions) of anti-CD49 and anti-CD28 (BD PharMingen, San Diego, USA) were also added to the cultures. Brefeldin A (Sigma, Castle Hill, Australia) was added after 2 hours (final concentration 5μg/mL). After overnight re-stimulation, CTLs were washed with flow buffer (1% FCS/PBS), centrifuged, counted and 0.5 million cells dispensed per tube for membrane and intracellular cytokine staining. A cocktail of fluorescent labelled membrane antibodies were added first to each tube (CD3 APC Cy7, CD8 PerCP PCy5.5, CCR7 PeCy7, CD45RA APC). After 15minutes incubation, cells were washed, fixed (Intraprep Kit, Beckman Coulter, Gladesville, Australia), and washed again prior to permeabilisation (Intraprep kit). Intracellular cytokine antibodies (IL-2 PE and IFNγ Alexa Fluor 488) were then added for 15 mins at RT. Cells were washed, resuspended in flow buffer and analysed on the FACSCanto flow cytometer.
CTLs were prepared, as stated above for overnight peptide re-stimulation, for Vbeta TCR repertoire staining. CD3 APC Cy7 and CD8 PerCP PCy5.5 were then added from the previous mentioned cocktail of surface membrane antibodies and the PE and FITC labelled Vbeta antibodies (Beckman Coulter, Gladesville, Australia) replacing the remaining membrane antibodies. Cells were then fixed and permeabilised after which IFNγ APC-A (BD) was added to each tube. Cells were washed, resuspended in flow buffer and analysed on the flow cytometer.
Cytotoxicity assays
Cytotoxicity was performed using the standard 4 hour 51Cr release assay.77 Target cells (EBV transformed B cells expressing the HLA allele of interest) were labelled with sodium 51Cr-labelled chromate (Perkin-Elmer, Melbourne, Australia) and pulsed with 2μg/mL HIV specific peptide over night at 37°C. After thorough washing, the B cell targets were admixed with peptide-specific CTLs at Effector:Target ratios (2:1, 10:1, 20:1) in triplicate wells of 96 well round-bottom plates. Supernatants were harvested after 4 hours, transferred to counting tubes and counted. Chromium labelled B-cells not pulsed with peptide were used as negative controls. 100% maximal lysis was determined by addition of 100μL of 0.5% TritonX 100 (Sigma, Castle Hill, Australia). Specific percent lysis was calculated using the formula ([counts per minute (cpm) sample - cpm spontaneous]/ [cpm 100% Lysis -cpm spontaneous])× 100. Peptide specific CTLs added to unpulsed target cells controlled for non-specific CTL lysis (median = 0, range 0 to 7% for unpulsed target cells). Where possible, two B cell lines were used to determine cytotoxicity.
HIV sequencing
Viral sequencing was performed on a single patient studied longitudinally. HIV-1 RNA was extracted from plasma samples and quantitated using the AmpliPrep/COBAS TaqMan HIV-1 version 1.5 according to manufacturer's instructions. Samples were analysed using the COBAS TaqMan 48 analyser with a linear dynamic range of 40-1×107 copies/ml. Standard bulk sequencing of HIV-1 Nef was based on a nested PCR of a first round 6 kb fragment, followed by shorter fragment amplification of region containing Nef. Direct sequencing was performed on PCR products using an ABI 3130XL analyser (Applied Biosystems, California, USA) and electropherograms were analyzed and edited using Assign™ (Connexio Genomics). The viral sequences in the longitudinal patient were aligned with a reference subtype A strain A-KE.Q23- 17 (GenBank accession number AF004885.1)
HLA class 1 typing
Genomic DNA was isolated from all study participants. PCR amplification of HLA-A, B and C genes was carried out using sequence specific primers. Products were resolved to 2 or 4 digit level resolution based on exon 2–3 sequence using standard sequence based typing (SBT) by DCI, Royal Perth Hospital.
