Summary
Aging is a major risk factor for chronic disease in the human population, but there is little human data on gene expression alterations that accompany the process. We examined human peripheral blood leucocyte in-vivo RNA in a large-scale transcriptomic microarray study (subjects aged 30 to 104 years). We tested associations between probe expression intensity and advancing age (adjusting for confounding factors), initially in a discovery set (n = 458), following-up findings in a replication set (n=240). We confirmed expression of key results by real-time PCR. Of 16,571 expressed probes, only 295 (2%) were robustly associated with age. Just six probes were required for a highly efficient model for distinguishing between young and old (Area Under the Curve in replication set; 95%). The focussed nature of age-related gene expression may therefore provide potential biomarkers of aging. Similarly, only 7 of 1065 biological or metabolic pathways were age-associated, in Gene Set Enrichment Analysis (GSEA), notably including the processing of messenger RNAs (mRNAs); (p<0.002, FDR q<0.05). This is supported by our observation of age-associated disruption to the balance of alternatively-expressed isoforms for selected genes, suggesting that modification of mRNA processing may be a feature of human aging.
Keywords: Aging, Gene expression, mRNA processing, Cell senescence, predictive model
Introduction
Advancing age is a major risk factor for many common diseases, including type 2 diabetes, cardiovascular disease and many cancers (Butler et al. 2008). However, aging is characterized by progressively rising heterogeneity, with some people becoming frail in their seventies while others remain fit into their nineties or even longer. Characterizing the changes underpinning the heterogeneity of aging processes at a molecular level has been a long held goal.
One theory of aging is that random and widespread unrepaired damage to DNA (and other molecules), accumulated over a lifetime may cause cellular senescence (Gensler & Bernstein 1981), but it has not been established whether such damage is associated with large scale alterations of gene expression in the aged human population. Alteration to highly sequence dependent processes such as mRNA processing (Cartegni et al. 2002) have been suggested in previous studies (Yannarell et al. 1977; Meshorer & Soreq 2002), but to date there are little human data to assess this empirically. Several age-related diseases are known to be caused by alterations in the splicing patterns of the mRNA transcripts, including the Hutchison Gilford progeria syndrome, where premature aging is caused by a synonymous mutation (G608G) in the Lamin A (LMNA) gene, which obstructs the normal post-translational processing of the protein product and leads to the premature aging phenotype (Eriksson et al. 2003). Similarly, alterations in the relative balance of alternatively-expressed microtubule-associated protein tau (MTAP) isoforms are a feature of Alzheimer’s disease-related tauopathies. (Chen et al. 2010)
Gene expression arrays provide a powerful technology for identifying age-related alteration to the levels of gene transcripts in a comprehensive genome-wide way. Identification of individual transcripts and functionally coherent gene sets that are under- or over-expressed with aging in humans would provide key insights into the mechanisms of aging processes and age-related disease (Zahn et al. 2006). This may provide a ‘biomarker signature’ for monitoring the effects of interventions to slow age related changes, in an easily accessible tissue, peripheral blood leucocytes.
A variety of age-related expression analyses in cell lines or stored cell material have been reported, although results have had limited reproducibility (de Magalhaes et al. 2009). This is likely to be due to the small sample sizes in previous studies and to the sensitivity of mRNA transcripts to variation in aspects of storage and handling (Min et al. 2010). It is clear that identifying robust changes in age-related gene expression in humans will depend on large numbers of samples collected with optimal sample handling, so that results reflect in-vivo mRNA expression. Blood-derived leucocytes are a relevant tissue for the study of in-vivo aging processes in humans, as ‘immunosenescence’ is well described (Gruver et al. 2007). Blood is likely to remain the principal accessible ‘live’ tissue for large-scale in-vivo expression studies and clinical analysis in humans. Blood-derived white cell transcriptome studies have already proved valuable in identifying signatures of major diseases and drug responses some with promising clinical applications (Dumeaux et al. 2010).
We used a well-characterized population representative cohort, the InCHIANTI aging study (Ferrucci et al. 2000), to examine transcriptome-wide alterations in gene expression associated with chronological age in samples from 698 individuals by microarray analysis. We predicted widespread transcriptomic alterations with advancing age, and that inflammatory or immune function genes would be prominent in our results given our choice of target tissue (peripheral white blood cells). We aimed to identify both the most deregulated individual transcripts, but also the most deregulated gene sets grouped into biological pathways.
We found that although the largest single-transcript associations in human peripheral blood leucocytes do indeed include genes involved in inflammation or immune function, widespread alterations in gene expression levels were not apparent. In fact only a very small proportion (295 of 16,571 transcripts; 2%) of transcripts demonstrated robust age-related differences in gene expression. Furthermore, a statistical model using just 6 of the top 25 transcripts was able to classify samples into ‘young’ or ‘old’ with high precision. These six transcripts may provide a biomarker set to monitor interventions aimed at slowing aging.
Gene Set Enrichment Analysis (GSEA) (a method to determine whether specific molecular or functional pathways are associated with a given trait) demonstrated that the pathways most disrupted by human aging include genes involved in messenger RNA splicing, polyadenylation and other post-transcriptional events. This was accompanied by specific changes in the ratios of expression between isoform-specific transcripts for selected genes in our extended analysis. Deregulation of mRNA processing pathways may comprise a mechanism involved in human cellular aging.
Results
A small proportion of transcripts demonstrate marked age-related expression differences
16,571 transcripts gave reliable signals above background (p≤0.01) in >5% of the sample population following QC of microarray data, and were selected for analysis of gene expression alterations with age. The cohort was divided into discovery and test sets, based on analytical batch. Individual probes were only considered to be associated with chronological age (with adjustment for major confounders) if they reached a False Discovery Rate (FDR) q<0.001 in the discovery set, and also replicated in the test set with an FDR q<0.05. Of 16,571 transcripts, we identified gene expression differences with age in only 2%; 360 probes in the discovery set, of which 295 replicated in our test set (see supplementary table S1 online). Fifty (18%) of differentially-expressed transcripts were up-regulated and 245 were down-regulated. Analysis of the extremes of the age distribution only (<50years (n=93) versus >80 years (n=232) yielded only 53 probes associated with age, supporting our observation that human aging is associated with large-scale alterations in the expression of only a small number of transcripts.
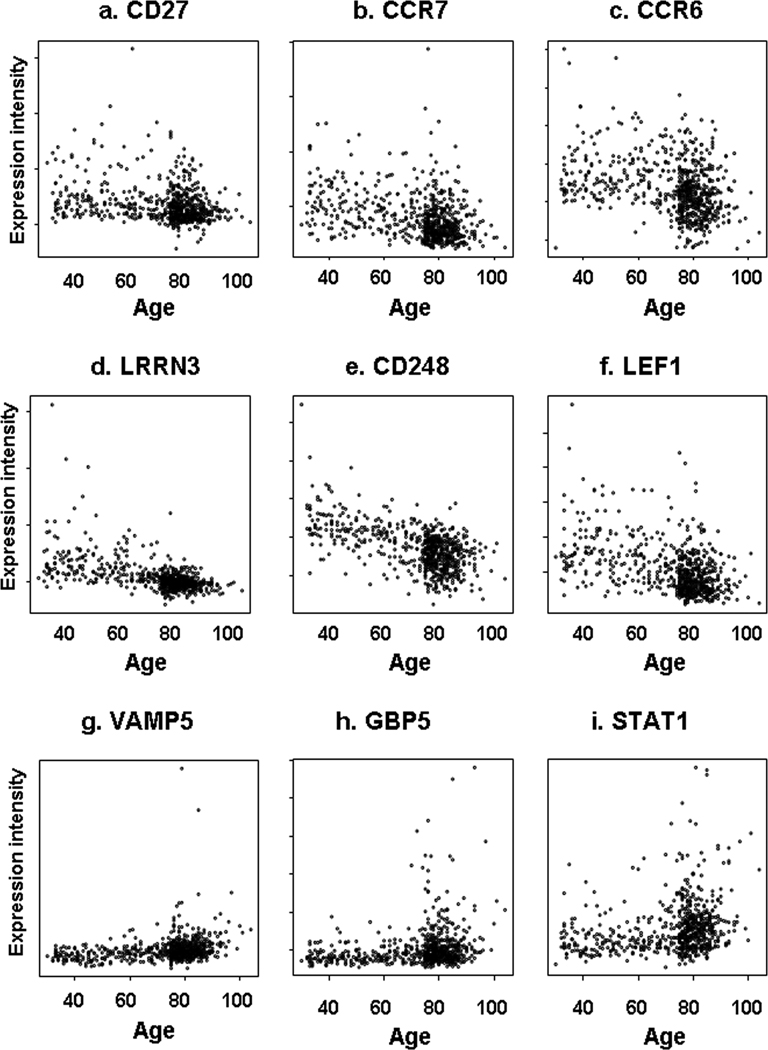
The CCR6 (chemokine receptor 6), CCR7 (chemokine receptor 7) and CD27 genes, previously associated with age and immunosenescence (Yung et al. 2007), were also found to be associated with age in our study. Plots of microarray signal intensity against increasing age (Cloud plots) for these, and the top 3 up- and down- regulated genes in our analysis; namely leucine-rich repeat neuronal protein 3 (LRRN3), endosialin (CD248), lymphoid enhancer-binding factor 1 (LEF1), vesicle-associated membrane protein 5 (VAMP5), guanylate binding protein 5 (GBP5) and signal transducer and activator of transcription 1 (STAT1) are presented in figure 1. The 25 most up-regulated and the 25 most down-regulated transcripts are shown in tables 1a and 1b. The most statistically significant association between age and gene expression was for the LRRN3 gene (p = 8.2 × 10−28), thought to be involved in MAPK activity and endocytosis.
