Abstract
Peroxisomes are ubiquitous organelles characterized by a protein-rich matrix surrounded by a single membrane. In filamentous fungi, peroxisomes are crucial for the primary metabolism of several unusual carbon sources used for growth (e.g. fatty acids), but increasing evidence is presented that emphasize the crucial role of these organelles in the formation of a variety of secondary metabolites. In filamentous fungi, peroxisomes also play a role in development and differentiation whereas specialized peroxisomes, the Woronin bodies, play a structural role in plugging septal pores. The biogenesis of peroxisomes in filamentous fungi involves the function of conserved PEX genes, as well as genes that are unique for these organisms. Peroxisomes are also subject to autophagic degradation, a process that involves ATG genes. The interplay between organelle biogenesis and degradation may serve a quality control function, thereby allowing a continuous rejuvenation of the organelle population in the cells.
Keywords: Filamentous fungi, Penicillin, Penicillium chrysogenum, Peroxisome, Secondary metabolism
Introduction
Peroxisomes are morphologically simple organelles that are present in virtually all eukaryotes. Initially, peroxisomes were characterized by the presence of one or more hydrogen peroxide-producing oxidoreductases together with catalase to decompose the hydrogen peroxide side product. However, at present various other functions have been ascribed to peroxisomes. These organelles play a key role in various anabolic and catabolic pathways and are essential in Trypanosomes, plants and mammals (South and Gould 1999; Guerra-Giraldez et al. 2002; Schumann et al. 2003). In humans, defects in peroxisome formation are associated with severe, inherited diseases called peroxisome biogenesis disorders (PBDs) some of which are lethal (Thoms et al. 2009). In plant oil seeds, peroxisomes may contain enzymes of the β-oxidation of fatty acids together with the key enzymes of the glyoxylate cycle (also termed glyoxysomes). In Trypanosomes, glycolysis enzymes are localized in peroxisomes (glycosomes). In fungi, these organelles are crucial for the metabolism of various unusual carbon sources (i.e. methanol, alkanes and purines). In filamentous fungi, peroxisomes in addition play a role in the formation of various secondary metabolites such as penicillins, polyketides and terpenes (Maggio-Hall et al. 2005; Saikia and Scott 2009; Imazaki et al. 2010). Moreover, in filamentous fungi peroxisomes can form Woronin bodies, protein-dense organelles that plug hyphal pores after hyphal wounding, thus preventing cytoplasmic leakage (Jedd 2011). This present review summarizes the current state of knowledge on the biology of peroxisomes in filamentous fungi, and highlights their role in the formation of secondary metabolites.
Biogenesis and degradation of peroxisomes in filamentous fungi
In fungi, peroxisomes generally proliferate by fission of pre-existing organelles or are formed from the endoplasmic reticulum (ER) (Fig. 1). In the filamentous fungus, Penicillium chrysogenum, peroxisomes arise predominantly in the hyphal tip (Meijer et al. 2010). As yet, it is however unknown whether this is a general phenomenon for filamentous fungi.
Fig. 1.
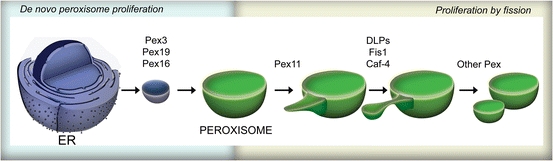
Hypothetical model of peroxisome biogenesis. Peroxisomes can form de novo from the ER. This step requires the function of a group of peroxisomal proteins, mainly Pex3, Pex16 and Pex19. Peroxisomes can also proliferate by fission of existing organelles. In this process a yet unknown signal activates Pex11 proteins that lead to elongation of the organelle. The tubulated organelle attracts GTPases from the dynamin like family (DLPs) that assemble into highly ordered multimers on the elongated organelle, causing actual membrane fission. The assembly of the DLPs complex requires Fis1 and Mdv1. Next, the newly formed organelle imports matrix and membrane proteins
The formation of peroxisomes is controlled by the function of specific proteins (peroxins), encoded by PEX genes (Kiel et al. 2006). Over 30 peroxins have been identified in fungal genomes, most of which are involved in matrix protein import and formation of the organelle membrane (Kiel et al. 2006). PEX genes are generally evolutionarily conserved; however, some of them are specific for filamentous fungi. Among these is Pex14/17, a protein that was recently identified to be involved in matrix protein import. In general, the docking machinery required for binding of the cytosolic receptor for newly synthesized matrix proteins at the peroxisome membrane, is composed of the conserved proteins Pex13 and Pex14. In yeast species, Pex17 is also part of this docking complex (Rucktaschel et al. 2010). However, filamentous fungi contain, in addition to Pex14, a unique peroxin Pex14/17 that seems to be a fusion protein between Pex14 and Pex17 (Kiel et al. 2006). Data from P. chrysogenum suggest that Pex14/17 may be part of the receptor docking complex. Interestingly, the C-terminal part of Pex14/17 that resembles yeast Pex17 is crucial for the function of Pex14/17, suggesting that Pex14/17 is filamentous fungi-specific form of Pex17 (Opalinski et al. 2010).
The biogenesis of the peroxisomal membrane in mammals requires Pex16 (South and Gould 1999). The PEX16 gene is also present in plants and all filamentous fungi, but absent in yeast species with the exception of Yarrowia lipolytica (Eitzen et al. 1997). In contrast to mammalian PEX16 deletion cells (pex16), Y. lipolytica pex16 cells contain peroxisomes. In this organism the protein may be involved in regulation of peroxisome proliferation (Guo et al. 2007). Also P. chrysogenum pex16 cells contain a decreased number of peroxisomes (L. Opalinski unpublished results). However, the molecular function of the protein in peroxisome biogenesis in filamentous fungi is unclear and requires further analysis.