Statistical Analysis
Summary data are typically presented as mean and standard deviation or median and range of the sample values as appropriate. These summaries do not take account of possible correlations induced by common host cells. However, paired differences for adapted vs nonadapted responses were analysed by mixed models accommodating repeated measures on the same individuals. The threshold for defining a “positive” response was chosen approximately as the 3rd standard deviation value based on fitting a half mean-zero-normal distribution to the negative responses relative to background, under the assumption that these represent only random non-response.
Supplementary Material
TABLE I. Selected HLA-associated neo-epitopes derived from a US HLA association study in which the neo-epitope corresponded to a published immunodominant epitope (25). Of nine examples shown here, all except the HLA-B*4201 association were examined in this study owing to insufficient numbers of individuals expressing HLA-B*4201 in the WAHCS. For each example, table columns from left to right indicate, selecting HLA allele, neo-epitope protein, codon positions and amino acid sequence, site of HLA association, p-value for association and specific amino acid substitution.
Acknowledgments
We thank all participants of the WA HIV Cohort Study and staff from the Department of Immunology, Royal Perth Hospital. We thank Dr Sonia Fernandez and Rom Krueger for help with flow cytometry, the adult AIDS clinical trial group (AACTG) and study teams of ACTG protocols 5142 and 5128 who were involved in the previous genetic study that formed the basis of this work. In particular we thank Dr David Haas (Department of Medicine, Center for Human Genetics Research, Vanderbilt University School of Medicine, Nashville USA, ACTG 5128), Dr Sharon Riddler (Graduate School of Public Health, University of Pittsburgh, USA, ACTG 5142 and Professor Richard Haubrich (Antiviral Research Centre, University of California, San Diego, USA). This work was supported by the Australian National Health and Medical Research Council (Program grant ID 384702), the National Institute of Allergy and Infectious Diseases (NIAID) RO1 AI060460 and was conducted as part of the Collaboration for AIDS Vaccine Discovery with support from the Bill & Melinda Gates Foundation. The authors have no conflicting financial interests. The content of this study is the responsibility of the authors and does not necessarily represent the official views of NIAID or the National Institutes of Health (US).
References
- 1.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, Ogunlesi AO, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 3.Goonetilleke N, Liu M, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov V, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viraemia in HIV-1 infection. J Exp Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, et al. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. PNAS. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, McNevin J, Cao J, Zhao H, Genowati I, Wong KS, et al. Selection on the human immunodeficiency virus type 1 proteome following primary infection. J Virol. 2006;80:9519–9529. doi: 10.1128/JVI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 8.Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, et al. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193:375–385. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 10.Draenert R, Le Gall S, Pfafferott KJ, Leslie AJ, Chetty P, Brander C, et al. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J Exp Med. 2004;199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen TM, Altfeld M, Yu XG, O'Sullivan KM, Lichterfeld M, Le Gall S, et al. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J Virol. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokomaku Y, Miura H, Tomiyama H, Kawana-Tachikawa A, Takiguchi M, Kojima A, et al. Impaired processing and presentation of cytotoxic-T-lymphocyte (CTL) epitopes are major escape mechanisms from CTL immune pressure in human immunodeficiency virus type 1 infection. J Virol. 