Figure 1. The expression of key genes with age.
The figure shows the association between probe signal intensity (arbitrary units; Y axis) and age (years; X axis) for three genes known to be associated with age; CD27, CCR6 and CCR7 (a–c), the top 3 down-regulated genes in our study; LRRN3, CD248 and LEF1 (d–f) and the top 3 up-regulated genes in our study; VAMP5, GBP1 and STAT1 (g–i)
Table 1.
| a. The most strongly down-regulated genes with advancing age, by discovery and replication set. | ||||||||
|---|---|---|---|---|---|---|---|---|
| DISCOVERY SET | REPLICATION SET | |||||||
| Number (by discovery p-value) |
Probe | Probe ID | age coefficient |
p-value | FDR q- value |
age coefficient |
p-value | FDR q- value |
| 1 | LRRN3 | ilmn_1773650 | −0.0058 | 9.20E-32 | 8.20E-28 | −0.0067 | 3.70E-16 | 2.20E-12 |
| 2 | CD248 | ilmn_1726589 | −0.003 | 3.40E-27 | 1.50E-23 | −0.0032 | 1.20E-17 | 1.40E-13 |
| 3 | LRRN3 | ilmn_2048591 | −0.0029 | 1.80E-23 | 5.40E-20 | −0.0023 | 1.30E-06 | 5.00E-04 |
| 4 | LEF1 | ilmn_2213136 | −0.0124 | 4.20E-16 | 9.30E-13 | −0.0131 | 7.60E-09 | 1.20E-05 |
| 5 | LEF1 | ilmn_1679185 | −0.0131 | 2.80E-15 | 5.00E-12 | −0.0146 | 4.20E-09 | 8.70E-06 |
| 6 | NELL2 | ilmn_1725417 | −0.0055 | 8.80E-15 | 1.30E-11 | −0.0056 | 2.70E-08 | 3.10E-05 |
| 7 | ABLIM1 | ilmn_1785424 | −0.0092 | 2.00E-14 | 2.50E-11 | −0.0089 | 5.90E-07 | 3.00E-04 |
| 8 | FAM102A | ilmn_2401779 | −0.0055 | 3.20E-14 | 3.60E-11 | −0.006 | 9.00E-09 | 1.30E-05 |
| 9 | CCR6 | ilmn_1690907 | −0.0024 | 2.40E-13 | 2.30E-10 | −0.0022 | 1.10E-05 | 2.30E-03 |
| 10 | ZSCAN18 | ilmn_1654946 | −0.004 | 7.10E-13 | 5.80E-10 | −0.0043 | 7.70E-08 | 6.70E-05 |
| 11 | BZW2 | ilmn_1676548 | −0.0016 | 7.80E-13 | 6.20E-10 | −0.001 | 2.30E-03 | 5.68E-02 |
| 12 | CCR7 | ilmn_1715131 | −0.0164 | 1.20E-12 | 8.10E-10 | −0.0228 | 5.80E-10 | 1.70E-06 |
| 13 | LTB | ilmn_2376204 | −0.0076 | 1.20E-12 | 8.40E-10 | −0.0084 | 2.10E-08 | 2.70E-05 |
| 14 | GRAP | ilmn_2264011 | −0.0091 | 1.60E-12 | 1.00E-09 | −0.0099 | 4.90E-07 | 3.00E-04 |
| 15 | HGFL | ilmn_1719986 | −0.011 | 2.10E-11 | 1.10E-08 | −0.0127 | 1.70E-06 | 6.00E-04 |
| 16 | PASK | ilmn_1667022 | −0.0041 | 2.90E-11 | 1.50E-08 | −0.0055 | 5.30E-07 | 3.00E-04 |
| 17 | KIAA1147 | ilmn_2355033 | −0.0044 | 3.10E-11 | 1.60E-08 | −0.005 | 2.10E-05 | 3.50E-03 |
| 19 | ABLIM1 | ilmn_2396672 | −0.0027 | 4.30E-11 | 2.00E-08 | −0.0034 | 1.70E-06 | 6.00E-04 |
| 20 | SYPL1 | ilmn_2377174 | −0.0038 | 6.00E-11 | 2.60E-08 | −0.0041 | 8.20E-05 | 8.10E-03 |
| 21 | CD27 | ilmn_1688959 | −0.0035 | 7.20E-11 | 2.90E-08 | −0.0034 | 1.40E-06 | 6.00E-04 |
| 22 | DQ894085 | ilmn_1680618 | −0.0048 | 7.40E-11 | 3.00E-08 | −0.0059 | 1.10E-07 | 8.40E-05 |
| 23 | DKC1 | ilmn_1671257 | −0.0033 | 1.60E-10 | 5.80E-08 | −0.0028 | 2.00E-04 | 1.47E-02 |
| 24 | FAIM3 | ilmn_1775542 | −0.0123 | 1.60E-10 | 5.90E-08 | −0.0126 | 2.60E-05 | 4.00E-03 |
| 25 | CDR2 | ilmn_1720270 | −0.0022 | 2.40E-10 | 8.60E-08 | −0.0029 | 7.40E-08 | 6.60E-05 |
| 26 | ATM | ilmn_2370825 | −0.0063 | 3.50E-10 | 1.20E-07 | −0.0063 | 3.60E-03 | 7.25E-02 |
| b. The most strongly up-regulated genes with advancing age, by discovery and replication set. | ||||||||
|---|---|---|---|---|---|---|---|---|
| DISCOVERY SET | REPLICATION SET | |||||||
| Number (by discovery p-value) |
Probe | Probe ID | age coefficient |
p-value | FDR q- value |
age coefficient |
p-value | FDR q- value |
| 18 | VAMP5 | ilmn_1809467 | 0.0092 | 3.70E-11 | 1.80E-08 | 0.0073 | 7.60E-05 | 7.60E-03 |
| 42 | GBP5 | ilmn_2114568 | 0.0107 | 4.60E-09 | 9.50E-07 | 0.0072 | 1.11E-02 | 1.21E-01 |
| 48 | STAT1 | ilmn_1691364 | 0.012 | 8.60E-09 | 1.60E-06 | 0.0133 | 2.80E-05 | 4.10E-03 |
| 51 | WARS | ilmn_1727271 | 0.009 | 1.50E-08 | 2.40E-06 | 0.0088 | 2.00E-04 | 1.41E-02 |
| 58 | WARS | ilmn_2337655 | 0.0093 | 2.10E-08 | 3.30E-06 | 0.009 | 5.00E-04 | 2.41E-02 |
| 74 | MT2A | ilmn_1686664 | 0.0131 | 6.30E-08 | 7.50E-06 | 0.014 | 7.00E-05 | 7.20E-03 |
| 77 | CTSA | ilmn_1719286 | 0.0049 | 8.40E-08 | 9.40E-06 | 0.0057 | 2.00E-04 | 1.46E-02 |
| 82 | SLC27A3 | ilmn_1719627 | 0.0034 | 9.60E-08 | 1.00E-05 | 0.0026 | 4.80E-03 | 8.42E-02 |
| 84 | FBXO6 | ilmn_1701455 | 0.0037 | 1.20E-07 | 1.20E-05 | 0.0024 | 3.45E-02 | 2.00E-01 |
| 95 | VCAN | ilmn_1687301 | 0.0079 | 1.60E-07 | 1.50E-05 | 0.0065 | 3.30E-03 | 6.93E-02 |
| 100 | EIF4G3 | ilmn_1775692 | 0.0017 | 2.10E-07 | 1.90E-05 | 0.0016 | 2.60E-03 | 6.08E-02 |
| 101 | DUSP3 | ilmn_1797522 | 0.0046 | 2.30E-07 | 2.00E-05 | 0.0045 | 2.80E-03 | 6.30E-02 |
| 114 | GZMH | ilmn_1731233 | 0.017 | 5.70E-07 | 4.30E-05 | 0.0216 | 8.10E-06 | 1.90E-03 |
| 123 | PSMB9 | ilmn_2376108 | 0.0082 | 8.90E-07 | 6.00E-05 | 0.0061 | 1.18E-02 | 1.25E-01 |
| 133 | TTC38 | ilmn_2059886 | 0.0029 | 1.20E-06 | 7.30E-05 | 0.0041 | 7.10E-05 | 7.30E-03 |
| 148 | SYT11 | ilmn_1717934 | 0.0046 | 2.10E-06 | 1.00E-04 | 0.0049 | 1.90E-03 | 5.08E-02 |
| 142 | FCGR1A | ilmn_2261600 | 0.0095 | 1.70E-06 | 1.00E-04 | 0.0075 | 1.01E-02 | 1.16E-01 |
| 157 | WIPI1 | ilmn_1781386 | 0.0014 | 2.90E-06 | 2.00E-04 | 0.0013 | 3.70E-03 | 7.31E-02 |
| 164 | P2RX7 | ilmn_1759326 | 0.0017 | 3.20E-06 | 2.00E-04 | 0.0022 | 1.00E-03 | 3.80E-02 |
| 184 | TTC38 | ilmn_1692464 | 0.0042 | 5.20E-06 | 2.00E-04 | 0.0066 | 2.00E-04 | 1.27E-02 |
| 192 | H3F3A | ilmn_2402930 | 0.0047 | 6.00E-06 | 2.00E-04 | 0.0034 | 3.42E-02 | 1.99E-01 |
| 171 | STOM | ilmn_1766657 | 0.0052 | 4.00E-06 | 2.00E-04 | 0.0054 | 1.90E-03 | 5.13E-02 |
| 176 | STAT1 | ilmn_1690105 | 0.0063 | 4.30E-06 | 2.00E-04 | 0.0073 | 2.00E-04 | 1.51E-02 |
| 182 | MT1A | ilmn_1691156 | 0.0077 | 4.80E-06 | 2.00E-04 | 0.0092 | 1.00E-03 | 3.71E-02 |
| 205 | B3GAT1 | ilmn_1761093 | 0.0012 | 8.80E-06 | 3.00E-04 | 0.0018 | 2.90E-06 | 9.00E-04 |
The gene and probe identities are given in each case. The age co-efficient, the significance p value and the false discovery rate (FDR) q value are also given.