Ascomycetous filamentous fungi contain Woronin bodies, protein-dense organelles that prevent cytoplasmic bleeding after hyphal wounding. The core protein of this structure is Hex1. The biogenesis of Woronin bodies occurs in peroxisomes in the apical compartment of the hyphae (Tey et al. 2005). This process initiates with the import of Hex1 into the organelle matrix, resulting in the formation of dense Hex1 aggregates. The luminal Hex1 core subsequently becomes surrounded by oligomers of the peroxisomal membrane protein WSC, leading to formation of nascent Woronin bodies (Liu et al. 2008). These structures are next positioned within the hyphae by Leashin protein (Ng et al. 2009). After association with the cell cortex, the Woronin bodies separate from the peroxisomes. This process is mediated by the general peroxisome fission machinery (Fig. 1) and is dependent of the function of Pex11 (Escano et al. 2009). In all species, including filamentous fungi, Pex11 is the major component of the peroxisome fission machinery that plays a crucial role in peroxisome proliferation (Kiel et al. 2005; Opalinski et al. 2011). The actual membrane fission step during Woronin body separation from the peroxisome also is dependent of the function of GTPases from the dynamin-like protein family (DLP), proteins that are also crucial for peroxisome proliferation. Interestingly, the Neurospora crassa orthologue of P. chrysogenum Pex14/17, Pex33, was demonstrated to be important for Woronin body formation.
The actual number of the peroxisomes that is present in the cell is tightly regulated by both biogenic and degradative processes. However, the organelles are also subject to quality control mechanism and a constitutive degradation process, most likely as a mechanism to maintain a healthy and juvenile organelle population to prevent cell damage (Aksam et al. 2007) (Fig. 2). Indeed in yeast, inhibition of organelle degradation leads to strong physiological disadvantages and may result in necrotic cell death (Aksam et al. 2007). The tight interplay between peroxisome biogenesis and selective peroxisome degradation also allows cells to adapt to rapidly changing environmental conditions.
Fig. 2.
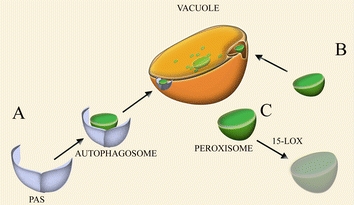
Turnover of peroxisomes. The major pathway of peroxisome degradation is autophagy. A In macropexophagy a pre-autophagosomal structure (PAS), probably formed from the ER, engulfs the peroxisome thus forming an autophagosome that fuses with the vacuole, delivering the organelle for degradation by vacuolar hydrolases. B In micropexophagy the peroxisome is directly engulfed by the vacuolar membrane and subsequently degraded in the vacuolar lumen. C Peroxisomes can be also degraded by the activity of 15-lipoxygenase (15-LOX) that disturbs the integrity of peroxisomal membrane
The role of peroxisomes in secondary metabolite formation
Introduction
Secondary metabolites comprise a diverse group of compounds, often of great biotechnological interest, that are produced by plants, animals, algae, fungi and bacteria. The formation of the secondary metabolites has implications for various cellular processes, including cell development, cellular defense and survival under nutrient limiting conditions (Keller et al. 2005; Schroeckh et al. 2009). Secondary metabolism often utilizes compounds and enzymes from primary metabolism and is characterized by the compartmentalization of the enzyme reactions into different subcellular structures. One of the cellular compartments that is of great significance in secondary metabolism is the peroxisome.
The role of peroxisomes in the β-lactam biosynthethic pathway
Peroxisomal enzymes are crucial in the synthesis of β-lactam antibiotics in filamentous fungi. The discovery of penicillins in 1928 by A. Fleming had a strong impact on therapeutic medicine development. Currently, over 20,000 antibiotics are known. Industrially, antibiotics are important components and the world market exceeds 24 billion Euro (Gavrilescu and Chisti 2005). Of the known antibiotics, β-lactams (i.e. penicillins) are the most important ones (Barber et al. 2004). These compounds are produced as secondary metabolites by filamentous fungi like P. chrysogenum and Aspergillus nidulans. During the late 1940s, the fungus Cephalosporium acremonium (renamed to Acremonium chrysogenum), that had the ability to produce different derivatives of β-lactam compounds designated cephalosporins, was isolated. The discovery of cephalosporins has initiated the development of a new group of clinically significant β-lactams. The success of β-lactams in the treatment of infectious diseases is related to their low toxicity for human/mammals paralleled by their high specificity. Regardless of the increase in the number of new antibiotics and the growing penicillin-resistance, β-lactams are still by far the most frequently used ones. The world-wide success of penicillin in the therapy of infectious diseases would not be achieved without the efforts that have gone into improving the efficiency of penicillin production. The production levels have been increased >40,000 times since the discovery of penicillin due to laborious classical strain improvement programs followed by genetic engineering approaches (the original Fleming’s P. notatum strain produced 1.2 μg/ml relative to 50,000 μg/ml by current industrial strains).
The biosynthetic pathways of penicillins and the related cephalosporins are compartmentalized to the cytosol and peroxisomes (for details on their formation see Fig. 3). The first two enzymatic reactions of β-lactam biosynthesis, catalysed by δ-(l-aminoadipoyl)-l-cysteinyl-d-valine synthase (ACVS) and isopenicillin N (IPN) synthase, occur in the cytosol and lead to the formation of IPN. The final steps in penicillin biosynthesis (in P. chrysogenum) take place in peroxisomes. Cephalosporins (produced by A. chrysogenum) are synthesized in the cytosol but require isomerization of IPN to penicillin N, a process that is thought to take place in peroxisomes. This assumption is based on recent data in P. chrysogenum, which revealed that the two enzymes involved in IPN isomerization are localized to the peroxisomal matrix. As the orthologs of these enzymes in A. chrysogenum contain putative peroxisomal targeting signals, peroxisomes are probably also important for cephalosporin synthesis in this organism (Kiel et al. 2009). The currently used β-lactam producing strains of P. chrysogenum are the result of many years of intensive strain improvement programs. The relative slow progress in improving penicillin production in the past few years lends support to the notion that these classical methods are close to having reached the maximum of what is possible this way. However, distinct improvements may be achieved by genetic engineering (see below).
