Abstract
Carboxysomes are metabolic modules for CO2 fixation that are found in all cyanobacteria and some chemoautotrophic bacteria. They comprise a semi-permeable proteinaceous shell that encapsulates ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) and carbonic anhydrase. Structural studies are revealing the integral role of the shell protein paralogs to carboxysome form and function. The shell proteins are composed of two domain classes: those with the bacterial microcompartment (BMC; Pfam00936) domain, which oligomerize to form (pseudo)hexamers, and those with the CcmL/EutN (Pfam03319) domain which form pentamers in carboxysomes. These two shell protein types are proposed to be the basis for the carboxysome’s icosahedral geometry. The shell proteins are also thought to allow the flux of metabolites across the shell through the presence of the small pore formed by their hexameric/pentameric symmetry axes. In this review, we describe bioinformatic and structural analyses that highlight the important primary, tertiary, and quaternary structural features of these conserved shell subunits. In the future, further understanding of these molecular building blocks may provide the basis for enhancing CO2 fixation in other organisms or creating novel biological nanostructures.
Keywords: Carboxysome, CO2 fixation, Bacterial microcompartment, Dark reactions, Cyanobacteria, Calvin cycle, Chemoautotroph
Introduction
Carboxysomes are metabolic modules for CO2 fixation that are found in all cyanobacteria and some chemoautotrophic bacteria (Badger and Price 2003; Cannon et al. 2001; Yeates et al. 2008). They are self-assembling, apparently icosahedral organelles of ~80–150 nm comprised entirely of protein (Schmid et al. 2006) (Fig. 1). Carboxysomes encapsulate a carbonic anhydrase (CA, Price et al. 1992), which converts bicarbonate to carbon dioxide, and most, if not all, cellular ribulose bisphosphate carboxylase oxygenase (RuBisCO) (Cannon and Shively 1983; Lichtle et al. 1995), the enzyme that catalyzes the first step in the Calvin–Benson cycle by combining CO2 and ribulose-1,5-bisphosphate (RuBP) to form two molecules of 3-phosphoglycerate (3PGA) (Fig. 2). Given that cyanobacteria carry out a large fraction of the total oxygenic photosynthesis on our planet, the carboxysome plays a significant role in the Earth’s primary production (Partensky et al. 1999; Whitman et al. 1998).
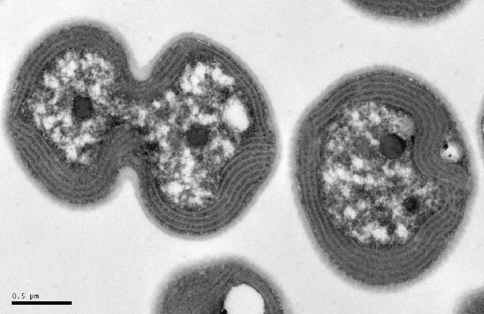
Fig. 1.
Transmission electron micrograph of Synechocystis sp. PCC6803 cells showing three carboxysomes. Image courtesy of Patrick Shih, UC Berkeley
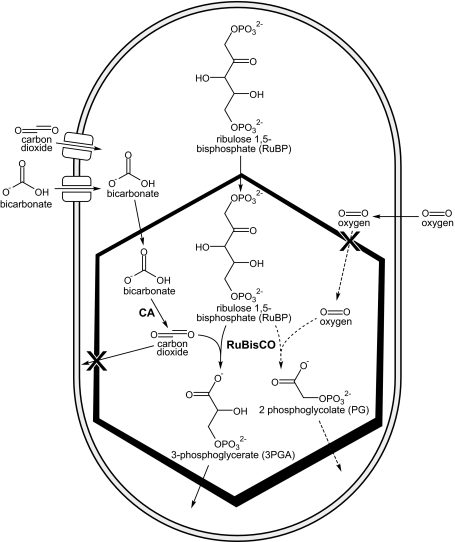
Fig. 2.
Schematic diagram of a cyanobacterial cell containing a carboxysome and depicting relevant metabolites that cross the cell membrane and carboxysome shell. The carboxysome-encapsulated reactions are shown. Those related to photorespiration catalyzed by RuBisCO in the presence of oxygen are shown in dashed lines
Structural and functional overview
Two types of carboxysome have been characterized: the α-carboxysome, which encapsulates Form IA RuBisCO, and the β-carboxysome, which encapsulates Form IB RuBisCO (Badger and Bek 2008; Tabita 1999). α-carboxysomes are found in Prochlorococcus and some marine Synechococcus species as well as in some chemoautotrophic bacteria. The β-carboxysomes are found in all other cyanobacteria, with the exception of an unusual marine species, UCYN-A (Tripp et al. 2010). In addition to differing in the encapsulated form of RuBisCO, α- and β-carboxysomes also differ in gene organization; components of the α-carboxysome are organized into an operon whereas the genes for the β-carboxysome components are generally more dispersed (Fig. 3).
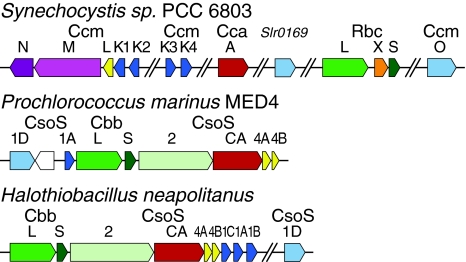
Fig. 3.