The gene and probe identities are given in each case. The age co-efficient, the significance p value and the false discovery rate (FDR) q value are also given.
It is possible that some of the effects we note may result from increases in age-related variation in inter-individual expression levels. However, the Breusch Pagan test of conditional heteroscedasticity did not provide evidence to support this hypothesis. The lowest p-value for the effect of age on residual expression variability failed to reach genome-wide significance before or after adjustment for age effects on mean expression (minimum p=3.2 × 10−5, FDR q=0.18). Therefore, these results provide no evidence for increased variability in inter-individual gene expression with age.
Six of the most deregulated 25 probes provide good discrimination between young and old
We then identified which of the 25 most strongly age-associated probes provide the best discrimination between a younger group (age < 65; n=107 in discovery set) and older subjects (age ≥ 75; n=308 in discovery set). We used multivariable logistic regression modelling to select probes that add significantly to classification (see methods, statistical analysis). Our final model included six genes; LRRN3, CD248, CCR6, GRAP (growth factor receptor-bound protein 2 related adaptor protein), VAMP5 and CD27, which together explained 63% of the age-group associated variation (supplementary table S2 online). The multivariable logistic regression model was used to generate an age-group classification for subjects in the independent replication set. The ROC curve (supplementary fig. 1 online) shows that this small subset of genes is sufficient to achieve exceptionally strong separation between older and younger subjects (ROC Area Under the Curve, AUC=96% in discovery set and 95% in validation set). The area under the curve (AUC) for alternative age-group comparisons within our replication set are 91% for < 60 v age >= 60 and 84% for the age 60–69 v age 70–79.
Key expression changes replicate when assayed by an alternative method; TaqMan Low Density Array (TLDA) analysis
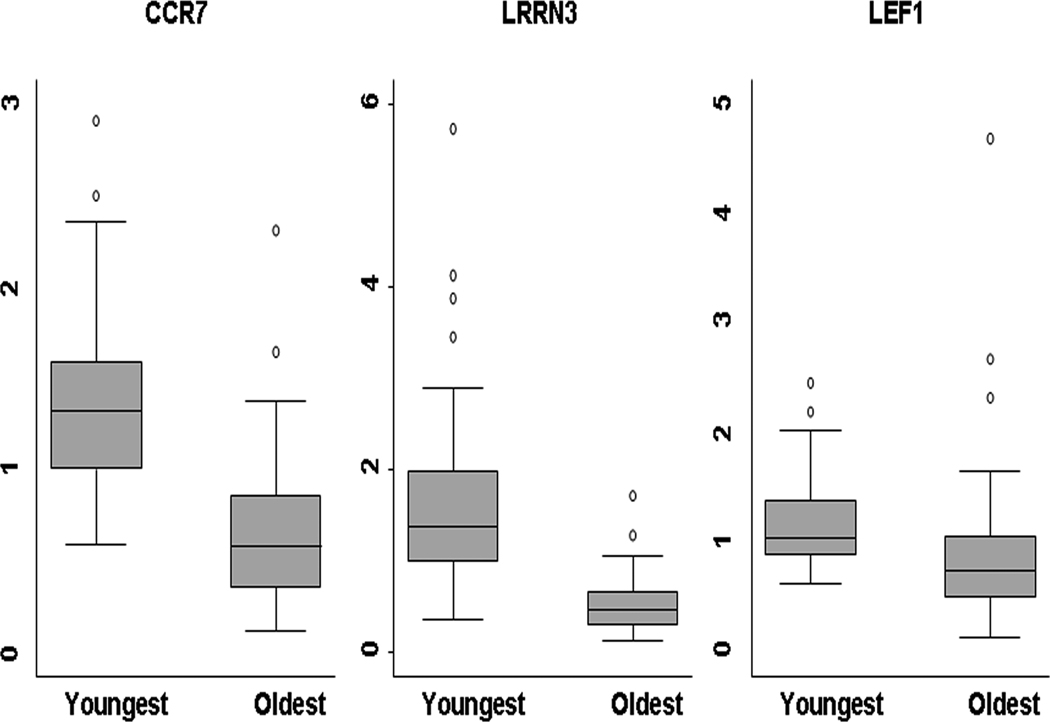
Taqman Low Density Array (TLDA) quantitative real-time PCR provides a robust independent laboratory technique to validate array results and provides more accurate estimates of effect sizes. We validated 27 of the most differentially-expressed genes identified by microarray analysis using TLDA quantitative real-time PCR. Of the 27 genes tested, 22 showed clear differences in gene expression with age (see supplementary table S3 online). Representative box plots (comparing data for 49 respondents aged 30 to 44 years and 50 respondents aged 85 to 104) for one gene previously associated with aging (CCR7) (Yung et al. 2007) and two of the top associations found in our study, LRRN3 and LEF1, are given in figure 2. Of those that did not replicate, two had no detectable expression (AKTIP and IGLL1) and three (RPS5, E2F5 and VEGFB) showed evidence of a trend for altered expression, but were not statistically significant. These transcripts were at the lower limit of statistical significance following Bonferroni correction for multiple testing (p~1 × 10−5) on the microarray data, indicating that some of the genes located at the limits of statistical significance may not represent genuine hits.
Figure 2. Box plots showing gene expression changes obtained by TLDA.
This figure demonstrates the difference in expression levels between young (30 – 44 years; n = 49) and old (85 – 104 years; n = 50 years) individuals as assessed by TLDA analysis. Gene expression levels expressed relative to the endogenous controls are given on the Y axis and the patient group (young or old) are given on the X-axis.
For our top gene (LRRN3) the mean expression intensity in the young sample (aged 30 to 44 yrs) was 1.65 (95%CI for the mean 1.36 – 1.94), compared to 0.53 (CI 0.44 – 0.62) in the older sample (85 – 104 yrs), a highly significant difference (p<0.0001) (supplementary table S3 online). This LRRN3 change represented a very large reduction i.e. the expression in the older sample is 32% of that in the young. In our TLDA validated genes, the expression in the older group ranged from 32% to 175% of that in the young.