Fig. 3.
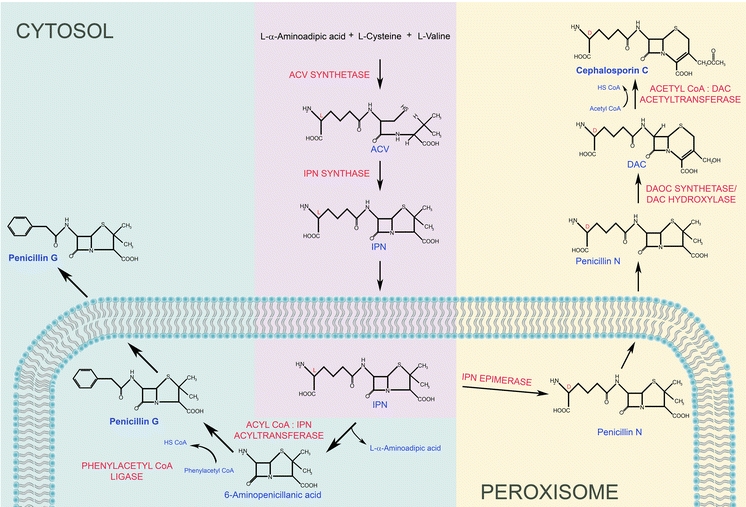
Hypothetical model of the compartmentalization of biosynthetic pathways involved in penicillin and cephalosporin C formation. The fungal β-lactam antibiotics derive from three amino acids (l-α-aminoadipic acid (l-α-AAA), l-cysteine, and l-valine, which are condensed via the function of ACV synthetase) into the tripeptide δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine (ACV) in the cytosol. Subsequently the bicyclic ring structure IPN is formed via oxidative ring closure from the linear ACV tripeptide by cytosolic IPN synthase. IPN is subsequently transported into the peroxisome by the CefP transporter. The penicillin and cephalosporin biosynthesis pathways diverge at this stage. Penicillin biosynthesis occurs in peroxisomes where the hydrophilic l-α-AAA side chain of IPN is exchanged for a hydrophobic acyl group of a side chain precursor by acyl coenzyme A (CoA):isopenicillin N acyltransferase (IAT). In the natural environment penicillin DF, F and K, containing hexenoic acid, ∆3-hexenoic acid and octenoic acid as a side chain, respectively, are produced. Penicillin G and V are synthesized upon supplementation of cultivation media with PAA or POA, respectively. Activation of the side chain precursor in the peroxisomal matrix requires phenylacetyl coenzyme A ligase (PCL) (Koetsier et al. 2009; Meijer et al. 2010). In A. chrysogenum, two enzymes are employed in the subsequent step of the cephalosporin biosynthesis pathway, IPN–CoA synthetase, IPN–CoA epimerase. These enzymes are the components of the IPN epimerase system and catalyse the isomerization of the l-α-AAA side chain of IPN to the d-enantiomer to produce penicillin N. Bioinformatic analysis of genes encoding two components of the IPN epimerase system, cefD1 and cefD2, revealed that both proteins contain putative peroxisomal targeting signals (Martin et al.). In P. chrysogenum the putative homologous proteins of the IPN epimerase system were found in the peroxisomal matrix supporting the hypothesis that this step of cephalosporin biosynthesis may occur in peroxisomes (Kiel et al. 2009). Moreover, recent studies have revealed the presence of two peroxisomal membrane proteins: CefP and CefM, implied for the translocation of the penicillin N intermediate. The completion of cephalosporin biosynthesis occurs in the cytosol, where bifunctional enzyme with expandase and hydroxylase activities converts penicillin N into deacetoxycephalosporin C and subsequently into deacetylcephalosporin C (DAC). The thiazolidine ring of penicillin is exchanged by the six-membered dihydrothiazine ring forming the cephem nucleus. In the final step, which is catalyzed by acetyl-CoA:DAC acetyltransferase (DAT), DAC is converted into cephalosporin C
Comparison of various P. chrysogenum strains from the strain improvement lineage has revealed that high penicillin producing strains contained increased volume fractions of peroxisomes relative to low producers (Fig. 4a) (van den Berg et al. 2008). Interestingly, proliferation of peroxisomes is also enhanced in A. nidulans grown at penicillin producing conditions, strengthening significance of peroxisomes in penicillin formation (Valenciano et al. 1998).
Fig. 4.
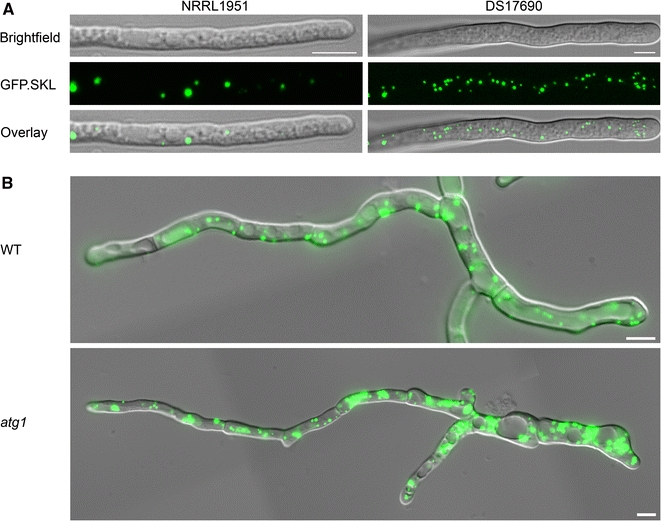
Peroxisome numbers are increased in high producing P. chrysogenum cells. a The original P. chrysogenum low producing strain (NRRL1951) and the high penicillin producer (DS17690), expressing GFP.SKL as a marker of peroxisomes, were grown for 40 h on penicillin production media and analyzed by CLSM. The high penicillin producing strain contains increased numbers of peroxisomes relative to the original strain NRRL1951. b P. chrysogenum WT cells and autophagy-deficient cells (atg1), expressing GFP.SKL, were grown for 5 days on penicillin production media and analyzed by fluorescence microscopy. In WT cells constitutive degradation of cell constituents occurs under penicillin producing conditions (seen as accumulation of GFP.SKL in the vacuolar lumen), leading to reduced peroxisome numbers relative to atg1 cells. Scale bars represent 5 μm
It is well documented that the presence of intact, functional peroxisomes is essential for efficient penicillin production. In cells that have aberrant peroxisomes or completely lack these organelles, penicillin productivity is strongly decreased (Meijer et al. 2010; Muller et al. 1992; Opalinski et al. 2010). Also in A. nidulans the absence of functional peroxisomes results in a significant decrease in penicillin titers (Sprote et al. 2009). Recently, the P. chrysogenum penicillin biosynthetic pathway has been introduced in the methylotrophic yeast Hansenula polymorpha and also in this organism efficient penicillin production was dependent of intact peroxisomes (Gidijala et al. 2009). Together, these data suggest that peroxisomes may create unique biochemical environments that facilitate optimal function of the enzymes of their production pathway.
