Abstract
Bacterial transcription mediated by RNA polymerase (RNAP) is a highly regulated process and RNAP action is modulated during the different phases of initiation, elongation and termination by proteins such as the Escherichia coli Nus transcription-factors. Here we discuss the structural interplay and the mechanistic role of the Nus-factors that are directly involved in the processivity of elongation, transcription:translation coupling and termination, as well as the varying effects of these proteins on transcription under the influence of additional signals.
Key words: transcription, initiation, elongation, termination, nus-factor, RNA polymerase
The Nus transcription-factors were originally identified as part of the E. coli phage λ N-protein-controlled antitermination system; hence, they were termed N-utilization substances.1 NusA, B, E and G are important for different levels of regulation within transcription and transcription:translation coupling. Their regulatory effect is often antithetic and depends on different external signals.
NusA
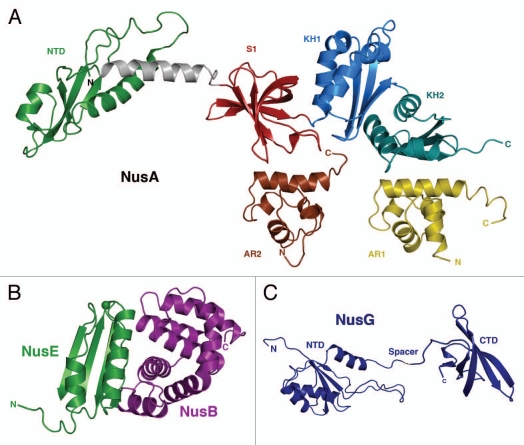
NusA is a highly conserved elongation factor identified in bacteria and archaea,2 which in E. coli consists of 495 amino acids (55 kDa) arranged in 6 domains (Fig. 1A). The amino-terminal domain (NTD; amino-acids 1–137) interacts with the RNAP close to the RNA exit channel.3 NusA-NTD is linked via a flexible helix to three RNA-binding subdomains, S1 (138–201), KH1 (202–276) and KH2 (277–344), forming the central SKK domain that binds single stranded RNA of the nascent transcript.4,5 C-terminal to this SKK domain follow the so-called acidic repeats 1 and 2 [AR1 (345–426), AR2 (427–495)], which so far could only be identified in E. coli.6 Although the latter domains share the same structural topology, they exhibit different functions. The only role for AR1 identified thus far in vitro is binding the λ protein N7 and the λN:AR1 complex may well be part of the λ antitermination complex in vivo; nevertheless, this could not be shown so far. AR2 has at least two distinct but interrelated functions: It masks the central SKK domain of NusA by forming an intramolecular interdomain complex, thus autoinhibiting RNA interaction of this domain;8,9 AR2 forms a complex with the C-terminal domain CTD of the α-subunit of RNAP (αCTD), which supposedly hinders this regulatory RNAP subunit from re-attaching to the UP-element (upstream promoter).8 Primarily, NusA supports intrinsic termination due to its interaction with the hairpin structures of pause and termination signals3,10 and it modulates p-dependent termination.11,12 In concert with the other Nus-factors, NusA enables the formation of stable ECs (elongation complexes), which leads to processive transcription and read-through of termination sites.13
Figure 1.
(A) Crystal-Structure of Thermotoga maritima NusA and solution NMR-structures of the two AR-domains of E. coli NusA. The NusA-NTD of T. maritima (green) is linked via a connecting helix (grey) to the central RNA-binding domains S1 (red), KH1 (blue) and KH2 (aquamarine) of NusA (PDB-ID: 1HH2).4 Additionally shown are the solution NMR-structures of AR1 (brown; PDB-ID: 1WCL) and AR2 (yellow; PDB-ID: 1WCH) domains,6 which so far could only been identified within E. coli. (B) Crystal-Structure of the E. coli NusB:NusE complex. NusE (green) and NusB (purple) form a tight complex (PDB-ID: 3D3B).17 The single sphere within NusE denotes Ser46, which replaces the ribosome binding loop 46–67 in the crystallized construct (see details in reference 17). (C) Solution NMR-Structure of NusG (NusG-NTD; PDB-ID: 2K06; NusG-CTD; PDB-ID: 2JVV).24
NusB
NusB (15.7 kDa) mainly acts as an auxiliary protein within transcriptional regulation, which consists of an all-helical fold with two perpendicular three-helix bundles.14 NusB is able to bind single stranded RNA.15,16 Additionally, NusB forms a stable complex with NusE (Fig. 1B),15 that, however, leads only to a slight rearrangement of its helix-bundles.17 This heterodimerization enhances significantly the affinity to RNA,16,18 because both proteins form an extended mosaic RNA-binding interface.17 Results of combined biophysical, mutational and genetic investigations indicate that this interaction is fine-tuned within the functional EC.16
NusE
NusE is an 11.7 kDa protein, which doubles as ribosomal protein S10 and exhibits a four-stranded antiparallel β-sheet that is backed by two α-helices on one side.17 Upon complex formation with NusB, helix α1 and irregular strand β2 bridge the two helical bundles of NusB and form a protein interaction surface of ∼1,700 Å2 without any evident structural rearrangement.17 In this heterodimer, NusE is the functionally active partner, which could be shown by its N-protein antitermination activity in the absence of NusB.17 In addition to its role in transcription, NusE is involved in translation as part of the 30S subunit of the ribosome, where it exhibits virtually the identical structure as in its NusB complex. NusE belongs to those proteins that finish ribosomal assembly.19 Binding to NusB ascertains a stable fold of NusE,17 and NusB is able to direct the heterodimer to the EC,17,20 where it becomes a central part of the EC. Whether NusB is important for the introduction of NusE into the ribosome and how this is achieved will be questions for future studies.