Gene Set Enrichment Analysis (GSEA) reveals that the major pathways affected by age in humans relate to post-transcriptional processing of messenger RNA transcripts
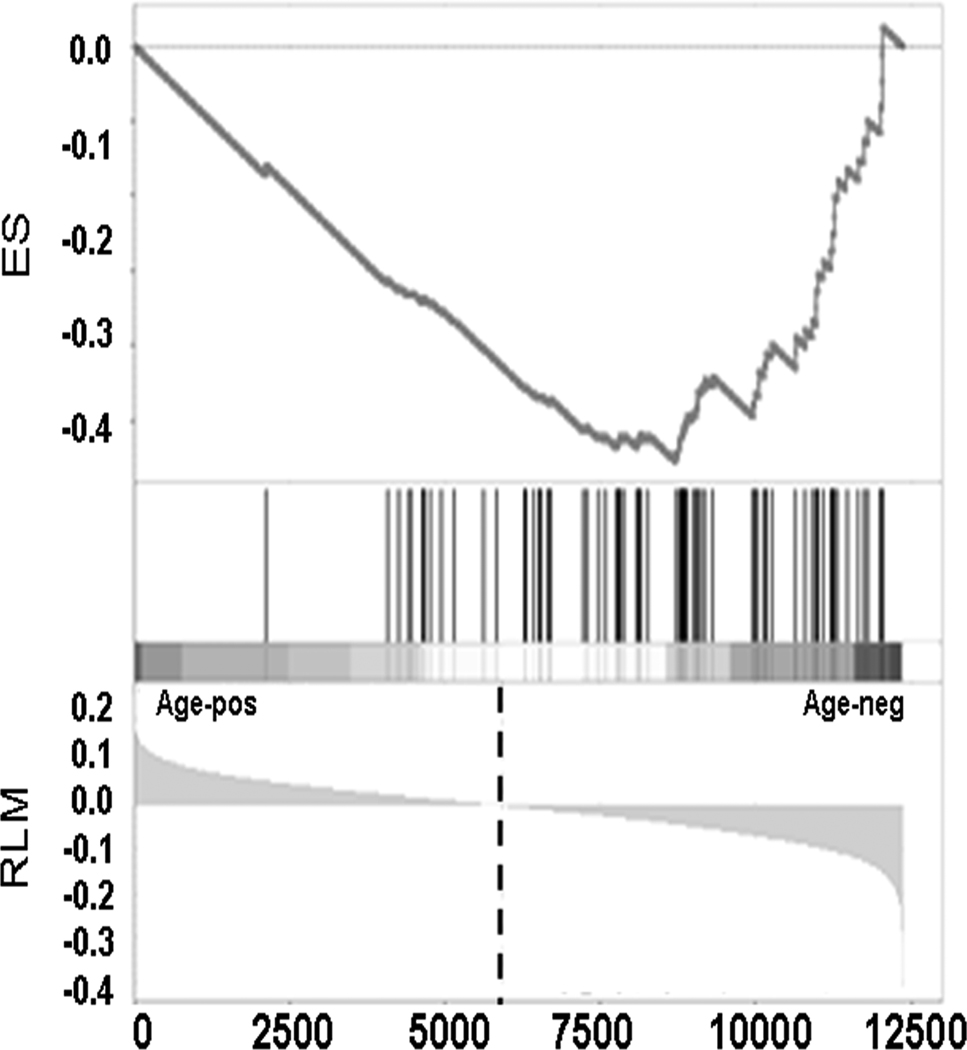
We applied GSEA (Subramanian et al. 2005) to identify pathways (rather than individual transcripts) that are associated with age. We examined 1065 predefined gene sets, grouped by (mainly Gene Ontology) classification of molecular function or biochemical processes. Of the 1065 pathways examined, only seven pathways were significantly associated with age, after accounting for the multiple statistical testing (FDR q-value<0.05) (table 2). These were: total RNA binding (nominal p-value<0.001), mRNA metabolic process (p<0.001), mRNA binding (p<0.001), RNA splicing (p<0.001), mRNA processing (p<0.001), ribonucleoprotein complex biogenesis and assembly (p<0.001) and chromatin assembly or disassembly (p=0.002). Figure 3 shows the enrichment plot for the RNA splicing pathway. Leading edge analysis (which identifies specific genes driving observed pathway associations) revealed that there was significant overlap between genes in these pathways, and that four pathways relate essentially to the same process; post-transcriptional processing of messenger RNA transcripts (supplementary fig. 2 online). The remaining pathways involve genes responsible for opening or closing the chromatin structure to allow (or disallow) transcription, and genes involved in the production and action of ribosomes, permitting translation of the processed messenger RNA transcript. GSEA also revealed two biological function gene sets possibly up-regulated in older people (extracellular region genes p=0.004) and ion transport genes p=0.008), although neither were significant when accounting for multiple testing (FDR q-value=0.197 and q-value=0.207 respectively).
Table 2.
Gene Set enrichment analysis plots for potential age associated gene sets, for molecular function or biochemical process pathways.
| Significance rank with age |
Pathway | Source of pathway definition |
Size (number of genes in pathway) |
NES | Nominal p-value |
FDR q- value |
|---|---|---|---|---|---|---|
| 1 | RNA binding | GO | 200 | −2.00092 | <0.001 | 0.023 |
| 2 | mRNA metabolic processes | GO | 73 | −1.92912 | <0.001 | 0.027 |
| 3 | mRNA binding | GO | 19 | −1.93444 | <0.001 | 0.029 |
| 4 | RNA splicing | GO | 75 | −1.95339 | <0.001 | 0.029 |
| 5 | mRNA processing | GO | 63 | −1.8974 | <0.001 | 0.03 |
| 6 | Ribonucleoprotein complex biogenesis/assembly |
GO | 68 | −1.89765 | <0.001 | 0.036 |
| 7 | Chromatin assembly or disassembly | GO | 20 | −1.95669 | 0.002 | 0.041 |
| 8 | RNA processing | GO | 136 | −1.98935 | <0.001 | 0.057 |
| 9 | Protein RNA complex assembly | GO | 56 | −1.82204 | 0.004 | 0.057 |
| 10 | DNA replication | GO | 73 | −1.82487 | 0.002 | 0.063 |
| 11 | DNA dependent DNA replication | GO | 37 | −1.77582 | 0.004 | 0.085 |
| 12 | Purine Metabolism | KEGG | 101 | −1.95911 | <0.001 | 0.089 |
| 13 | Organelle lumen | GO | 346 | −1.66622 | 0.002 | 0.091 |
| 14 | Negative regulation of programmed cell death |
GO | 110 | −1.71244 | <0.001 | 0.099 |
Size refers to the number of genes in the pathway. NES refers to the normalised enrichment score for each gene. The false discovery rate (FDR) is given by the q value and the statistical significance by the p-value. GO refers to the Gene ontology subset and KEGG to the Kyoto encyclopedia of genes and genomes.
Figure 3. Gene Set enrichment analysis plot for the RNA splicing pathway.
The Y axis gives the enrichment score for an association with age in the top panel and the ranked list matrix in the bottom panel. The X-axis refers to the rank of each gene in the ordered dataset. Each vertical line in the central portion of the figure refers to one gene within the pathway. The position of each line relative to the central dashed line indicates whether the gene is positively or negatively correlated with age. Positive correlations with age (age-pos) locate to the left of the figure and negative correlations with age (age-neg) locate to the right side of the figure. The dashed line represents the null point, where a gene demonstrating no positive or negative correlation would appear. RLM = Ranked List Metric, ES = Enrichment score.
Evidence of age related changes in isoform ratios with advancing age
Given the prevalence of genes involved in mRNA processing in our GSEA results, we sought to determine whether the pattern of alternatively-expressed isoforms might therefore be disrupted with age. Of the ten genes fulfilling our criteria for study, we found evidence of disruption to splicing patterns in seven of these (actin binding LIM protein 1 [ABLIM1], signal transducer and activator of transcription 1 [STAT1], CD79a molecule, immunoglobulin-associated alpha [CD79A], heat shock 60kDa protein 1 [HSPD1], clusterin associated protein 1 [CLUAP1], lysosomal-associated membrane protein 2 [LAMP2] and SON DNA binding protein [SON]), and the eighth, caspase 8 (CASP8) was near significant at p = 0.097 (supplementary fig. 3 online; table 3). The ABLIM1 gene codes for four alternatively processed reference sequence isoforms, NM_002313.5, NM_001003407.1, NM_001003408.1 and NM_006720.3. Comparison of the amount of NM_006720.3 relative to the other three isoforms reveals a progressive increase with age (correlation co-efficient 0.0009; p= <0.0001; supplementary fig. 3 online). The STAT1, CD79A, HSPD1, CLUAP1, LAMP2 and SON genes exhibit similar disturbances to isoform ratios with advancing age (table 3).