Interestingly, not only the presence of intact peroxisomes but also their ultimate volume fraction is important for efficient antibiotic production in filamentous fungi. Direct evidence for this came from manipulation of the level of Pex11 (Opalinski et al. 2011). Pex11-induced peroxisome proliferation in P. chrysogenum was associated with a 2–3 fold increased penicillin productivity in a laboratory strain. Interestingly, this increase was not due to variations in the levels of peroxisome-borne penicillin biosynthetic enzymes, since these were unaffected (Kiel et al. 2005). Similar positive results on penicillin production were observed when Pex14/17 was overproduced in the P. chrysogenum wild-type strain (NRRL1951) (Opalinski et al. 2010). Since the efficiency of protein import into peroxisomes in P. chrysogenum is very high, the observed elevated penicillin productivity upon PEX14/17 overexpression may be related to the enhanced peroxisome volume fractions, similar as in PEX11 overexpression cells, in this strain rather than enhanced protein import rates. The number of peroxisomes may also be stimulated by manipulation of degradative processes. Inhibition of autophagy, the main cellular process that recycles peroxisomes, resulted in a two-fold increase in peroxisome numbers in P. chrysogenum associated with significantly increased penicillin production rates (Bartoszewska et al. 2011).
Interestingly, not all conditions that induce peroxisome proliferation are associated with enhanced penicillin production rates. For instance, both oleate (added as carbon source) and overproduction of dynamin-like protein Dnm1 in P. chrysogenum stimulated peroxisome proliferation but had a negative effect on penicillin production (Meijer et al. 2010).
Taken together, these data suggest that peroxisome numbers are important under conditions that they do not interfere with overall cellular metabolism (e.g. metabolism of oleic acid) or homeostasis of other organelles (Dnm1 is involved also in fission of mitochondria).
The role of peroxisomes in the formation of other secondary metabolites
Recent data revealed several additional metabolic and non-metabolic functions of peroxisomes. During contact of filamentous fungi from Aspergillus species with plant seeds, fungi synthesize and produce seed-contaminating polyketide mycotoxins, such as aflatoxin and sterigmatocystin. The basic building block for the formation of fungal polyketides, acetyl-CoA, can be formed in various catabolic pathways, among others by peroxisomal β-oxidation of fatty acids (Maggio-Hall et al. 2005). Interestingly, the first stable intermediate compound in the synthesis of aflatoxin, norsolorinic acid (NOR), accumulates in peroxisomes of NOR-accumulating mutants. NOR is produced by a complex of polyketide synthase (PKS) and fatty acid synthase (FAS) (Maggio-Hall et al. 2005). Although PKS proteins do not posses clear peroxisome-targeting signals and have not yet been shown to be localized in peroxisomes, the accumulation of intermediates of polyketide biosynthesis in these organelles suggests that peroxisomes, besides supplying acetyl-CoA for polyketide synthesis, may also be directly involved in compartmentalization of the enzymes of polyketide biosynthethic pathway. This hypothesis requires further investigations.
The pathogenic strains of the fungus Alternaria alternata produce toxins that cause severe plant diseases after host infection. The AK-toxin that is involved in fungal infection of Japanese pear is synthesized by products of the recently identified AKT genes. Interestingly, all proteins encoded by AKT genes contain a C-terminal peroxisome targeting signal type I (PTS1) and were shown to be localized to the peroxisome matrix (Tanaka et al. 1999, 2000; Imazaki et al. 2010). In the A. alternata PEX6 deletion strain, Akt proteins were mislocalized to cytosol, which was accompanied by the reduction of conidiation and inability to grow on fatty acids. Interestingly, PEX6 mutant cells were not able to produce AK-toxin and to infect the plant host, highlighting the importance of peroxisomes for plant invasion and tissue colonization by the phytopathogenic fungus A. alternata (Imazaki et al. 2010).
The filamentous fungus Penicillium paxilli produces the indole-diterpene toxin paxilline that is the selective and reversible blocker of the smooth muscle high conductance Ca2+ activated K+ channels. The initial step in biosynthesis of paxilline is the formation of geranylgeranyl diphosphate (GGPP) from isopentenyl diphosphate (IPP) and farnesyl diphosphate (FPP) catalysed by GGPP synthases PaxG (Saikia et al. 2008). The paxilline-producer P. paxilli contains two GGPP synthases: PaxG and GgsA. The analysis of the sequence of PaxG from P. paxilli has revealed that the C-terminus of this enzyme contains a putative PTS1 (GRV). Green fluorescent protein (GFP) fused with the C-terminal tripeptide GRV from PaxG was efficiently imported into peroxisomes, suggesting that PaxG is a peroxisome-borne enzyme. Indeed, paxilline formation appeared to be dependent of the location of PaxG in peroxisomes (Saikia and Scott 2009).