NusG
NusG is an essential bacterial regulator of the RNAP with a size of 20.5 kDa. Proteins homologous to NusG can be found in archaea21 and eukaryotes.22 With its two domain structure and a flexible linker between the transiently interacting domains,23 NusG is able to interact with different partners and therefore is predestined to function as a linker protein (Fig. 1C).24 The NusG-NTD, residues 1–116, consists of four central β-strands forming an anti-parallel β-sheet surrounded by three α-helices and is linked to the CTD, residues 123–181, which itself forms a β-barrel composed of five anti-parallel β-strands.24 The NusG-NTD is supposed to directly interact with the RNAP through a hydrophobic patch on its surface25 and NusG-CTD is able to bind either to NusE20 to enable transcription:translation coupling or to the ρ-factor20 to support ρ-dependent termination. NusG versatile role within transcription regulation is reflected by its ability to modulate the EC in different ways: NusG is able to enhance the elongation rate, an effect probably due to its ability to suppress transcriptional pausing,26 in turn attributable to its promotion of forward translocation of the RNAP;27 NusG is also important for activation of ρ in vivo and in vitro for most ρ-dependent termination events in order to maintain transcriptional boundaries within the bacterial genome.12
Elongation Complexes
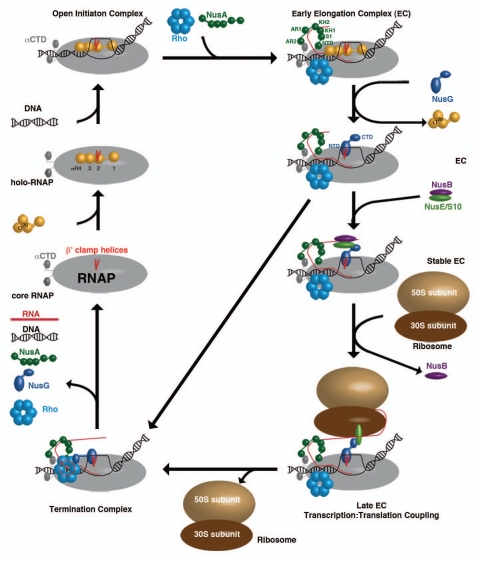
E. coli core RNAP consists of five subunits (α2, β′, β and ω) of roughly 400 kDa molecular mass (Fig. 2).28 The β and β′-subunits form the active site, whereas the NTDs of the two α subunits ascertain correct RNAP assembly. The ω subunit supports this assembly, but is not essential for RNAP functionality.28 To enable DNA binding, a σ-factor has to associate to form the holo-RNAP, which is able to move along the double-stranded DNA to recognize promoter sequences.28 Once bound to these, an open initiation complex is formed that induces bending and unwinding of the DNA to form a transcription bubble.29 During RNA synthesis, several transcription regulators bind to the RNAP to ensure a stable and processive EC. NusA is among the first proteins to interact with RNAP after initiation.30 The NusA NTD binds close to the RNA exit channel of RNAP3 and forms a stable RNAP complex via NusA-AR2:αCTD-RNAP domain interaction.9 The AR2:αCTD complex in all likelihood prevents αCTD from reattaching to DNA,8 a process that would lead to a stalled transcription complex. Recent Chromatin Immunoprecipitation (ChIP)-chip analysis30 and in vivo assays31 showed that the ρ-termination factor already collocates with elongating RNAP at this early stage and that ρ stays bound to the RNAP throughout the whole transcription circle,30–32 an interaction that is independent of the presence of the nascent RNA chain.31 On a structural level, nothing is known yet about the ρ:RNAP interaction, but the architecture of this crucial linkage is part of several ongoing research programs. With progressing elongation, the affinity of the σ-factor to RNAP gradually decreases, accompanied by displacement of σR2 (region 2) from its binding site at the β′-clamp-helices of RNAP by the NTD of NusG.25 The formation of this σ-less EC is an important checkpoint within the transcription circle. If no additional factors associate with the EC at this stage, the RNAP is unable to effectively transcribe the whole DNA, it only produces short abortive RNAs,33 and presumably the NusG-CTD is able to bind ρ that will be loaded onto the nascent RNA and initiate the formation of a termination complex, leading directly to complete complex disassembly. This premature termination is avoided by binding of the NusB:NusE heterodimer to single stranded RNA,16–18 an interaction crucial for processive transcription.16 This interaction allows direct binding between NusE and NusG-CTD and enables coupling of the EC to ribosomes.20
Figure 2.