Table 3.
Analysis of the effect of age on the expression of alternatively-spliced isoforms.
| Gene | id | isoform | Mean | se | Comparison | Coeff | CI | CI | p-value |
|---|---|---|---|---|---|---|---|---|---|
| ABLIM1 | |||||||||
| 1 | ilmn_1731610 | NM_006720.3 | 102.29 | 6.21 | |||||
| 2 | ilmn_2396672 | all | 115.61 | 10.49 | 1 / 2 | 0.0009 | 0.0006 | 0.0012 | <0.0001 |
| 3 | ilmn_1785424 | all | 209.54 | 53.73 | 1 / 3 | 0.0032 | 0.0027 | 0.0037 | <0.0001 |
| STAT1 | |||||||||
| 4 | ilmn_1777325 | NM_007315.3 | 317.9793 | 145.709 | |||||
| 5 | ilmn_1691364 | NM_139266.2 | 715.9618 | 329.8048 | 4 / 5 | −0.0026984 | −0.0033706 | −0.0020263 | 0.0033706 |
| CD79A | |||||||||
| 6 | ilmn_2410371 | NM_001783.3 | 94.26653 | 4.791344 | |||||
| 7 | ilmn_1734878 | all | 338.5959 | 213.6811 | 6 / 7 | 0.0028078 | 0.0021338 | 0.0034817 | 0.0021338 |
| HSPD1 | |||||||||
| 8 | ilmn_1784367 | NM_002156.42 | 96.76617 | 3.201362 | |||||
| 9 | ilmn_1766713 | all | 142.1247 | 19.87898 | 8 / 9 | 0.0012785 | 0.0008516 | 0.0017055 | 0.0008516 |
| CASP8 | |||||||||
| 10 | ilmn_1750596 | NM_024793.1 | 100.70 | 3.63 | |||||
| 11 | ilmn_2242491 | NM_024793.1 | 10361.48 | 1983.51 | 7 / 8 | 0.0000 | 0.0000 | 0.0000 | <0.0001 |
| LAMP2 | |||||||||
| 12 | ilmn_1752351 | NM_013995.2 | 238.35 | 52.02 | 12 / 14 | −0.0004 | −0.0011 | 0.0002 | 0.208 |
| 13 | ilmn_2279961 | NM_013995.2 | 261.87 | 83.04 | 12 / 13 | −0.0016 | −0.0030 | −0.0002 | 0.027 |
| 14 | ilmn_1673282 | NM_002294.2 | 354.04 | 97.12 | |||||
| MAX | |||||||||
| 15 | ilmn_1695633 | NM_145116.1 | 93.97 | 4.54 | 15 / 16 | −0.0001 | −0.0003 | 0.0002 | 0.578 |
| 16 | ilmn_1802457 | NM_145114.1 | 107.29 | 3.99 | 15 / 17 | −0.0002 | −0.0004 | 0.0001 | 0.155 |
| 17 | ilmn_1706546 | NM_145113.1 | 108.31 | 5.55 | 16 / 17 | −0.0001 | −0.0004 | 0.0002 | 0.423 |
| SON | |||||||||
| 18 | ilmn_1663090 | NM_032195.1 | 106.31 | 4.13 | |||||
| 19 | ilmn_2247664 | NM_138927.1 | 200.96 | 45.44 | 18 / 19 | 0.0017 | 0.0011 | 0.0023 | <0.0001 |
| WTAP | |||||||||
| 20 | ilmn_1734544 | NM_004906.3 | 238.35 | 52.02 | |||||
| 21 | ilmn_2279339 | NM_004906.3 | 261.87 | 83.04 | 20 / 22 | 0.0003 | −0.0002 | 0.0009 | 0.208 |
| 22 | ilmn_1657618 | NM_152857.1 | 354.04 | 97.12 | 21 / 22 | 0.0015 | −0.0014 | 0.0044 | 0.305 |
The ratio of alternatively expressed isoforms of specific genes in relation to increasing age is given in the table above. Association with age is calculated by logistic regression. Id = probe identity, isoform = transcript identity, Mean = mean ratio, se = standard error, comparison indicates which transcripts were used for the analysis, coeff = correlation coefficient, CI = confidence intervals. The analysis was carried out on 689 individuals.
In a separate analysis, we sought to provide extra evidence for disruption of splicing with increasing age using a more accurate quantitative technique, real-time PCR. We selected alternatively spliced genes from the top 250 genes most associated with age, for which commercial probes were available. Of 8 genes tested in the fifty youngest and fifty oldest individuals, we found evidence for disruption to the ratio of alternatively expressed isoforms in 3 (EFNA1, GPR18 and VCAN; p = 0.05, 0.05 and 0.02–0.04 respectively depending on the comparison), with the 4th (BCL11B) being very near significance at p=0.053 (supplementary figure 4 online, table 4).
Table 4.
Genes showing disruption to the balance of alternatively-spliced isoforms by real-time PCR.
| Gene | id | isoform | Mean | se | Comparison | Coeff | CI | CI | p value |
|---|---|---|---|---|---|---|---|---|---|
| BCL11B | |||||||||
| 1 | hs01102259_m1 | NM_138576.2 | 1.058325 | 0.349844 | |||||
| 2 | hs01112109_m1 | NM_022898.1 | 1.062152 | 0.36903 | 1 over 2 | 0.00125 | −0.0000175 | 0.0025182 | 0.053 |
| EFNA1 | |||||||||
| 3 | hs00358887_m1 | NM_004428.2 | 1.376142 | 1.084887 | |||||
| 4 | hs01020895_m1 | NM_182685.1 | 1.315693 | 1.144735 | 3 over 4 | −0.00394 | −0.0078545 | −0.0000253 | 0.049 |
| GPR18 | |||||||||
| 5 | hs00245542_m1 | NM_001098200.1 | 1.070176 | 0.407375 | |||||
| 6 | hs01649814_m1 | NM_005292.3 | 1.081336 | 0.446335 | 5 over 6 | 0.00189 | 3.60E-06 | 0.0037757 | 0.05 |
| VCAN | |||||||||
| 7 | hs01007937_m1 | NM_001164097.1 | 1.098376 | 0.559999 | |||||
| 8 | hs01007941_m1 | NM_001126336.2 | 1.086235 | 0.457961 | 7 over 8 | 0.002647 | 0.0001322 | 0.0051619 | 0.039 |
| 9 | hs01007943_m1 | NM_001164098.1 | 1.23125 | 0.845863 | 9 over 7 | −0.01048 | −0.0193924 | −0.0015691 | 0.022 |
| 10 | hs01007944_m1 | NM_004385.4 | 1.210006 | 0.92149 | 9 over 10 | −0.00778 | −0.0154129 | −0.0001444 | 0.046 |
| ANXA7 | |||||||||
| 11 | hs00244504_m1 | NM_004034.2 | 1.066305 | 0.388382 | 12 over 11 | ||||
| 12 | hs00559416_m1 | NM_001156.3 | 1.929157 | 8.420153 | 11 over 12 | −0.00219 | −0.0057466 | 0.0013578 | 0.223 |
| CXCR5 | |||||||||
| 13 | hs00540527_s1 | NM_001716.3 | 1.116763 | 0.540786 | 14 over 13 | ||||
| 14 | hs00540548_s1 | NM_032966.1 | 1.100177 | 0.497535 | 13 over 14 | −0.0011 | −0.0026285 | 0.0004256 | 0.156 |
| IGL6ST | |||||||||
| 15 | hs01006741_m1 | NM_002184.3 | 1.068342 | 0.397245 | 16 over 15 | ||||
| 16 | hs01006742_m1 | NM_175767.2 | 1.898088 | 5.478007 | 15 over 16 | 0.004691 | −0.0045713 | 0.0139524 | 0.317 |
| PUM1 | |||||||||
| 17 | hs00982771_m1 | NM_001020658.1 | 1.031583 | 0.253846 | |||||
| 18 | hs00985111_m1 | NM_014676.2 | 1.091318 | 0.995023 | 17 over 18 | 0.001596 | −0.0003754 | 0.0035682 | 0.111 |
The ratio of alternatively expressed isoforms of specific genes as determined by real-time PCR is correlated with age in the table above. Association with age is calculated by logistic regression. Id = probe identity, isoform = transcript identity, Mean = mean ratio, se = standard error, comparison indicates which comparisons were made for the analysis, coeff = correlation coefficient, CI = confidence intervals. The analysis was carried out on 100 individuals.