Likely, the atlas of peroxisome functions is still far from complete. Consequently, peroxisomes may be important for the production of other secondary metabolites that are yet not linked with these organelles. This may be exemplified by the analysis of the proteome of glyoxysomes from N. crassa that has revealed the presence of nitropropane dioxygenase in the organelle matrix, suggesting that peroxisomes may play a role in detoxification of nitroalkanes in filamentous fungi (Managadze et al. 2010a, b). Interestingly, recent findings suggest that peroxisomes may be also important for the organization of the cytoskeleton in filamentous fungi, as well as for the development of the appressorium in plant infecting fungi (Fujihara et al. 2010; Zekert et al. 2010).
In general, compartmentalization of biosynthetic pathways is common in secondary metabolism (Roze et al. 2010). Since peroxisomes allow compartmentalization of efficient metabolite formation, peroxisomes could possibly serve as a host compartment for redirection of other, non-peroxisomal biosynthetic pathways as a tool to improve secondary metabolite production. Alternatively, introduction of specialized enzymes into peroxisomes may lead to efficient formation of derivative compounds. For instance, introduction of the cefE gene (encoding deacetoxycephalosporin C synthetase) from Streptomyces clavuligerus, or cefEF (encoding expandase/hydroxylase) from A. chrysogenum and cmcH (encoding carbamoyltransferase) from S. clavuligerus in P. chrysogenum allowed production of cephalosporins and cephalomycins, respectively. The adipoyl side-chain can be removed from the precursor via enzymatic catalysis in peroxisomes, allowing for cheaper and targeted production of variety of novel semi-synthetic antibiotics that may be of value in overcoming the increasing microbial resistance.
β-Oxidation pathways that operates in P. chrysogenum in peroxisomes and mitochondria are involved in side chain precursor (adipate) consumption (van den Berg et al. 2008). Therefore, a new way of improving production of novel carbonylated cephem antibiotics may be based on downregulation of this catabolic pathway.
Transport of β-lactams and their intermediates across the peroxisomal membrane
Compartmentalization of β-lactam biosynthesis requires transport of intermediates and products through biological membranes (see Fig. 3). The peroxisomal membrane bilayer is characterized by its relatively small width and low protein/P-lipid ratio. As protons, acetyl-CoA and NADP(H) cannot freely diffuse through the organelle membrane they require the function of peroxisomal transporters. However, relatively little is yet known on the specific requirements for transport of β-lactams and its intermediates. Given the relative low amount of integral membrane proteins, as suggested from freeze-fracture images (Fig. 5), it is tempting to speculate that the penicillin end product and its intermediates are transported via generalized transporter proteins, i.e. multidrug efflux pumps belonging to the major facilitator superfamily. This view is supported by the finding of Gidijala et al. who showed that introduction of the entire P. chrysogenum penicillin biosynthetic pathway in the methylotrophic yeast H. polymorpha led to an efficient secretion of the penicillin produced (Gidijala et al. 2009). However, in industrial production strains specific penicillin transporters may have evolved or homologous ones up-regulated. Interestingly, CefT, a member of the major facilitator superfamily, is not essential for cephalosporin biosynthesis in A. chrysogenum but promotes the production of this compound upon amplification of the cefT gene (Ullan et al. 2002). α-Amino adipate, a penicillin precursor, requires general amino acid permeases for transport over the cell membrane (Trip et al. 2004). However, also free diffusion may play a role, i.e. in the import of the penicillin side chain precursors phenyloacetic acid (PAA) and phenoxyacetic acid (POA) that can reach the peroxisome matrix via diffusion across the peroxisomal membrane (Evers et al. 2004). Transport of ATP, required for penicillin biosynthesis inside the peroxisome, may require the adenine nucleotide transporter Ant1p (Palmieri et al. 2001). Recent data suggest that transport of IPN in the organelle of A. chrysogenum to form penicillin N for cephalosporin synthesis is mediated by CefP (Ullan et al. 2010) while export of the penicillin N formed to the cytosol requires the peroxisomal transporter CefM (Teijeira et al. 2009) (Fig. 3).
Fig. 5.
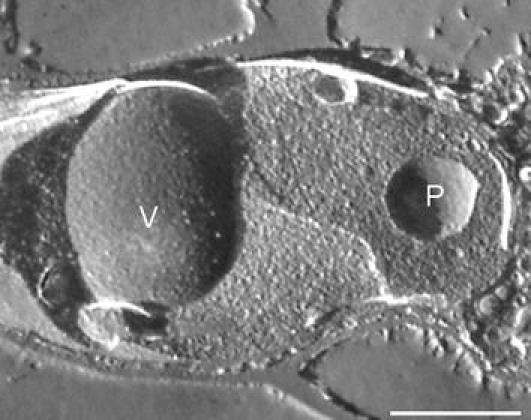
Freeze etch replica of P. chrysogenum. Details of a freeze etch replica of a P. chrysogenum Wisconsin 54-1255 cell grown for 2 days on penicillium induction medium, showing the smooth concave fracture face of a peroxisomal membrane (P). The vacuolar membrane (V) is covered with particles representing membrane proteins. Scale bar represents 5 μm
Future perspectives
Secondary metabolism very often employs enzymes and utilizes compounds from primary metabolic pathways. Because of their highly versatile nature, it may not be surprising that peroxisomes play vital roles in the production of secondary metabolites. The large variety of peroxisome functions suggest that peroxisomes can be important for production of other secondary metabolites that are yet not linked with those organelles. The significance of peroxisomes for efficient penicillin production in filamentous fungi suggests that compartmentalization of biosynthetic pathway to this specific compartment can be beneficial for efficiency of the biosynthetic process. This suggest that peroxisomes could serve as a host compartment for redirection of other, non-peroxisomal biosynthetic pathways as a strategy to improve the efficiency of the compound production.