2004;78:1324–1332. doi: 10.1128/JVI.78.3.1324-1332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlsson AC, Iversen AKN, Chapman JM, de Oliviera T, Spotts G, McMichael AJ, et al. Sequential Broadening of CTL Responses in Early HIV-1 Infection Is Associated with Viral Escape. PLoS ONE. 2007;2:e225. doi: 10.1371/journal.pone.0000225. doi:10.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feeney ME, Tang Y, Roosevelt KA, Leslie AJ, McIntosh K, Karthas N, et al. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J Virol. 2004;78:8927–8930. doi: 10.1128/JVI.78.16.8927-8930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, et al. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 17.Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, Carlson J, et al. Evidence of Differential HLA Class I-Mediated Viral Evolution in Functional and Accessory/Regulatory Genes of HIV-1. PLoS Pathog. 2007;3:e94. doi: 10.1371/journal.ppat.0030094. Erratum in: PLoS Pathog. 10: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brumme ZL, Tao I, Szeto S, Brumme CJ, Carlson JM, Chan D, et al. Human leucocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS. 2008;22:1277–1286. doi: 10.1097/QAD.0b013e3283021a8c. [DOI] [PubMed] [Google Scholar]
- 19.Rousseau CM, Daniels MG, Carlson JM, Kadie C, Crawford H, Prendergast A, et al. HLA Class-I Driven Evolution of Human Immunodeficiency Virus Type 1 Subtype C Proteome: Immune Escape and Viral Load. J. Virol. 2008;82:6434–6446. doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson JM, Brumme ZL, Rousseau CM, Brumme CJ, Matthews P, Kadie C, et al. Phylogenetic dependency networks: inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput Biol. 2008;4:e1000225. doi: 10.1371/journal.pcbi.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang YE, Li B, Carlson JM, Streeck H, Gladden AD, Goodman R, et al. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J Virol. 2009;83:1845–1855. doi: 10.1128/JVI.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, et al. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef Proteins. PloS ONE. 2009;4:e6687. doi: 10.1371/journal.pone.0006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, et al. Adaptation of HIV-1 to human leucocyte antigen class 1. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avila-Rios S, Ormsby CE, Carlson JM, Valenzuela-Ponce H, Blanco-Heredia J, Garrido-Rodriguez D, et al. Unique features of HLA-mediated HIV evolution in a Mexican cohort: a comparative study. Retrovirology. 2009;6:72. doi: 10.1186/1742-4690-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.John M, Heckerman D, James I, Park LP, Carlson JM, Chopra A, et al. Adaptive interactions between HLA and HIV-1: highly divergent selection imposed by HLA class 1 molecules with common supertype motifs. J Immunol. 2010;184:4368–4377. doi: 10.4049/jimmunol.0903745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen TM, Yu XG, Kalife ET, Reyor LL, Lichterfeld M, John M, et al. De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1 infection. J Virol. 2005;79:12952–12960. doi: 10.1128/JVI.79.20.12952-12960.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turnbull EL, Lopes AR, Jones NA, Cornforth D, Newton P, Aldam D, et al. HIV-1 epitope -specific CD8+ T cell responses strongly associate with delayed disease progression cross-recognise epitope variants efficiently. J Immunol. 2006;176:6130–6146. doi: 10.4049/jimmunol.176.10.6130. [DOI] [PubMed] [Google Scholar]