Three examples of carboxysome gene clusters for a β-carboxysome (top) of Synechocystis PCC 6803 and two α-carboxysomes (bottom), from the cyanobacterium Prochlorococcus marinus MED4 and from a chemoautotroph Halothiobacillus neapolitanus. Parallel diagonal lines denote large genomic segments between genes. Single-domain BMC proteins are colored dark blue; tandem-domain BMC proteins are colored light blue. Pentameric carboxysome shell proteins are colored yellow. Homologous proteins are colored similarly. Rbc and Cbb are the locus tags for RuBisCO in β- and α-carboxysomes, respectively
There are several differences in the complement of genes that are necessary for carboxysome formation. In addition to encapsulating RuBisCO, the α-carboxysome contains an unusual β-CA (Sawaya et al. 2006) for the conversion of bicarbonate to carbon dioxide and yet to be characterized structural protein, CsoS2 (Baker et al. 1999). A β-CA is also encapsulated in the β-carboxysome of some cyanobacteria (So et al. 2002).
All β-carboxysome gene clusters encode two proteins, CcmM and CcmN (Ludwig et al. 2000), that are also thought to play a catalytic and/or organizational role in the carboxysome interior. CcmM contains 3–5 repeats of the RuBisCO small subunit domain in its C-terminus, while the N-terminal domain is homologous to a γ-type CA (Cot et al. 2008; Long et al. 2007). This domain has been shown to be catalytically active in an organism that lacks the β-CA ortholog (Peña et al. 2010). CcmM has also been shown to interact with the RuBisCO large subunit (RbcL), the proteins of the shell, CcmN, and the CA CcaA (Cot et al. 2008; Long et al. 2007, 2010).
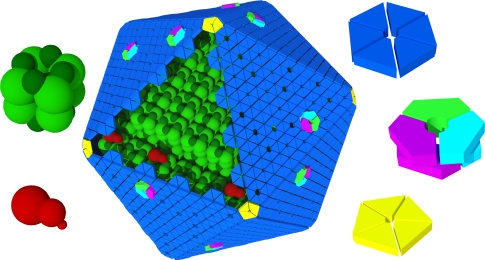
The carboxysome shell is comprised mainly of small (~100 amino acid) proteins (Cannon and Shively 1983) (Figs. 3, 4a) that contain the bacterial microcompartment (BMC) domain (Pfam00936); these are thought to form the flat facets of the shell (Fig. 5) (Kerfeld et al. 2005; Tsai et al. 2007). In addition, one or two small, well-conserved proteins containing the Pfam03319 domain (Figs. 3, 4b) form pentamers that are thought to introduce curvature to the shell by forming the vertices (Cai et al. 2009; Tanaka et al. 2008) (Fig. 5). The complement of shell protein genes differs between the two types of carboxysome in terms of number of paralogs, gene order, and primary structure, but each type contains more than one paralog of the BMC domain and at least one copy of the Pfam03319 domain (Fig. 3). Also of note is the presence in all carboxysome-containing organisms of genes encoding one or two proteins with two fused BMC domains, also known as tandem BMC proteins (Figs. 3, 5).
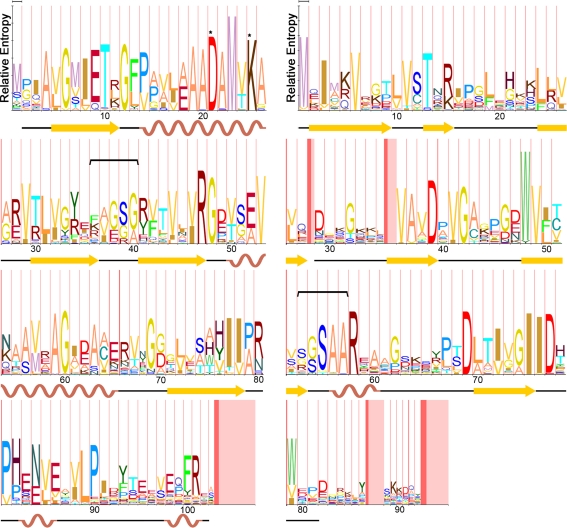
Fig. 4.
a Hidden Markov model (HMM)-logo for all unique single-domain carboxysome BMC shell proteins (CcmK1, CcmK2, CcmK3, CcmK4, CsoS1A, CsoS1B, and CsoS1C). Secondary structure of CcmK2 [Protein Data Bank (PDB) ID: 2A1B] is mapped to the corresponding positions on the logo. A horizontal bracket marks the residues lining the pore, and asterisks mark residues located at the edge of each monomer in the known structures. b HMM-logo for all Pfam03319 proteins in carboxysomes (CcmL, CsoS4A, and CsoS4B). Secondary structure of CsoS4A (PDB:2RCF) is mapped to the corresponding positions on the logo. A horizontal bracket marks the residues lining the pore. For both logos, the width of the vertical red bars is proportional to the frequency of an insertion at that position in the model. The width of the subsequent vertical pink bar is proportional to the length of that insertion [Figures prepared using MUSCLE (Edgar 2004), HMMER 3.0 (Eddy 1998), and LogoMat-M (Schuster-Bockler et al. 2004)]
Fig. 5.