The presence of intact peroxisomes is crucial for efficient penicillin production in filamentous fungi. Interestingly, also the enhanced number of peroxisome volume fraction correlates with increased penicillin production rates. The molecular basis of this phenomenon is still not clear and needs further studies. It seems that the manipulation of the properties of peroxisomal membrane by activity of Pex11 protein is beneficial for penicillin production rates. It needs to be re-investigated whether the changes in the peroxisomal membrane properties influence function of peroxisome localized transporters and/or facilitate free diffusion of metabolites through peroxisomal membrane (affecting membrane fluidity and permeability).
The identification and detailed characterization of peroxisomal membrane components that are involved in metabolite and protein transport across membrane can open new possibilities for directed strain improvement. The enhanced transport through cell membranes may be achieved by directed adjustments of transporter protein levels and/or manipulation of their structures/activities as well as heterologous expression of specific transporters. Alternatively, the compounds produced inside peroxisomes could be directly secreted to the medium by application of the newly engineered peroxicretion route (Sagt et al. 2009).
A characteristic feature of filamentous fungi is the differentiation of cells along the hyphae. These structures are comprised from actively growing regions (apical cells), metabolic active non-growing regions (sub-apical cells) in conjunction with the older late-apical compartments that are composed of degenerating, highly vacuolated cells. Structured models of fungal growth indicate that secondary metabolites are produced in sub-apical, vacuolated hyphal compartments that are still metabolic active (Paul and Thomas 1996). Therefore, to improve performance of fermentation, the effort should be made to delay deterioration of those hyphal elements. Two cellular processes are involved in self-digestion events of old portions of the hyphae: autolysis and autophagy (White et al. 2002; Zustiak et al. 2008). Autolysis employs activation of several hydrolytic enzymes and it is affected by aging, programmed cell death, development, nutrient limitations, and numerous other factors. Despite its importance in biotechnological processes, autolysis is a poorly understood element of fungal life. Autophagy is a highly conserved mechanism by which cell components (organelles, proteins) are degraded and recycled in the vacuolar lumen. In general, this mechanism is beneficial for the cell allowing survival at nutrient starvation conditions and maintenance of cellular homeostasis, but under specific conditions can also participate in cell death. In filamentous fungi autophagy was shown to be crucial for nutrient recycling under starvation conditions (Bartoszewska and Kiel 2010). A constitutive autophagic turnover of cellular components, including peroxisomes, was observed in late sub-apical compartments of P. chrysogenum under penicillin production conditions. The inhibition of this process resulted in distinct delay of deterioration of those hyphal elements leading to increased microbody numbers accompanied by elevated antibiotic production (Fig. 4b) (Bartoszewska et al. 2011). Thus, to improve performance of fungi in production process, emphasis may also be placed on prolonging the viability (and thus the function) of these organisms.
Acknowledgments
This project was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Aksam EB, Koek A, Kiel JA, Jourdan S, Veenhuis M, van der Klei IJ. A peroxisomal ion protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells. Autophagy. 2007;3:96–105. doi: 10.4161/auto.3534. [DOI] [PubMed] [Google Scholar]
- Barber MS, Giesecke U, Reichert A, Minas W. Industrial enzymatic production of cephalosporin-based beta-lactams. Adv Biochem Eng Biotechnol. 2004;88:179–215. doi: 10.1007/b99261. [DOI] [PubMed] [Google Scholar]
- Bartoszewska M, Kiel JA (2010) The role of macroautophagy in development of filamentous fungi. Antioxid Redox Signal. doi:10.1089/ars.2010.3528 [DOI] [PubMed]
- Bartoszewska M, Kiel JA, Bovenberg RA, Veenhuis M, van der Klei IJ. Autophagy deficiency promotes beta-lactam production in Penicillium chrysogenum. Appl Environ Microbiol. 2011;77:1413–1422. doi: 10.1128/AEM.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen GA, Szilard RK, Rachubinski RA. Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane peroxin. J Cell Biol. 1997;137:1265–1278. doi: 10.1083/jcb.137.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escano CS, Juvvadi PR, Jin FI, Takahashi T, Koyama Y, Yamashita S, Maruyama J, Kitamoto K. Disruption of the Aopex11-1 gene involved in peroxisome proliferation leads to impaired Woronin body formation in Aspergillus oryzae. Eukaryot Cell. 2009;8:296–305. doi: 10.1128/EC.00197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers ME, Trip H, van den Berg MA, Bovenberg RA, Driessen AJ. Compartmentalization and transport in beta-lactam antibiotics biosynthesis. Adv Biochem Eng Biotechnol. 2004;88:111–135. doi: 10.1007/b99259. [DOI] [PubMed] [Google Scholar]
- Fujihara N, Sakaguchi A, Tanaka S, Fujii S, Tsuji G, Shiraishi T, O’Connell R, Kubo Y. Peroxisome biogenesis factor PEX13 is required for appressorium-mediated plant infection by the anthracnose fungus Colletotrichum orbiculare. Mol Plant Microbe Interact. 2010;23:436–445. doi: 10.1094/MPMI-23-4-0436. [DOI] [PubMed] [Google Scholar]