Discussion
In this study we present the first assessment of age-related alterations in gene expression in a large population-based cohort. Contrary to our expectations, we found relatively few age-associated transcripts, but these did include several genes associated with inflammation or immune senescence, as expected. Gene Set Enrichment Analysis (GSEA) indicated that only a very small number of molecular or biological function pathways were robustly associated with advancing age, comprising mainly genes involved in the processing and maturation of messenger RNA transcripts. This finding was supported by subsequent observation of age-associated differences in the balance of alternatively-expressed isoforms, as suggested by previous studies (Yannarell et al. 1977; Meshorer & Soreq 2002). Our results suggest that modification of messenger RNA (mRNA) processing may comprise an important feature of human aging.
Very few genes (295 of 16,571; 2% of transcripts identified) demonstrated strong and robust associations with advancing age in our study. Power calculations based on the method of Hsieh (Hsieh et al. 1998) indicate that the relatively large sample available (n=458 in discovery set) is sufficient to detect effect sizes for transcripts statistically ‘explaining’ as little as 5.3% of the variation in age. Thus if we have not detected other genes due to ‘noisy’ data, individual effect sizes for such genes are likely to be of limited biological significance.
The transcripts with the largest effect sizes comprise a set of peripherally expressed biomarkers of the aging process, some substantially stronger than those currently known. Transcripts such as CCR6, CCR7 and CD27, previously reported to be involved in immune senescence (Yung et al. 2007), were also associated with age in our study (figure 1). As predicted, genes showing the largest differential in expression with age were involved in inflammatory responses, or immune function (de Magalhaes et al. 2009). Genes related to the human immune system, e.g. LRRN3, CD248, LEF1 (lymphoid enhancer-binding factor 1), CCR6, CCR7, CD27 and LTB (lymphotoxin beta - TNF superfamily, member 3) expressed lower levels of mRNA transcripts with increasing age in our cohort, which is in agreement with other studies in human peripheral lymphocytes (Hong et al. 2008). This is not unexpected given that our study tissue was white blood cells. Reduced expression of these genes with age implies that the mechanisms involved are likely to mark the process of “immunosenescence”, characterised by reduced chemotactic migration of the immune mediator cells, lowered activation and differentiation of lymphocytes and macrophages with reduced synthesis of immunoglobulins and increased apoptosis of immune cells. Our finding of a strong association of lower CCR7 gene expression with advancing age validates the findings from Yung et al. (Yung et al. 2007).
Our findings should also be considered in the context of age-related changes in lymphocyte composition. Some age-associated transcripts may be expressed in different white cell subtypes; the CD28, CCR7 and GZMH genes are differentially expressed in CD8+CD28+ T-cells compared with CD8+CD28− T-cells, the ratio of which is know to alter with age (Lazuardi et al. 2009). Some of the alterations in gene expression may therefore derive from differences in the composition of the lymphocyte population in older people. What remains to be seen is whether any of alterations in lymphocyte composition are themselves attributable to the altered gene expression patterns we have demonstrated.
Several other factors have been implicated in aging or longevity, including alterations to nutrient sensing pathways, such as insulin or TOR signalling. Oxidative stress/DNA repair, inhibition of respiration, reproductive system signaling and telomere related mechanisms (Kenyon 2010). Accordingly, genes representing some of these processes are evident amongst the top 100 associations we have identified. These include the PASK, FOXO1, DKC1 and MYC transcripts, involved in energy sensing, insulin signalling, telomere maintenance and ribosomal biogenesis respectively (Gu et al. 2009; Schlafli et al. 2009; Dai et al. 2010; Smith et al. 2010).
The association of individual gene transcripts with age does not account for correlations between markers. We developed a bi-class discriminant model which identified a set of six transcripts within the top 25 strongest associations which form a potential predictive signature of chronological aging (supplementary table S2; supplementary fig. 1 online). This model was exceptionally good at classifying young and old, (the area under ROC curve was 96% in our discovery set, and this was only slightly diminished [95%] in the independent replication set). The six genes in the predictive group comprise genes involved in immunity or inflammation (LRRN3, CD27, CCR6) but also include some that are involved in the maintenance or development of muscle tissue (VAMP5; myobrevin) or in vascularisation (CD248, endosialin). Future versions of this discriminant model might attempt to disentangle ‘aging’ from age-related disease effects: however, doing so is conceptually and practically difficult, but the relative robustness of the model for different age-group comparisons suggests that the transcript set is not merely distinguishing disease-free, from diseased, individuals.
Gene Set Enrichment Analysis (GSEA) revealed a striking restriction in the number of pathways that were associated with age in our dataset; only 7 of 1065 molecular or biological function pathways are robustly associated with age. Four of these pathways identified are involved in mRNA processing (namely mRNA binding, mRNA processing, mRNA processing reactome and RNA splicing). Three of the age-associated pathways we identified (RNA binding, RNA splicing and ribosome biogenesis and assembly) were also noted in previous GSEA analyses in mice (Southworth et al. 2009). Differences between our results and the studies may arise from differences between species in the aging process, or from differences in the tissue-specificity of changes.
RNA processing is the mechanism by which the initial RNA products transcribed from genes are prepared for eventual translation. This includes removal of introns and addition of the poly-A tail and 5’ Cap structures (Keene 2010). These processes occur simultaneously and ensure diversity of the mRNA transcriptome, and determine stability and half-life of the mRNA transcripts (Fong & Bentley 2001). In our GSEA analysis, transcripts responsible for all aspects of mRNA processing were present in the core-enriched fraction of the pathway (supplementary table S4 online), with a surprising amount of overlap between the groups (supplementary fig. 2 online). The remaining pathways relate to the accessibility of the chromatin to transcription factors (chromatin assembly and disassembly) and to the processes that surround the translation of the mRNA transcripts (RNA binding and ribonucleoprotein complex biogenesis and assembly), which is in keeping with previous observations of a reduction in rates of protein synthesis with age in humans and animal species (Ballard & Read 1985; Kennedy & Kaeberlein 2009).
Our observation that disruption to the proteins involved in mRNA processing appears to occur without widespread alterations in gene expression levels, may indicate that whilst aged leukocytes in-vivo may be expressing most genes at comparable levels to those found in younger cells, there may be differences in the relative balance of splice products produced or increases in the occurrence of aberrantly spliced transcripts. In seven of ten alternatively spliced genes we studied, we found disruptions to the patterns of isoform expression with increasing age, and a near-significant result in one further case (table 3). We also noted variation in splicing patterns with age by real-time PCR, finding alteration to the balance of alternatively expressed isoforms in 3 of 8 (38%) alternatively spliced genes, with the 4th very near significance. Our observation of very modest effects even in a very limited cohort of 100 people (7% to 37% alteration in the ratio of isoforms)suggests that the true level of splicing disruption may be higher than we report and warrants a more detailed, targeted study.
Whether or not the processing of a particular transcript is disrupted will depend on many factors, not least how many different sequence elements are necessary to ensure the usage of a particular splice junction. Presumably, highly regulated transcripts with weaker splice sites that are very dependent on additional sequence factors (such as exon splicing enhancers (Cartegni et al. 2002)) may be more susceptible to age-accumulated DNA and RNA damage and thus more likely to show disruption of splicing and other related processes with age. Interestingly, two of the most deregulated splicing proteins in our microarray data; SRFR6 (SRp55) and SRFS1 (ASF/SF2) (p = 1.3 × 10−6 and 0.0001 respectively), were members of the SRFS (Splicing Factor, Arginine/Serine-Rich) family of splicing factors, which are key players in maintaining the plasticitiy of the transcriptome by regulation of alternative splicing (Valcarcel & Green 1996). This is particularly interesting since these proteins are key ligands for the exon splicing enhancer (ESE) motifs which regulate splice site choice in development and differentiation (Cartegni et al. 2002). Their deregulated expression during the aging process may therefore manifest as a reduction in the adaptive capacity of the transcriptome.
Disruption to the mRNA processing machinery may also lead to an increase in the occurrence of unusual or aberrant splicing products, which of course would not be present on the microarray chip. This would not be an unexpected finding in an aging organism, given that they may arise from mutations in the DNA sequence elements that control splice site usage, or from alterations in the RNA transcript itself, which is very susceptible to damage by oxidative and other insult (Kong & Lin 2010). Some of the proteosomal components that intercept and neutralise aberrant proteins produced from such transcripts were up-regulated in our data (PMSB9; FDR q value = 0.008, p = 6 × 10−5, PMSB10; q= 0.005, p = 0.0008).