Schematic model of the α-carboxysome assembly containing RuBisCO small (dark green) and large (green) subunits and carbonic anhydrase (red). The shell is composed of hexamers (blue), pseudohexamers (light blue, magenta, and light green), and pentamers (yellow)
The structures of the BMC domain: a key building block of the carboxysome shell
The first structures determined from the carboxysome shell were the CcmK2 and CcmK4 proteins from the carboxysome of the β-cyanobacteria Synechocystis sp. PCC6803 (Kerfeld et al. 2005). The structures revealed that the BMC domain forms hexamers with a disk-like shape, giving each a concave and a convex side (Fig. 6). Packing of the hexamers in some of the crystal forms immediately suggested a model for the underlying architecture of the carboxysome shell: the shell proteins formed a two-dimensional layer similar to hexagonal tiles (Fig. 5). CcmK2 formed a uniform layer with all hexamer faces oriented in the same direction whereas CcmK4, in one of two crystal forms, formed a layer with strips of hexamers alternating between convex and concave orientations (Kerfeld et al. 2005).
Fig. 6.
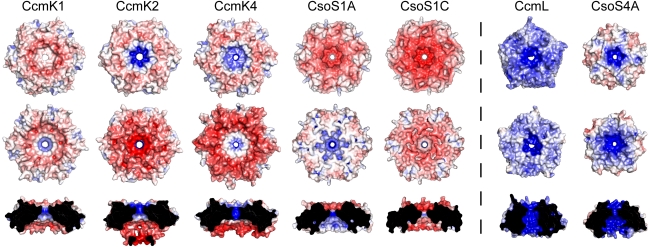
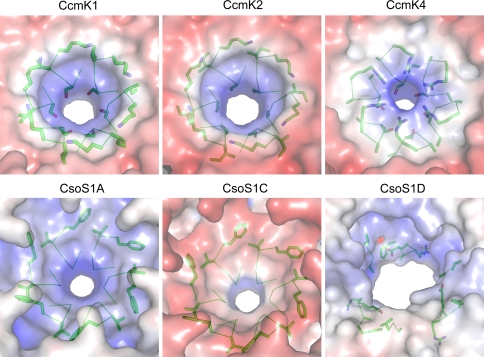
Electrostatic comparison of structurally characterized single-domain BMC [PDB:3BN4 (CcmK1), 2A1B (CcmK2), 2A10 (CcmK4), 2G13 (CsoS1A), 3H8Y (CsoS1C)] proteins and pentameric shell proteins [PDB:2QW7 (CcmL), 2RCF (CsoS4A)]. Convex (top), concave (middle), and pore cross-section (bottom) views are shown for each structure. Red denotes negative charge; blue denotes positive charge [Figure generated with APBS Plug-in (Baker et al. 2001) for PyMOL]
Crystal structures of the CsoS1A (Tsai et al. 2007) and CsoS1C (Tsai et al. 2009) proteins from the α-carboxysome of Halothiobacillus neapolitanus have also been determined. These displayed the same concave/convex sidedness and uniformly oriented layer formation as observed for CcmK2. Despite a high degree of sequence homology between CsoS1A and CsoS1C (97% identity), a comparison of the electrostatics of these structures shows a difference in the charge distribution on the concave faces (Fig. 6). There is a single amino acid substitution between CsoS1A and CsoS1C at position 97 (from Glu to Gln) that apparently accounts for this difference in electrostatic potential.
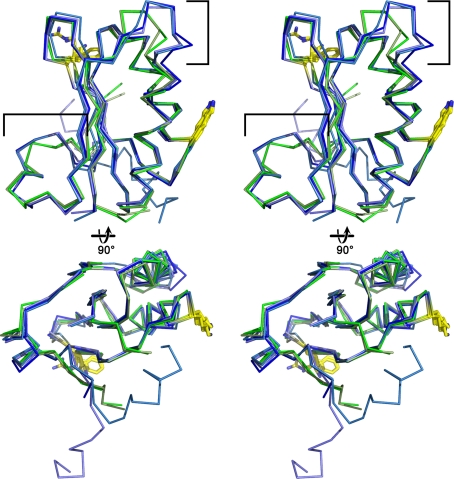
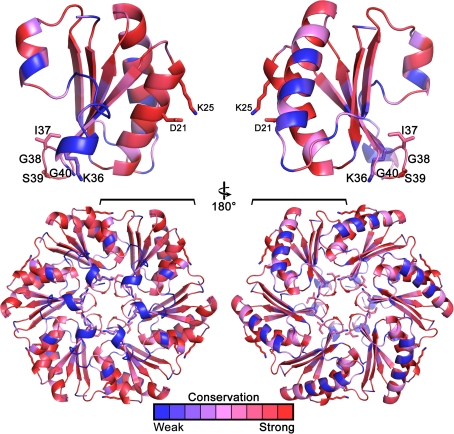
A superposition of all the single-domain carboxysome BMC protein structures show they share a conserved fold [root mean square deviation (RMSD) range of 0.36–0.71 Å over 66–86 C-α atoms] with only slight differences between the Cso-type homologs from the α-carboxysomes and the Ccm-type homologs from the β-carboxysome (Fig. 7). The differences are in the loop connecting the second α-helix to the fourth β-strand and in the length of the third α-helix. In the hexamers, these differences result in slight variations in the convex surfaces and monomer–monomer interactions, respectively. From structure, as well as sequence alignments, one can identify the residues that are structurally conserved and important to the hexamer–hexamer interactions. For example, the absolutely conserved D-X-X-X-K (Fig. 4a, 8) motif located at the hexamer edges forms the interface between two hexamers. A less conserved R-P-H-X-N (Fig. 4a) at the hexamer edges also contributes to the interface between two adjacent hexamers.