- 28.Yu XG, Addo MM, Rosenberg ES, Rodriguez WR, Lee PK, Fitzpatrick CA, et al. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8 T-cell responses following acute HIV-1 infection. J Virol. 2002;76:8690–8701. doi: 10.1128/JVI.76.17.8690-8701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brumme ZL, Brumme CJ, Carlson J, Streeck H, John M, Eichbaum Q, et al. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency virus type 1 (HIV-1) Gag, Pol, and Nef cytotoxic lymphocyte epitopes in acute/early HIV-1 infection. J Virol. 2008;82:9216–9227. doi: 10.1128/JVI.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goulder PJ, Altfeld MA, Rosenberg ES, Nguyen T, Tang Y, Eldridge RL, et al. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J Exp Med. 2001;193:181–94. doi: 10.1084/jem.193.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker KC, Bednarek MA, Hull LK, Utz U, Cunningham B, Zweerink HJ, et al. Sequence motifs important for peptide binding to the human MHC class 1 molecule, HLA-A2. J. Immunol. 1992;149:3580–3587. [PubMed] [Google Scholar]
- 32.Altfeld M, Addo MM, Eldridge RL, Yu XG, Thomas S, Khatri A, et al. Vpr is preferentially targeted by CTL during HIV-1 infection. Immunol. 2001;167:2743–2752. doi: 10.4049/jimmunol.167.5.2743. 2001. [DOI] [PubMed] [Google Scholar]
- 33.Altfeld M, Allen TM, Yu XG, Johnston MN, Agrawal D, Korber BT, et al. HIV-1 superinfection despite broad CD 8 T-cell responses containing replication of the primary virus. Nature. 2002;420:434–439. doi: 10.1038/nature01200. [DOI] [PubMed] [Google Scholar]
- 34.Leslie A, Price DA, Mkhize P, Bishop K, Rathod A, Day C, et al. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J Immunol. 2006;177:4699–4708. doi: 10.4049/jimmunol.177.7.4699. [DOI] [PubMed] [Google Scholar]
- 35.Cao J, McNevin J, Holte S, Fink L, Corey L, McElrath MJ. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon secreting CD8+ T cells in primary HIV-1 infection. J Virol. 2003;77:6867–6878. doi: 10.1128/JVI.77.12.6867-6878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernardin F, Bernardin F, Kong D, Peddada L, Baxter-Lowe LA, Delwart E. Human immunodeficiency virus mutations during the first month of infection are preferentially found in known cytotoxic T-lymphocyte epitopes. J Virol. 2005;79:11523–11528. doi: 10.1128/JVI.79.17.11523-11528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–774. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 38.Allen TM, Altfeld M, Geer SC, Kalife EF, Moore C, O'Sullivan KM, et al. Selective escape from CD8+ T-cell responses represents a major driving force of Human Immunodeficiency Virus Type-1 (HIV-1) sequence diversity and reavels constraints on HIV-1 evolution. J Virol. 2005;79:13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korber B,C, Brander BF, Haynes R, Koup JP, Moore BD, Walker BD, et al. HIV molecular Immunology. Volume 1. Los Alamos National Laboratory, Theoretical Biology and Biophysics; Los Alamos, New Mexico: 2006/2007. [Google Scholar]
- 40.Goulder PJ, Watkins DI. Impact of MHC class 1 diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–30. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA, Pantaleo G. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol Rev. 2006;211:236–254. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- 42.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors; implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 43.Inwoley A, Recordon-Pinson P, Dupuis M, Gaston J, Genete M, Minga A, et al. Cross-Clade conservation of HIV Type 1 Nef immunodominant regions recognised by CD8 T cells of HIV Type 1 CRF02_AG-Infected Ivorian (West Africa) AIDS Res & Hum Retroviruses. 2005;21:602–628. doi: 10.1089/aid.2005.21.620. [DOI] [PubMed] [Google Scholar]
- 44.Barbour JD, Grant RM. The role of viral fitness in HIV pathogenesis. Curr HIV/AIDS Rep. 2005;2:29–34. doi: 10.1007/s11904-996-0006-1. [DOI] [PubMed] [Google Scholar]
- 45.Kent SJ, Fernandez CS, Dale CJ, Davenport MP. Reversion of immune escape HIV variants upon transmission: insights into effective viral immunity. Trends Microbiol. 2005;13:243–246. doi: 10.1016/j.tim.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 46.Woodman Z, Williamson C. HIV molecular epidemiology: transmission and adaptation to human populations. Curr Opin HIV AIDS. 2009;4:247–252. doi: 10.1097/COH.0b013e32832c0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klenerman P, Zinkernagel RM. Original antigenic in impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature. 1998;394:482–485. doi: 10.1038/28860. [DOI] [PubMed] [Google Scholar]