There is a growing body of evidence that results from peripheral blood white cells are also relevant to less accessible tissues (Tang et al. 2003; Twine et al. 2003; Achiron et al. 2004). In some reports, the majority of changes appeared to be in genes with little tissue specific regulation, suggesting that most of the transcriptomic alterations with age might be generalised (Rodwell et al. 2004; de Magalhaes et al. 2009). This is supported by other evidence, where age-related expression differences were compared in brain, kidney and muscle, in a population of 81 human subjects, and found to be conserved in each of these tissues (Zahn et al. 2007). The age-related transcriptomic signature is also relatively stable; of the 50 top associations reported by Hong et al (Hong et al. 2008), 32 of the 42 represented in our data also showed age-related differences, at least at p=<0.05 (data available from authors), despite the differences expected comparing our in-vivo leukocyte mRNA to mRNA from stored isolated lymphocytes. Lymphocytes are also the most appropriate tissue for the study of immune senescence, which is key to many chronic disease processes, including inflammation, and autoimmune alterations with age (Desai et al. 2010)
Possible limitations of our study analysis include the deliberate absence of accounting for specific disease. This is because an increased susceptibility to disease is intrinsic to any definition of aging, and thus accounting for disease would risk controlling out the associations of interest (and also poses major practical difficulties with undiagnosed disease being common at older ages). Similarly, our study does not account for variation in the transcriptome of different blood cell subtypes. However, as noted above our results are consistent with those from isolated lymphocytes (Hong et al. 2008), and our approach avoids disruption to in-vivo expression patterns.
In conclusion, we present the first genome-wide assessment of age-related in-vivo leukocyte gene expression profiles in a large population based study. As aging is associated with random damage to DNA (Gensler & Bernstein 1981), we tested the hypothesis that this would result in widespread deregulation of gene expression. Instead, we found that human aging is associated with a small number of focussed changes, mainly in individual genes associated with immune cell function. The major pathways associated with older age in humans were mainly involved with the processing of the primary RNA transcripts into mature mRNAs, an observation supported by our finding of age-related changes in the relative expression of alternatively-expressed isoforms of example loci. We suggest that disruption to messenger RNA processing may comprise an important feature of aging in the human population.
Experimental Procedures
Ethics Statement
Ethical permission was granted by the Instituto Nazionale Riposo e Cura Anziani institutional review board in Italy. Participants gave informed consent to participate.
Cohort details
The InCHIANTI study (Ferrucci et al. 2000) is a population-based, prospective epidemiological study of factors affecting aging, in the Chianti area (Tuscany) of Italy. The participants were originally enrolled in 1998–2000, and were interviewed and examined every three years. The recent 9-year follow-up exam involved 733 participants. Characteristics of the study cohort are given in table 5.
Table 5.
Characteristics of the discovery and replication samples. Clinical characteristics and demographics of the study cohort are given.
| Discovery Sample | Replication Sample | Pooled Sample | ||||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| Age (quartiles) | ||||||
| 30yrs – 68yrs | 121 | 26.42 | 59 | 24.58 | 180 | 25.79 |
| 69yrs – 78yrs | 141 | 30.79 | 72 | 30 | 213 | 30.52 |
| 79yrs – 82yrs | 91 | 19.87 | 43 | 17.92 | 134 | 19.2 |
| 83yrs – 104yrs | 105 | 22.93 | 66 | 27.5 | 171 | 24.5 |
| Gender | ||||||
| Men | 211 | 46.07 | 102 | 42.5 | 313 | 44.84 |
| Women | 247 | 53.93 | 138 | 57.5 | 385 | 55.16 |
| Site | ||||||
| Greve | 188 | 41.05 | 154 | 64.17 | 342 | 49 |
| Bagno a Ripoli | 270 | 58.95 | 86 | 35.83 | 356 | 51 |
| Education | ||||||
| Nothing | 65 | 14.19 | 29 | 12.08 | 94 | 13.47 |
| Elementary | 199 | 43.45 | 123 | 51.25 | 322 | 46.13 |
| Secondary | 55 | 12.01 | 38 | 15.83 | 93 | 13.32 |
| High school | 62 | 13.54 | 25 | 10.42 | 87 | 12.46 |
| University / Professional |
77 | 16.81 | 25 | 10.42 | 102 | 14.61 |
|
Pack years smoked (lifetime) |
||||||
| None | 249 | 54.37 | 130 | 54.17 | 379 | 54.30 |
| <20 | 97 | 21.18 | 56 | 23.33 | 153 | 21.92 |
| 20–39 | 71 | 15.50 | 30 | 12.50 | 101 | 14.47 |
| 40+ | 35 | 7.64 | 20 | 8.33 | 55 | 7.88 |
| missing | 6 | 1.31 | 4 | 1.67 | 10 | 1.43 |
| Waist Circumference (cm) | ||||||
| Mean (sd) | 458 | 95.6 (12.4) | 240 | 94.8 (11.5) | 698 | 95.3 (12.1) |
RNA collection and extraction
Peripheral blood specimens preserving in-vivo RNA expression were collected at the 9 year follow-up (2008/9), using the PAXgene technology to preserve levels of mRNA transcripts as they were at the point of collection (Debey-Pascher et al. 2009). RNA was extracted from peripheral blood samples using the PAXgene Blood mRNA kit (Qiagen, Crawley, UK) according to the manufacturer’s instructions.
Whole Transcriptome Scan
Whole genome expression profiling of the samples was conducted using the Illumina Human HT-12 microarray (Illumina, San Diego, USA) as previously described (Zeller et al. 2010). Pre-processing of microarray data is described in the supplementary methods.
Statistical analysis
Our dataset was subdivided on the basis of hybridization batch: a discovery dataset containing approximately two-thirds of the full sample (n=458 individuals), and an independent replication set (n=240 individuals). This is a form of test-set cross-validation, a common and well-established approach when assessing performance of classification models (Lubomirski et al. 2007).
We calculated median centered gene expression levels on the log2 scale to ensure maximum overlap of profiles without altering their variance, in line with methodology used in previous studies (Idaghdour et al. 2010). The relationship between gene expression and chronological age was tested using a linear regression model with (log-transformed) gene expression level as the dependent variable, chronological age (recorded at RNA extraction) as an explanatory variable, and with adjustment for potential confounders. Separate regression models were fitted for each of the full set of 16,571 probes which passed QC in the discovery dataset. We used the false discovery rate (FDR) to account for multiple testing, applying an FDR cut off of q ≤0.001 to select probes expressing differentially with age. Transcripts selected at the screening stage in the discovery set were tested for association with age in the replication dataset using the same model specification. The development of a bi-class discriminant model to identify the transcripts explaining the majority of the association with age is described in the supplementary methods.
All our analyses were adjusted for the following potential confounding factors on gene expression: gender; lifetime pack-years smoked (in five categories: none, less than 20 years, 20 to 39 years, 40 plus years, and missing); waist circumference (as a continuous trait); highest level of education attained (in five categories: none, elementary, secondary, high school, and university / professional); and study site (individuals were drawn from a rural village [Greve] and an urban population [Bagno a Ripoli]). We also controlled for potential hybridization and/or amplification batch effects in all our analyses (supplementary table S5 online).
TaqMan Low Density Array (TLDA) validation of microarray results
A subset of the cohort comprising the oldest men and women (85 – 104 years, n=50), was compared with one comprising the youngest men and women (30–44y years, n=49) in this analysis. Total RNA (100ng) was reversed transcribed in 20µl reactions using the Superscript III VILO kit (Invitrogen, Paisley, UK), according to the manufacturer’s instructions. RNA samples were then used for TLDA analysis. A list of target genes is given in supplementary table S6 (online). Each 32-gene set included four endogenous control genes which had been empirically validated as being unaffected by age on the basis of the microarray results; 18S, GUSB, PPIA and IDH3B. Reaction conditions are described in the supplementary methods online.
Pathway analysis (GSEA)
Gene Set Enrichment Analysis (GSEA was performed to assess pathways or pre-defined gene sets associated with chronological age according to the method of Subramanian et al, (Subramanian et al. 2005). “Enrichment statistic” and “Metric for ranking gene” parameters were configured to “Weighted” and “Pearson” respectively. One thousand random permutations of the phenotype label were used to calculate the empirical p-values of each pathway. The gene sets with a nominal of p-value < 0.01 and false discovery rate (FDR) of ≤ 25% were considered as potential associated gene sets as previously described (Subramanian et al. 2005). Gene symbols and the Illumina annotation file were used to collapse 16,571 probes to 12357 genes by taking the median intensity of probes representing each gene.
Molecular or biological function pathways and Gene Ontology (GO) gene sets were selected from the molecular signature database (MSigDB) (http://www.broadinstitute.org/gsea/msigdb/index.jsp). After filtering gene sets to those with a minimum of 15 and a maximum of 500 gene set size, 294, 439, 209 and 123 gene sets from the Canonical pathways, Biological Process Ontology gene sets, Molecular Function Ontology gene sets and Cellular Components Ontology gene sets were used in the analysis, respectively.