Fig. 7.
Stereo images of superpositioned single-domain BMC monomers from the β- (blue shades) and α- (green shades) carboxysomes. The upper pair is viewed from the convex side of the protein, whereas the bottom view is rotated clockwise 90° about the x-axis from the upper view. One pore residue (Arg from CcmK4, Lys from CcmK1 and CcmK2, Phe from CsoS1A and CsoS1C) and the conserved Lys found at the edge of the hexamer are shown in yellow sticks. The regions flanked by brackets are those that display the largest structural differences between the Cso and CcmK type shell proteins
Fig. 8.
Conservation of all unique single-domain carboxysome BMC shell proteins mapped onto the structure of CcmK2 (PDB: 2A1B). Key residues are shown in sticks and labeled (Figure prepared using the Consurf (Ashkenazy et al. 2010) server and PyMOL)
The primary structures of CsoS1B, CcmK1, and CcmK4 contain a C-terminal extension of ~10 residues compared to their paralogs. A comparison of the structures of CcmK2 and CcmK4 from Synechocystis sp. PCC6803 reveals that the additional C-terminal residues of CcmK4 form an α helix. In CcmK2 a short, five residue helix occludes the depression in the concave face of the hexamer; in CcmK4 the additional C-terminal residues form an extended helix that folds back on the edge of the hexamer, leaving the concave side unobstructed (Figs. 6, 7). The structure of CcmK1 is missing its C-terminal 17 residues (Tanaka et al. 2009), but based on sequence similarity to the C-terminus of CcmK4 it could likewise be helical. This C-terminal extension may offer clues to the as yet unknown orientation of the shell proteins with regard to which side faces the cytosol. If facing the interior of the carboxysome, the disposition of this helix may be important for interacting with encapsulated proteins. A second hypothesis is that the orientation of the helix might act as a switch that can change the propensity for incorporation of the shell protein into an assembling shell (Kerfeld et al. 2005).
Pentameric proteins of the carboxysome shell
Representative structures of proteins containing the Pfam03319 domain have been solved from both the α- and β-carboxysome (Tanaka et al. 2008). CsoS4A is one of two paralogs (CsoS4A and B) in the α-carboxysome, and CcmL is the only protein with a Pfam03319 domain found among the β-carboxysome genes (Fig. 3). CcmL and CsoS4A have been structurally characterized (Tanaka et al. 2008); both form pentamers and have a pronounced concave/convex sidedness similar to the hexamers. In contrast to the hexameric shell proteins, the electrostatic potential of these proteins is predominantly positive (Fig. 6). The structures of CcmL and CsoS4A can be superimposed with an RMSD of 0.74 Å over 58 C-α atoms. The largest difference between the primary structures of these two proteins is in the region corresponding to an 8–10 amino acid loop on the concave face of the pentamer that seems to influence the charge of the concave face. A similar difference is seen between the paralogs CsoS4A and CsoS4B. In this region CsoS4B has more positively charged residues than CsoS4A.
The pores
Based on the current models of carboxysome function and structure, pores in the shell protein hexamers provide conduits for the flux of metabolites; bicarbonate ions and RuBP diffuse in and 3PGA to diffuses out, while preventing the leakage of CO2 from the interior (Dou et al. 2008). The shell also prevents oxygen from diffusing in, reducing unwanted photorespiration by RuBisCO (Marcus et al. 1992). As the shell localizes CA and RuBisCO together, the overall rate of CO2 fixation by RuBisCO is enhanced; effectively, the carboxysome provides a focal point for the carbon concentrating mechanism (CCM) (Fig. 2).
A key characteristic of carboxysome shell proteins is a narrow (~4–7 Å diameter; Kerfeld et al. 2005) central pore that is formed at the 5- and 6-fold axis of symmetry by a loop in the hexamers and pentamers, respectively. Residues forming this loop tend to be conserved among paralogs; for example, these residues are K-I-G-S and R-(A/V)-G-S in CcmK2 and CcmK4, respectively (Table 1). Such differences in residues flanking the pore likely influence the flux of metabolites into or out of the carboxysome by influencing the size and charge of the pore. All of the pores of structurally characterized carboxysome shell proteins are positively charged at the narrowest point (Fig. 9); presumably this provides a favorable attractive force for negatively charged metabolites such as bicarbonate. At the same time, a charged pore would not attract molecules lacking a dipole moment, such as CO2 and oxygen (Fig. 9).