- 48.Derby M, Alexander-Miller M, Tse R, Berzofsky J. High-avidity cytotoxic T lymphocyte clones with a disparity between functional avidity and MHC tetramer staining. J Immunol. 2001;166:1690–1697. [Google Scholar]
- 49.Bennett MS, Ng HL, Dagarag M, Ali A, Yang OO. Epitope-dependant avidity thresholds for cytotoxic T-lymphocyte clearance of virus-infected cells. J. Virol. 2007;81:4973–4980. doi: 10.1128/JVI.02362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Speiser DE, Kyburz D, Stubi U, Hengartner H, Zinkernagel RM. Discrepancy between in vitro measurable and in vivo virus neutralizing cytotoxic T cell reactivities. Low T cell receptor specificity and avidity sufficient for in vitro proliferation or cytotoxicity to peptide-coated target cells but not for in vivo protection. J Immunol. 1992;149:972–980. [PubMed] [Google Scholar]
- 51.Alexander-Miller MA, Leggatt GR, Berzofsky JA. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc Natl Acad Sci U S A. 1996;93:4102–4107. doi: 10.1073/pnas.93.9.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Messaoudi I, Guevara Patino JA, Dyall R, Le Maoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in Immune defense. Science. 2002;298:1797–1800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 53.O'Connor DH, Allen TM, Vogel TU, Jing P, DeSouza IP, Dodds E, et al. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat Med. 2002;8:493–399. doi: 10.1038/nm0502-493. [DOI] [PubMed] [Google Scholar]
- 54.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belyakov IM, Kuznetsov VA, Kelsall B, Klinman D, Moniuszko M, Lemon M, et al. Impact of vaccine-induced mucosal high-avidity CD8+ CTLs in delay of AIDS viral dissemination from mucosa. Blood. 2006;107:3258–3264. doi: 10.1182/blood-2005-11-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Price DA, Sewell AK, Dong T, Tan R, Goulder PJ, Rowland-Jones SL, et al. Antigen-specific release of beta-chemokines by anti-HIV-1 cytotoxic T lymphocytes. Curr Biol. 1998;8:355–358. doi: 10.1016/s0960-9822(98)70138-1. [DOI] [PubMed] [Google Scholar]
- 57.Betts MR, Price DA, Brenchley JM, Loré K, Guenaga FJ, Smed-Sorensen A, et al. The functional profile of primary human antiviral CD8+ T cell effector and memory subsets of CD8+ T cells. J Immunol. 2004;172:6407–6417. doi: 10.4049/jimmunol.172.10.6407. [DOI] [PubMed] [Google Scholar]
- 58.Almeida JR, Sauce D, Price DA, Papagno L, Shin SY, Moris A, et al. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113:6351–6360. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dzutsev AH, Belyakov IM, Isakov DV, Margulies DH, Berzofsky JA. Avidity of CD8 T cells sharpens immunodominance. Int Immunol. 2007;19:497–507. doi: 10.1093/intimm/dxm016. [DOI] [PubMed] [Google Scholar]
- 60.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Appay V, Douek D, Price DA. CD8+ T cell efficacy in vaccination and disease. Nat Med. 2008;14:623–628. doi: 10.1038/nm.f.1774. [DOI] [PubMed] [Google Scholar]
- 62.Jenkins MR, La Gruta NL, Doherty PC, Trapani JA, Turner SJ, Waterhouse NJ. Visualizing CTL activity for different CD8+ effector T cells supports the idea that lower TCR/epitope avidity may be advantageous for target cell killing. Cell Death Differ. 2009;16:537–542. doi: 10.1038/cdd.2008.176. [DOI] [PubMed] [Google Scholar]
- 63.Harari A, Cellerai C, Enders FB, Köstler J, Codarri L, Tapia G, et al. Skewed association of polyfunctional antigen-specific CD8 T cell populations with HLA-B genotype. Proc Nat Acad Sci USA. 2007;104:16233–16238. doi: 10.1073/pnas.0707570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feeney ME, Tang Y, Pfafferott K, Roosevelt KA, Draenert R, Trocha A, et al. HIV-1 viral escape in infancy followed by emergence of a variant-specific CTL response. J Immnol. 2005;174:7524–7530. doi: 10.4049/jimmunol.174.12.7524. [DOI] [PubMed] [Google Scholar]