- Gavrilescu M, Chisti Y. Biotechnology—a sustainable alternative for chemical industry. Biotechnol Adv. 2005;23:471–499. doi: 10.1016/j.biotechadv.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Gidijala L, Kiel JA, Douma RD, Seifar RM, van Gulik WM, Bovenberg RA, Veenhuis M, van der Klei IJ. An engineered yeast efficiently secreting penicillin. PLoS One. 2009;4:e8317. doi: 10.1371/journal.pone.0008317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Giraldez C, Quijada L, Clayton CE. Compartmentation of enzymes in a microbody, the glycosome, is essential in Trypanosoma brucei. J Cell Sci. 2002;115:2651–2658. doi: 10.1242/jcs.115.13.2651. [DOI] [PubMed] [Google Scholar]
- Guo T, Gregg C, Boukh-Viner T, Kyryakov P, Goldberg A, Bourque S, Banu F, Haile S, Milijevic S, San KH, Solomon J, Wong V, Titorenko VI. A signal from inside the peroxisome initiates its division by promoting the remodeling of the peroxisomal membrane. J Cell Biol. 2007;177:289–303. doi: 10.1083/jcb.200609072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imazaki A, Tanaka A, Harimoto Y, Yamamoto M, Akimitsu K, Park P, Tsuge T. Contribution of peroxisomes to secondary metabolism and pathogenicity in the fungal plant pathogen Alternaria alternata. Eukaryot Cell. 2010;9:682–694. doi: 10.1128/EC.00369-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami MS, Barreiro C, Garcia-Estrada C, Martin JF. Proteome analysis of the penicillin producer Penicillium chrysogenum: characterization of protein changes during the industrial strain improvement. Mol Cell Proteomics. 2010;9:1182–1198. doi: 10.1074/mcp.M900327-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G. Fungal evo-devo: organelles and multicellular complexity. Trends Cell Biol. 2011;21:12–19. doi: 10.1016/j.tcb.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Kiel JA, van der Klei IJ, van den Berg MA, Bovenberg RA, Veenhuis M. Overproduction of a single protein, Pc-Pex11p, results in 2-fold enhanced penicillin production by Penicillium chrysogenum. Fungal Genet Biol. 2005;42:154–164. doi: 10.1016/j.fgb.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Kiel JA, Veenhuis M, van der Klei IJ. PEX genes in fungal genomes: common, rare or redundant. Traffic. 2006;7:1291–1303. doi: 10.1111/j.1600-0854.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- Kiel JA, van den Berg MA, Fusetti F, Poolman B, Bovenberg RA, Veenhuis M, van der Klei IJ. Matching the proteome to the genome: the microbody of penicillin-producing Penicillium chrysogenum cells. Funct Integr Genomics. 2009;9:167–184. doi: 10.1007/s10142-009-0110-6. [DOI] [PubMed] [Google Scholar]
- Koetsier MJ, Jekel PA, van den Berg MA, Bovenberg RA, Janssen DB. Characterization of a phenylacetate-CoA ligase from Penicillium chrysogenum. Biochem J. 2009;417:467–476. doi: 10.1042/BJ20081257. [DOI] [PubMed] [Google Scholar]
- Liu F, Ng SK, Lu Y, Low W, Lai J, Jedd G. Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J Cell Biol. 2008;180:325–339. doi: 10.1083/jcb.200705049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio-Hall LA, Wilson RA, Keller NP. Fundamental contribution of beta-oxidation to polyketide mycotoxin production in planta. Mol Plant Microbe Interact. 2005;18:783–793. doi: 10.1094/MPMI-18-0783. [DOI] [PubMed] [Google Scholar]
- Managadze D, Wurtz C, Wiese S, Meyer HE, Niehaus G, Erdmann R, Warscheid B, Rottensteiner H. A proteomic approach towards the identification of the matrix protein content of the two types of microbodies in Neurospora crassa. Proteomics. 2010;10:3222–3234. doi: 10.1002/pmic.201000095. [DOI] [PubMed] [Google Scholar]
- Managadze D, Wurtz C, Wiese S, Schneider M, Girzalsky W, Meyer HE, Erdmann R, Warscheid B, Rottensteiner H. Identification of PEX33, a novel component of the peroxisomal docking complex in the filamentous fungus Neurospora crassa. Eur J Cell Biol. 2010;89:955–964. doi: 10.1016/j.ejcb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Martin JF, Ullan RV, Garcia-Estrada C. Regulation and compartmentalization of beta-lactam biosynthesis. Microb Biotechnol. 2010;3:285–299. doi: 10.1111/j.1751-7915.2009.00123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer WH, Gidijala L, Fekken S, Kiel JA, van den Berg MA, Lascaris R, Bovenberg RA, van der Klei IJ. Peroxisomes are required for efficient penicillin biosynthesis in Penicillium chrysogenum. Appl Environ Microbiol. 2010;76:5702–5709. doi: 10.1128/AEM.02327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller WH, Bovenberg RA, Groothuis MH, Kattevilder F, Smaal EB, van der Voort LH, Verkleij AJ. Involvement of microbodies in penicillin biosynthesis. Biochim Biophys Acta. 1992;1116:210–213. doi: 10.1016/0304-4165(92)90118-e. [DOI] [PubMed] [Google Scholar]
- Ng SK, Liu F, Lai J, Low W, Jedd G. A tether for Woronin body inheritance is associated with evolutionary variation in organelle positioning. PLoS Genet. 2009;5:e1000521. doi: 10.1371/journal.pgen.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalinski L, Kiel JA, Homan TG, Veenhuis M, van der Klei IJ. Penicillium chrysogenum Pex14/17p—a novel component of the peroxisomal membrane that is important for penicillin production. FEBS J. 2010;277:3203–3218. doi: 10.1111/j.1742-4658.2010.07726.x. [DOI] [PubMed] [Google Scholar]