Table 1.
List of structurally characterized BMC-domain proteins from the carboxysome and their dimensions
| Pfam00936 protein | Carboxysome type | Hexamer diameterb (Å) | Hexamer edge lengthc (Å) | Pore residues | Pore diameter (Å) |
|---|---|---|---|---|---|
| CsoS1A [2G13] | α | 72 | 36 | FVGG | 4 |
| CsoS1C [3H8Y] | α | 72 | 36 | FVGG | 4 |
| CcmK1 [3BN4] | β | 75 | 37 | KIGS | 4.8 (5.5) |
| CcmK2 [2A1B] | β | 75 | 35 | KIGS | 5.5 (7) |
| CcmK4 [2A10] | β | 75 | 37 | RAGS | 4 |
| CsoS1Da [3F56] | α | 72 | 36 | ERAF | 12.5 (14) |
PDB IDs of the listed structures are in brackets. aCsoS1D is a tandem BMC-domain protein; values for the dimensions of the pseudohexamer are reported. b Hexamer diameter was measured from one vertex to its opposite vertex. c Hexamer edge length was measured from one vertex to its shared edge vertex. Dimensions of the hexamers were measured using PyMOL (DeLano 2002), and all pore diameters were measured for this study using HOLE (Smart et al. 1996). Previously published pore diameters are in parenthesis if the difference was >0.5 Å between this analysis and published values
Fig. 9.
Electrostatic comparison of pores from structurally characterized BMC shell proteins, viewed from the concave side. Pore residues are shown as green sticks. Red denotes negative charge; blue denotes positive charge
The pores of the pentamers are also narrow with diameters of ~5 and ~3.5 Å for CcmL and CsoS4A, respectively. They are also positively charged, even more so than the hexamers (Fig. 6). At its narrowest point, the pore for CcmL is formed by R-G-S-A-A and CsoS4A’s is formed by G-S-S-A-A (Table 2). Although the pore residues of carboxysome Pfam03319 orthologs are not as well conserved as their hexameric counterparts, sequence comparison reveals some conservation, with a pore motif of X-(G/S)-S-A-A (Fig. 4b).
Table 2.
List of structurally characterized pentameric Pfam03319 domain-containing proteins from the carboxysome and their dimensions
| Pfam03319 protein | Carboxysome type | Pentamer diametera (Å) | Pentamer edge lengthb (Å) | Pore residues | Pore diameter (Å) |
|---|---|---|---|---|---|
| CcmL [2QW7] | β | 58 | 36 | RGSAA | 5 |
| CsoS4A [2RCF] | α | 57 | 34 | GSSAA | 3.5 |
PDB IDs of the listed structures are in brackets. a Pentamer diameter was measured from one vertex to its opposite edged. b Pentamer edge length was measured from one vertex to its shared edge vertex. Dimensions of the pentamers were measured using PyMOL (DeLano 2002), and all pore diameters for this study were measured using HOLE (Smart et al. 1996)
Tandem BMC proteins
Among the genes encoding components of both the α- and β-carboxysomes are some containing fusions of BMC domains (Fig. 3): CsoS1D in the α-carboxysome and CcmO and a CsoS1D ortholog (slr0169 in Synechocystis sp. PCC6803) in the β-carboxysome. In 2009, the first structure of a tandem BMC protein was determined, CsoS1D of Prochlorococcus marinus MED4 (Klein et al. 2009). This protein was not predicted to contain two BMC domains; the N-terminal domain lacks obvious sequence similarity to any other BMC domain. However, the α-carbon backbones of the two domains superimpose with an RMSD of 1.27 Å over 95 atoms; guided by a structure-based sequence alignment, the domains are 18% identical. CsoS1D forms trimers resulting in pseudohexamers that are similar in dimensions to hexameric shell proteins (Table 1), with pronounced concave and convex sides (Fig. 9). The edges of the pseudohexamers contain the conserved D-X-X-X-K edge motif and CsoS1D could be readily fitted into existing models of the facets of the α-carboxysome shell (Fig. 5) (Klein et al. 2009).
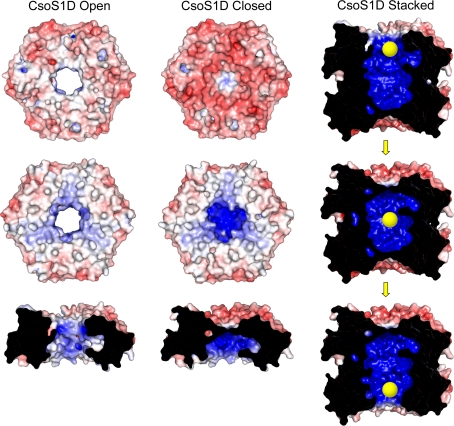
The structure of CsoS1D also provided the first evidence that pores in the carboxysome shell could be gated, potentially providing a mechanism for regulating metabolite flux across the shell. In the CsoS1D trimers, conformational changes in the absolutely conserved pore loop residues Glu120 and Arg121 (Fig. 9) result in either a relatively large open pore of ~14 Å diameter or an occluded pore (Fig. 10). The large size of the CsoS1D pore, which would allow for free passage of RuBP, likely requires gating to prevent the loss of important metabolites or infiltration of inhibitory species.