- 65.Ueno T, Idegami Y, Motozono C, Oka S, Takiguchi M. Altering effects of antigenic variations in HIV-1 on antiviral effectiveness of HIV-specific CTLs. J Immunol. 2006;178:5513–5523. doi: 10.4049/jimmunol.178.9.5513. [DOI] [PubMed] [Google Scholar]
- 66.Draenert R, Verrill CL, Tang Y, Allen TM, Wurcel AG, Boczanowski M, et al. Persistent recognition of autologous virus by high-avidity CD8 T cells in chronic, progressive human immunodeficiency virus type 1 infection. J Virol. 2004;78:630–641. doi: 10.1128/JVI.78.2.630-641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, et al. Comprehensive epitope analysis of human immunodeficiency virus type-1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrated broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Rosa R, Fernandez CS, Smith MZ, Batten CJ, Alcantara S, Peut V, et al. Control of Viremia and prevention of AIDS following immunotherapy of SIV-Infected Macaques with peptide-pulsed blood. PLoS Pathogens. 2008;4(5):e1000055. doi: 10.1371/journal.ppat.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 70.Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007;13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- 71.Korber BT, Letvin NL, Haynes BF. T cell vaccines for Human immunodeficiency virus, the virus with a thousand faces. J Virol. 2009;83:8300–8314. doi: 10.1128/JVI.00114-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pohling J, Zipperlen K, Hollett NA, Gallant ME, Grant MD. Human immunodeficiency virus type 1-specific CD8+ T cell subset abnormalities in chronic infection persist through effective antiretroviral therapy. BMC Infect Dis. 2010;10:129. doi: 10.1186/1471-2334-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Casazza JP, Betts MR, Picker LJ, Koup RA. Decay Kinetics of human immunodeficiency virus-specific CD8+ T cells in peripheral blood after initiation of highly active antiretroviral therapy. J Virol. 2001;75:6508–6516. doi: 10.1128/JVI.75.14.6508-6516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horton H, Frank I, Baydo R, Jalbert E, Penn J, Wilson S, et al. Preservation of T cell proliferation restricted by protective HLA alleles is critical for immune control of HIV-1 infection. J Immunol. 2006;177:7406–7415. doi: 10.4049/jimmunol.177.10.7406. [DOI] [PubMed] [Google Scholar]
- 75.Kroger CJ, Amoah S, Alexander-Miller MA. Cutting edge: Dendritic cells prime a high avidity CTL response independent of the level of presented antigen. J Immunol. 2008;180:5784–5788. doi: 10.4049/jimmunol.180.9.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keane NM, Price P, Stone SF, John M, Murray RJ, French MA. Assessment of immune function by lymphoproliferation underestimates lymphocyte functional capacity in HIV patients treated with highly active antiretroviral therapy. AIDS Res and Hum Retroviruses. 2000;16:1991–1996. doi: 10.1089/088922200750054729. [DOI] [PubMed] [Google Scholar]
- 77.Foley BA, De Santis D, Van Beelen E, Lathbury LJ, Christiansen FJ, Witt CS. The reactivity of Bw4+ HLA-B and HLA-A alleles with KIR3DL1: implications for patient and donor suitability for haploidentical stem cell transplantations. Blood. 2009;112:435–443. doi: 10.1182/blood-2008-01-132902. [DOI] [PubMed] [Google Scholar]
- 78.Decrion AZ, Varin A, Drobacheff C, Estavoyer JM, Herbein G. A subset of functional effector-memory CD8+ T lymphocytes in human immunodeficiency virus-infected patients. Immunology. 2007;121:405–415. doi: 10.1111/j.1365-2567.2007.02589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE I. Selected HLA-associated neo-epitopes derived from a US HLA association study in which the neo-epitope corresponded to a published immunodominant epitope (25). Of nine examples shown here, all except the HLA-B*4201 association were examined in this study owing to insufficient numbers of individuals expressing HLA-B*4201 in the WAHCS. For each example, table columns from left to right indicate, selecting HLA allele, neo-epitope protein, codon positions and amino acid sequence, site of HLA association, p-value for association and specific amino acid substitution.