- Opalinski L, Kiel JA, Williams C, Veenhuis M, van der Klei IJ. Membrane curvature during peroxisome fission requires Pex11. EMBO J. 2011;30:5–16. doi: 10.1038/emboj.2010.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Rottensteiner H, Girzalsky W, Scarcia P, Palmieri F, Erdmann R. Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J. 2001;20:5049–5059. doi: 10.1093/emboj/20.18.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul GC, Thomas CR. A structured model for hyphal differentiation and penicillin production using Penicillium chrysogenum. Biotechnol Bioeng. 1996;51:558–572. doi: 10.1002/(SICI)1097-0290(19960905)51:5<558::AID-BIT8>3.3.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Roze LV, Chanda A, Linz JE. Compartmentalization and molecular traffic in secondary metabolism: a new understanding of established cellular processes. Fungal Genet Biol. 2010;48:35–48. doi: 10.1016/j.fgb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucktaschel R, Girzalsky W, Erdmann R. Protein import machineries of peroxisomes. Biochim Biophys Acta. 2010;1808:892–900. doi: 10.1016/j.bbamem.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Sagt CM, ten Haaft PJ, Minneboo IM, Hartog MP, Damveld RA, van der Laan JM, Akeroyd M, Wenzel TJ, Luesken FA, Veenhuis M, van der Klei IJ, de Winde JH. Peroxicretion: a novel secretion pathway in the eukaryotic cell. BMC Biotechnol. 2009;9:48. doi: 10.1186/1472-6750-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia S, Scott B. Functional analysis and subcellular localization of two geranylgeranyl diphosphate synthases from Penicillium paxilli. Mol Genet Genomics. 2009;282:257–271. doi: 10.1007/s00438-009-0463-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia S, Nicholson MJ, Young C, Parker EJ, Scott B. The genetic basis for indole-diterpene chemical diversity in filamentous fungi. Mycol Res. 2008;112:184–199. doi: 10.1016/j.mycres.2007.06.015. [DOI] [PubMed] [Google Scholar]
- Schroeckh V, Scherlach K, Nutzmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C. AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA. 2003;100:9626–9631. doi: 10.1073/pnas.1633697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South ST, Gould SJ. Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol. 1999;144:255–266. doi: 10.1083/jcb.144.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprote P, Brakhage AA, Hynes MJ. Contribution of peroxisomes to penicillin biosynthesis in Aspergillus nidulans. Eukaryot Cell. 2009;8:421–423. doi: 10.1128/EC.00374-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Tsuge T. Structural and functional complexity of the genomic region controlling AK-toxin biosynthesis and pathogenicity in the Japanese pear pathotype of Alternaria alternata. Mol Plant Microbe Interact. 2000;13:975–986. doi: 10.1094/MPMI.2000.13.9.975. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Shiotani H, Yamamoto M, Tsuge T. Insertional mutagenesis and cloning of the genes required for biosynthesis of the host-specific AK-toxin in the Japanese pear pathotype of Alternaria alternata. Mol Plant Microbe Interact. 1999;12:691–702. doi: 10.1094/MPMI.1999.12.8.691. [DOI] [PubMed] [Google Scholar]
- Teijeira F, Ullan RV, Guerra SM, Garcia-Estrada C, Vaca I, Martin JF. The transporter CefM involved in translocation of biosynthetic intermediates is essential for cephalosporin production. Biochem J. 2009;418:113–124. doi: 10.1042/BJ20081180. [DOI] [PubMed] [Google Scholar]
- Tey WK, North AJ, Reyes JL, Lu YF, Jedd G. Polarized gene expression determines Woronin body formation at the leading edge of the fungal colony. Mol Biol Cell. 2005;16:2651–2659. doi: 10.1091/mbc.E04-10-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S, Gronborg S, Gartner J. Organelle interplay in peroxisomal disorders. Trends Mol Med. 2009;15:293–302. doi: 10.1016/j.molmed.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Trip H, Evers ME, Kiel JA, Driessen AJ. Uptake of the beta-lactam precursor alpha-aminoadipic acid in Penicillium chrysogenum is mediated by the acidic and the general amino acid permease. Appl Environ Microbiol. 2004;70:4775–4783. doi: 10.1128/AEM.70.8.4775-4783.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullan RV, Casqueiro J, Banuelos O, Fernandez FJ, Gutierrez S, Martin JF. A novel epimerization system in fungal secondary metabolism involved in the conversion of isopenicillin N into penicillin N in Acremonium chrysogenum. J Biol Chem. 2002;277:46216–46225. doi: 10.1074/jbc.M207482200. [DOI] [PubMed] [Google Scholar]
- Ullan RV, Teijeira F, Guerra SM, Vaca I, Martin JF. Characterization of a novel peroxisome membrane protein essential for conversion of isopenicillin N into cephalosporin C. Biochem J. 2010;432:227–236. doi: 10.1042/BJ20100827. [DOI] [PubMed] [Google Scholar]
- Valenciano S, De Lucas JR, van der Klei IJ, Veenhuis M, Laborda F. Characterization of Aspergillus nidulans peroxisomes by immunoelectron microscopy. Arch Microbiol. 1998;170:370–376. doi: 10.1007/s002030050655. [DOI] [PubMed] [Google Scholar]
- van den Berg MA, Albang R, Albermann K, Badger JH, Daran JM, Driessen AJ, Garcia-Estrada C, Fedorova ND, Harris DM, Heijne WH, Joardar V, Kiel JA, Kovalchuk A, Martin JF, Nierman WC, Nijland JG, Pronk JT, Roubos JA, van der Klei IJ, van Peij NN, Veenhuis M, von Dohren H, Wagner C, Wortman J, Bovenberg RA. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotechnol. 2008;26:1161–1168. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- White S, McIntyre M, Berry DR, McNeil B. The autolysis of industrial filamentous fungi. Crit Rev Biotechnol. 2002;22:1–14. doi: 10.1080/07388550290789432. [DOI] [PubMed] [Google Scholar]
- Wurtz C, Schliebs W, Erdmann R, Rottensteiner H. Dynamin-like protein-dependent formation of Woronin bodies in Saccharomyces cerevisiae upon heterologous expression of a single protein. FEBS J. 2008;275:2932–2941. doi: 10.1111/j.1742-4658.2008.06430.x. [DOI] [PubMed] [Google Scholar]
- Zekert N, Veith D, Fischer R. Interaction of the Aspergillus nidulans microtubule-organizing center (MTOC) component ApsB with gamma-tubulin and evidence for a role of a subclass of peroxisomes in the formation of septal MTOCs. Eukaryot Cell. 2010;9:795–805. doi: 10.1128/EC.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zustiak MP, Pollack JK, Marten MR, Betenbaugh MJ. Feast or famine: autophagy control and engineering in eukaryotic cell culture. Curr Opin Biotechnol. 2008;19:518–526. doi: 10.1016/j.copbio.2008.07.007. [DOI] [PubMed] [Google Scholar]