Fig. 10.
Electrostatic comparison of the two trimers of the tandem BMC-domain protein CsoS1D (PDB:3F56) and modeled representation of the “air-lock” mechanism for metabolite movement through the protein. Convex (top), concave (middle), and pore cross-section (bottom) views are shown for each of the two structures on the left. The top and bottom images of the “air-lock” mechanism are generated from the same solved stacked structure from two different orientations. The middle image is a hypothetical model generated in PyMOL by structurally aligning a copy of a closed trimer over the open trimer in the stacked structure. Red denotes negative charge and blue denotes positive charge
Interestingly, in two independent crystal structures, the CsoS1D trimers stacked to form a dimer of trimers (Fig. 10). The two trimers were rotated ~60° with respect to each other so that the C-terminal domain of a subunit in the upper trimer interacted with the N-terminal domain of a subunit in the lower trimer. The dimerization was across the concave face of each trimer, resulting in a large cavity of 13,613 Å3. Additional biophysical analyses that support the potential biological relevance for the dimer of trimers include a buried surface area of 6,573 Å2 and a shape correlation value of 0.70 (range of 0–1, 1 being a perfect fit and 0 being no interaction) between the two trimers (Klein et al. 2009). The cavity could, like the pore gating, influence the flux of larger metabolites (e.g., RuBP, 3PGA) into and out of the carboxysome in a manner analogous to an airlock. For example, the trimer facing the cytosol would open to accept a metabolite and then close; subsequently, the trimer facing the carboxysome interior would open to allow for release of the metabolite from the cavity (Fig. 10).
An ortholog to CsoS1D, with the locus tag slr0169 in Synechocystis sp. PCC6803, has also been identified in all β-carboxysome-containing cyanobacteria (Klein et al. 2009). It is ~200 amino acids in length and lacks ~50 N-terminal residues that are present in the α-cyanobacterial CsoS1D homologs. slr0169 contains the conserved Glu and Arg residues (Glu69, Arg70) responsible for gating the CsoS1D pore as well as the universally conserved edge Lys residues in the N- and C-terminal domains (Lys108, Lys212) for interacting with other hexamers to incorporate into the shell (Cai et al. in press).
A second ~200 amino acid BMC-domain protein is found only in low-light adapted strains of Prochlorococcus and some marine Synechococcus species. This protein, dubbed CsoS1E, is similar to CsoS1D in that it has a C-terminal BMC domain, but its N-terminus lacks homology to any other known domain. A remarkable feature of CsoS1E is its high-isoelectric point of 10.3, with positively charged residues concentrated in the N-terminal half of the protein. Further structural studies are needed to determine whether this N-terminus will form a BMC domain much like the cryptic N-terminal BMC domain of CsoS1D. Finally, CcmO represents another type of tandem BMC-domain protein that is present in all the β-cyanobacteria. It is ~260 amino acids in length and appears to be a fusion of two CcmK-like proteins. It is known to be essential for carboxysome formation in the β-cyanobacteria (Marco et al. 1994).
Variability of shell composition
Both the α- and β-carboxysome shells are composed of multiple paralogs of single BMC-domain proteins. The reason for this redundancy is unknown. One hypothesis is that the carboxysome shell composition might be altered by a change in the environment; this is consistent with the observation that the size of the pore varies among paralogs. Alternatively, hexamers could form from more than one paralog, resulting in hetero-hexamers. By modulating the shell protein composition, selectivity for metabolites may be increased or decreased based on the charge or size differences present at the pores of the shell subunits. This could help to increase the organism’s fitness in a wider variety of growth conditions.
Some evidence for modulation of shell protein expression under different conditions has come from transcriptome analysis of the β-cyanobacterium Synechocystis sp. PCC6803, where the expression of the CsoS1D ortholog, slr0169 is greater under high-light and low-carbon stresses and clusters with other carboxysome shell components (Cai et al. in press; Eisenhut et al. 2007).
Conclusions and future prospects
Structural information for the building blocks of the carboxysome shell is rapidly accumulating. With the current knowledge, several convincing models of the protein interactions involved in forming the carboxysome have been built (Cot et al. 2008; Iancu et al. 2007; Long et al. 2007; Tanaka et al. 2008) and attractive hypotheses regarding the metabolic flux and function of the shell have been posited (Dou et al. 2008; Fridlyand et al. 1996). An area that needs more attention is the structural characterization and analysis of the interactions among the encapsulated proteins (CsoS2, CcmN and CcmM and RuBisCO). Also, little is known about the assembly of the carboxysome and the dynamics of the shell. More sophisticated imaging methods and/or gene expression analysis under controlled growth conditions may give a better idea as to the composition of the carboxysome shell.