Examination of age related changes in microarray isoform ratios with advancing age
We examined the relative balance of alternatively expressed isoforms of selected genes for evidence of disruption to splicing patterns. We first examined the top 50 genes robustly associated with age (table S1) identified as candidates for study, but only four of them; ABLIM1, CD79A, STAT1 and HSPD1, showed evidence both of alternative splicing, and expression identified by suitable probes within the expression data available to us. We then selected further genes for study based on the following criteria: the presence of >5 alternative probes for the same gene, and the presence of >2 reference isoforms for the gene. The probe sequences were then mapped back onto the specific transcript sequences to determine which probe signals to use for analysis. By these methods, we identified ten genes for study: ABLIM1, CD79A, STAT1, HSPD1, CASP8, CLUAP1, LAMP2, MAX, SON and WTAP. We then carried out a logistic regression of the ratio of signal deriving from isoform-specific probes according to age. Data were adjusted for gender.
TaqMan Low Density Array (TLDA) assessment of disruption to mRNA splicing
A subset of the cohort comprising the oldest men and women (85 – 104 years, n=50), was compared with one comprising the youngest men and women (30–44y years, n=50) in this analysis. Total RNA (100ng) was reversed transcribed in 20µl reactions using the Superscript III VILO kit (Invitrogen, Paisley, UK), according to the manufacturer’s instructions. RNA samples were then used for TLDA analysis for transcripts of the ANXA7, BCL11B, CXCR5, EFNA1, GPR18, IL6ST, PUM1 and VCAN genes, which were selected on the basis that they were alternatively-expressed isoforms of genes in the top 250 associations with age which produced a minimum of 2 isoforms. Endogenous control genes were IDH3B and GUSB, which were identified as the most stable controls by the GeNORM function of the StatMiner TLDA analysis software (Integromics, UK). Reaction conditions are described in the supplementary methods online.
Supplementary Material
Acknowledgements
We thank the many people who contributed to the InCHIANTI study, including all of the anonymous participants. This study was supported in part by the Intramural Research Program, National Institute on Aging and the U.S. National Institutes of Health. We thank Luke Pilling for extra statistical analyses. William Henley completed this work while seconded to PenCLARHC which is funded by the National Institute of Health Research (NIHR). The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Footnotes
Author Contributions
LWH oversaw the validation experiments, interpreted the data and co-wrote the manuscript.
DH carried out the microarray experiments.
WH oversaw the statistical analysis, carried out the multivariable regression analysis and contributed to the manuscript..
AW carried out initial preprocessing of the data.
AH carried out the analysis of differential splicing with age
RBS carried out the TLDA validation of the microarray results.
HY carried out the Gene Set Enrichment analysis.
AD contributed to the manuscript.
AM oversaw the collection and extraction of the RNA samples.
JMG aided in design of the InCHIANTI study and contributed to the manuscript.
TF contributed to the manuscript.
SB oversees the InCHIANTI study and contributed to the manuscript.
AS oversaw the microarray experiments.
LF organized the sample cohort and contributed to the manuscript.
DM managed the project, interpreted the data, co-wrote the manuscript and contributed funding.
References
- Achiron A, et al. Blood transcriptional signatures of multiple sclerosis: unique gene expression of disease activity. Ann Neurol. 2004;55:410–417. doi: 10.1002/ana.20008. [DOI] [PubMed] [Google Scholar]
- Ballard FJ, et al. Changes in protein synthesis and breakdown rates and responsiveness to growth factors with ageing in human lung fibroblasts. Mech Ageing Dev. 1985;30:11–22. doi: 10.1016/0047-6374(85)90055-7. [DOI] [PubMed] [Google Scholar]
- Butler RN, et al. New model of health promotion and disease prevention for the 21st century. BMJ. 2008;337:a399. doi: 10.1136/bmj.a399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, et al. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Chen S, et al. MAPT Isoforms: Differential Transcriptional Profiles Related to 3R and 4R Splice Variants. J Alzheimers Dis. 2010 doi: 10.3233/JAD-2010-101155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, et al. Ribosomal protein L11 associates with c-Myc at 5 S rRNA and tRNA genes and regulates their expression. J Biol Chem. 2010;285:12587–12594. doi: 10.1074/jbc.M109.056259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, et al. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debey-Pascher S, et al. RNA stabilization of peripheral blood and profiling by bead chip analysis. Methods Mol Biol. 2009;496:175–210. doi: 10.1007/978-1-59745-553-4_13. [DOI] [PubMed] [Google Scholar]
- Desai A, et al. Leukocyte function in the aging immune system. J Leukoc Biol. 2010;87:1001–1009. doi: 10.1189/jlb.0809542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumeaux V, et al. Deciphering normal blood gene expression variation--The NOWAC postgenome study. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000873. e1000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci L, et al. Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc. 2000;48:1618. doi: 10.1111/j.1532-5415.2000.tb03873.x. [DOI] [PubMed] [Google Scholar]
- Fong N, et al. Capping, splicing, and 3' processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensler HL, et al. DNA damage as the primary cause of aging. Q Rev Biol. 1981;56:279–303. doi: 10.1086/412317. [DOI] [PubMed] [Google Scholar]
- Gruver AL, et al. Immunosenescence of ageing. Journal of Pathology. 2007;211:144–156. doi: 10.1002/path.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, et al. Dyskerin, telomerase and the DNA damage response. Cell Cycle. 2009;8:6–10. doi: 10.4161/cc.8.1.7265. [DOI] [PubMed] [Google Scholar]
- Hong MG, et al. Transcriptome-wide assessment of human brain and lymphocyte senescence. PLoS One. 2008;3:e3024. doi: 10.1371/journal.pone.0003024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh FY, et al. A simple method of sample size calculation for linear and logistic regression. Stat Med. 1998;17:1623–1634. doi: 10.1002/(sici)1097-0258(19980730)17:14<1623::aid-sim871>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Idaghdour Y, et al. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat Genet. 2010;42:62–67. doi: 10.1038/ng.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. Minireview: global regulation and dynamics of ribonucleic Acid. Endocrinology. 2010;151:1391–1397. doi: 10.1210/en.2009-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, et al. Hot topics in aging research: protein translation, 2009. Aging Cell. 2009;8:617–623. doi: 10.1111/j.1474-9726.2009.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kong Q, et al. Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell Mol Life Sci. 2010;67:1817–1829. doi: 10.1007/s00018-010-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazuardi L, et al. Microarray analysis reveals similarity between CD8+CD28− T cells from young and elderly persons, but not of CD8+CD28+ T cells. Biogerontology. 2009;10:191–202. doi: 10.1007/s10522-008-9167-1. [DOI] [PubMed] [Google Scholar]
- Lubomirski M, et al. A consolidated approach to analyzing data from high-throughput protein microarrays with an application to immune response profiling in humans. J Comput Biol. 2007;14:350–359. doi: 10.1089/cmb.2006.0116. [DOI] [PubMed] [Google Scholar]
- Meshorer E, et al. Pre-mRNA splicing modulations in senescence. Aging Cell. 2002;1:10–16. doi: 10.1046/j.1474-9728.2002.00005.x. [DOI] [PubMed] [Google Scholar]
- Min JL, et al. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genomics. 2010;11:96. doi: 10.1186/1471-2164-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodwell GE, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlafli P, et al. The PAS-domain kinase PASKIN: a new sensor in energy homeostasis. Cell Mol Life Sci. 2009;66:876–883. doi: 10.1007/s00018-009-8699-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Jr., et al. Calorie restriction: what recent results suggest for the future of ageing research. Eur J Clin Invest. 2010;40:440–450. doi: 10.1111/j.1365-2362.2010.02276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth LK, et al. Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000776. e1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, et al. Blood genomic expression profile for neuronal injury. J Cereb Blood Flow Metab. 2003;23:310–319. doi: 10.1097/01.WCB.0000048518.34839.DE. [DOI] [PubMed] [Google Scholar]
- Twine NC, et al. Disease-associated expression profiles in peripheral blood mononuclear cells from patients with advanced renal cell carcinoma. Cancer Res. 2003;63:6069–6075. [PubMed] [Google Scholar]
- Valcarcel J, et al. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- Yannarell A, et al. Age-dependence of nuclear RNA processing. Mech Ageing Dev. 1977;6:259–264. doi: 10.1016/0047-6374(77)90026-4. [DOI] [PubMed] [Google Scholar]
- Yung R, et al. The effect of aging and caloric restriction on murine CD8+ T cell chemokine receptor gene expression. Immun Ageing. 2007;4:8. doi: 10.1186/1742-4933-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, et al. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genetics. 2006;2:1058–1069. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller T, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.