When the first structural characterization of carboxysome shell proteins was reported, it was pointed out that proteins with homology to carboxysome shell proteins are widespread among bacteria (Kerfeld et al. 2005). Collectively, these are known as BMCs. Our bioinformatic analyses of all sequenced bacterial genomes that have a BMC-domain homolog have yielded a phyletically and functionally diverse set of BMCs, a majority of which have yet to be characterized (Kerfeld et al. 2010). Approximately 20% of bacteria with genomic sequence data have open reading frames (ORFs) coding for BMC-domain proteins. The distribution of BMC shell proteins across the bacterial phyla has been suggested to be the product of horizontal gene transfer. Inferences can be made as to the function of unknown BMC operons using a “guilt-by-association” analysis of the putative operon, where the enzymes near known BMC-domain homologs and a Pfam03319 homolog are analyzed and an encapsulated metabolism proposed. Most of the functionally uncharacterized BMCs belong to heterotrophic organisms. An interesting observation from comparison of the genomes of Rhodopseudomonas palustris strains, which can grow autotrophically, is that only strain BisB18 contains a BMC gene cluster, and it is associated with a glycyl-radical enzyme but not RuBisCO.
Two types of heterotrophic BMCs are well characterized. Studies of the propanediol utilization (pdu) BMC and the ethanolamine utilization (eut) BMC mostly in Salmonella typhimurium LT2 have yielded other important clues involving the structure, function, and assembly of microcompartment shells (Crowley et al. 2008; Parsons et al. 2008; Sagermann et al. 2009). Surprisingly, several of the pdu single BMC-domain proteins and those of the β-carboxysome are very similar and share the same pore residues although they are encapsulating completely different enzymatic reactions. Another curious observation from the eut microcompartment is that the oligomeric state of the Pfam03319 homolog EutN (Tanaka et al. 2008; Wunderlich et al. 2004) is a hexamer and not a pentamer as in the CcmL and CsoS4A structures. Thus, the possibility that carboxysome shell proteins may display quasi-equivalency like viral capsid proteins, where the protein can be either a hexamer or a pentamer, cannot be ruled out. Since BMCs were first observed, their resemblance to viral capsids has been pointed out (Gantt and Conti 1969; Shively et al. 1973). Although microcompartments are larger than viral capsids, they can be modeled as icosahedra. However, an evolutionary link between microcompartments and viral capsids, from either sequence or structural data, has not been established.
Acknowledgments
We thank Fei Cai, Annette Salmeen, Gustaf Sandh, and William Greenleaf for helpful discussions. We also thank Patrick Shih for the transmission electron micrograph image. This work was supported by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357. The JGI is under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract number DE-AC02-05CH11231, Lawrence Livermore National Laboratory under contract number DE-AC52-07NA27344.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- RuBisCO
Ribulose bisphosphate carboxylase/oxygenase
- RuBP
Ribulose-1,5-bisphosphate
- 3PGA
3-Phosphoglycerate
- CA
Carbonic anhydrase
- HMM
Hidden Markov model
- PDB
Protein Data Bank
- BMC
Bacterial microcompartment
- RMSD
Root mean square deviation
- CCM
Carbon concentrating mechanism
- ORF
Open reading frame
- pdu
Propanediol utilization
- eut
Ethanolamine utilization
References
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38 Suppl:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Bek EJ. Multiple rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot. 2008;59:1525–1541. doi: 10.1093/jxb/erm297. [DOI] [PubMed] [Google Scholar]
- Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- Baker SH, Lorbach SC, Rodriguez-Buey M, Williams DS, Aldrich HC, Shively JM. The correlation of the gene csoS2 of the carboxysome operon with two polypeptides of the carboxysome in Thiobacillus neapolitanus. Arch Microbiol. 1999;172:233–239. doi: 10.1007/s002030050765. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS ONE. 2009;4:e7521. doi: 10.1371/journal.pone.0007521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Sandh G, Kerfeld CA (in press) Bioinformatic identification and structural characterization of a new carboxysome shell protein. In: Burnap RL, Vermaas W (eds) Functional genomics and evolution of photosynthetic systems
- Cannon GC, Shively JM. Characterization of a homogenous preparation of carboxysomes from Thiobacillus neapolitanus. Arch Microbiol. 1983;134:52–59. doi: 10.1007/BF00429407. [DOI] [Google Scholar]
- Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl Environ Microbiol. 2001;67:5351–5361. doi: 10.1128/AEM.67.12.5351-5361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cot SS-W, So AK-C, Espie GS. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J Bacteriol. 2008;190:936–945. doi: 10.1128/JB.01283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley CS, Sawaya MR, Bobik TA, Yeates TO. Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure. 2008;16:1324–1332. doi: 10.1016/j.str.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL molecular graphics system, version 1.3, Schrödinger, LLC
- Dou Z, Heinhorst S, Williams EB, Murin CD, Shively JM, Cannon GC. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J Biol Chem. 2008;283:10377–10384. doi: 10.1074/jbc.M709285200. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, von Wobeser EA, Jonas L, Schubert H, Ibelings BW, Bauwe H, Matthijs HCP, Hagemann M (2007) Long-term response towards inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. doi:10.1104/pp.107.103341 [DOI] [PMC free article] [PubMed]
- Fridlyand L, Kaplan A, Reinhold L. Quantitative evaluation of the role of a putative CO2-scavenging entity in the cyanobacterial CO2-concentrating mechanism. Biosystems. 1996;37:229–238. doi: 10.1016/0303-2647(95)01561-2. [DOI] [PubMed] [Google Scholar]
- Gantt E, Conti SF. Ultrastructure of blue-green algae. J Bacteriol. 1969;97:1486–1493. doi: 10.1128/jb.97.3.1486-1493.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol. 2007;372:764–773. doi: 10.1016/j.jmb.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- Kerfeld CA, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu Rev Microbiol. 2010;64:391–408. doi: 10.1146/annurev.micro.112408.134211. [DOI] [PubMed] [Google Scholar]
- Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol. 2009;392:319–333. doi: 10.1016/j.jmb.2009.03.056. [DOI] [PubMed] [Google Scholar]
- Lichtle C, Thomas JC, Spilar A, Partensky F. Immunological and ultrastructural characterization of the photosynthetic complexes of the prochlorophyte Prochlorococcus (Oxychlorobacteria) J Phycol. 1995;31:934–941. doi: 10.1111/j.0022-3646.1995.00934.x. [DOI] [Google Scholar]
- Long BM, Badger MR, Whitney SM, Price GD. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple RubisCO complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem. 2007;282:29323–29335. doi: 10.1074/jbc.M703896200. [DOI] [PubMed] [Google Scholar]
- Long BM, Tucker L, Badger MR, Price GD. Functional cyanobacterial b-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol. 2010;153:285–293. doi: 10.1104/pp.110.154948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M, Sultemeyer D, Price GD. Isolation of ccmKLMN genes from the marine cyanobacterium Synechococcus sp. PCC7002 and evidence that CcmM is essential for carboxysome assembly. J Phycol. 2000;36:1109–1118. doi: 10.1046/j.1529-8817.2000.00028.x. [DOI] [Google Scholar]
- Marco E, Martinez I, Ronen-Tarazi M, Orus I, Kaplan A. Inactivation of ccmO in synechococcus sp. strain PCC 7942 results in a mutant requiring high levels of CO2. Appl Environ Microbiol. 1994;60:1018–1020. doi: 10.1128/aem.60.3.1018-1020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus Y, Berry J, Pierce J. Photosynthesis and photorespiration in a mutant of the cyanobacterium Synechocystis PCC-6803 lacking carboxysomes. Planta. 1992;187:511–516. doi: 10.1007/BF00199970. [DOI] [PubMed] [Google Scholar]
- Parsons JB, Dinesh SD, Deery E, Leech HK, Brindley AA, Heldt D, Frank S, Smales CM, Lunsdorf H, Rambach A, et al. Biochemical and structural insights into bacterial organelle form and biogenesis. J Biol Chem. 2008;283:14366–14375. doi: 10.1074/jbc.M709214200. [DOI] [PubMed] [Google Scholar]
- Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña KL, Castel SE, de Araujo C, Espie GS, Kimber MS. Structural basis of the oxidative activation of the carboxysomal gamma-carbonic anhydrase, CcmM. Proc Natl Acad Sci USA. 2010;107:2455–2460. doi: 10.1073/pnas.0910866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Coleman JR, Badger MR. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1992;100:784–793. doi: 10.1104/pp.100.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagermann M, Ohtaki A, Nikolakakis K. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci USA. 2009;106:8883–8887. doi: 10.1073/pnas.0902324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Cannon GC, Heinhorst S, Tanaka S, Williams EB, Yeates TO, Kerfeld CA. The structure of beta-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J Biol Chem. 2006;281:7546–7555. doi: 10.1074/jbc.M510464200. [DOI] [PubMed] [Google Scholar]
- Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Bockler B, Schultz J, Rahmann S. HMM logos for visualization of protein families. BMC Bioinform. 2004;5:7. doi: 10.1186/1471-2105-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. doi: 10.1016/S0263-7855(97)00009-X. [DOI] [PubMed] [Google Scholar]
- So AK-C, John-McKay M, Espie GS. Characterization of a mutant lacking carboxysomal carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Planta. 2002;214:456–467. doi: 10.1007/s004250100638. [DOI] [PubMed] [Google Scholar]
- Tabita FR. Microbial ribulose 1, 5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth Res. 1999;60:1–28. doi: 10.1023/A:1006211417981. [DOI] [Google Scholar]
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Sawaya MR, Phillips M, Yeates TO. Insights from multiple structures of the shell proteins from the beta-carboxysome. Protein Sci. 2009;18:108–120. doi: 10.1002/pro.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp HJ, Bench SR, Turk KA, Foster RA, Desany BA, Niazi F, Affourtit JP, Zehr JP. Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature. 2010;464:90–94. doi: 10.1038/nature08786. [DOI] [PubMed] [Google Scholar]
- Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007;5:e144. doi: 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y, Sawaya MR, Yeates TO. Analysis of lattice-translocation disorder in the layered hexagonal structure of carboxysome shell protein CsoS1C. Acta Crystallogr D. 2009;65:980–988. doi: 10.1107/S0907444909025153. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich Z, Acton TB, Liu J, Kornhaber G, Everett J, Carter P, Lan N, Echols N, Gerstein M, Rost B, et al. The protein target list of the Northeast Structural Genomics Consortium. Proteins. 2004;56:181–187. doi: 10.1002/prot.20091. [DOI] [PubMed] [Google Scholar]
- Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]