The different ECs within a transcription cycle consisting of RNAP (grey), β′ clamp helices (red), σ70 (yellow), DNA (black), Rho-hexamer (light-blue), NusA (dark-green), RNA (red), NusG (dark-blue), NusB (purple), NusE (light-green), 50S ribosomal subunit (sand) and 30S ribosomal subunit (brown).
The rate of transcription could be shown to be ribosome-dependent as this rate depends on the translation rate, leading to the model that RNAP waits for the leading ribosome to be loaded onto RNAP,34 presumably by the NusE:NusG interaction,20 and a train of ribosomes, the so-called polysome, is able to follow. This flexibility to adjust the transcriptional yield to their translational needs34 is unique to bacteria, as both processes are spatially connected in contrast to the situation in eukaryotic cells, where transcription and translation progress in different cellular compartments. At the end of bacterial operons, ribosomes encounter nonsense codons and the leading ribosome, as well as the ones following, is removed from the RNA. This would ascertain that the mRNA can quickly accumulate the required length12 for ρ interaction, as ρ needs to bind to single stranded RNA of about 80–90 nucleotides in length to be able to use its inherent ATP hydrolysis to translocate along the RNA,31 and that the freed NusG-CTD is able to bind ρ to promote termination. Nevertheless, not much is known about the structural and functional details of the ρ termination mechanism, the effect of NusG binding, and the process by which NusG-CTD leaves NusE to bind ρ. Presently, a model for the termination mechanism proposes that, upon interaction with RNA and binding to NusG, ρ uses its intrinsic ATP-dependent helicase-activity to slide along the RNA in 3′-direction into the RNA exit channel.31 This triggers significant widening of the channel, which leads to a collapse of the transcription bubble. This process is supported by a weakening of the RNA:DNA hybrid due to an adenine-rich DNA-stretch and results in the disassembly of the whole EC and in the components being at disposal for a new round of transcription.
Concluding Remarks
The weakest link in the chain of information on the transcription process in E. coli at present is the structure of the RNAP itself. E. coli RNAP could not be crystallized yet, probably due to large insertions in its β- and β′-subunits, which are in close proximity to important regulatory sites like the β′-clamp-helices and the β-lid-domain.28 Therefore, most of the models for RNAP interactions are based on a high-resolution structure from Thermus thermophilus35 and a recently published structural model of E. coli RNAP,36 which awaits structural and functional validation. Knowledge about the effects induced by proteins like NusG and NusA that interact directly with RNAP is very limited. Specially, little is known about the recently identified direct ρ-binding to RNAP that occurs upon initiation to elongation transition.31 Indeed, in many cases, not even the interacting domains of RNAP have been clearly identified so far. Structural details of the ρ:NusG interaction and the ρ termination mechanism are still not known. Questions also remain on the influence of the ribosome on transcription—do ribosomes just load onto RNAP and push it along the DNA, or do ribosomes induce structural changes in the RNAP and thus act antitermination factor-like to prevent transcriptional pausing, as proposed by Roberts?37 Taken together, the model for the different states of transcription will undergo significant refinement as future work addressing these important questions leads to more detailed understanding of transcription regulation and transcription:translation coupling.
Acknowledgments
Funding by the Deutsche Forschungge-meinschaft to PR (Ro617/16-1;18-1) is gratefully acknowledged.
Abbreviations
- RNAP
RNA polymerase
- Nus
N-utilization substance
- E. coli
Escherichia coli
- NTD
amino-terminal domain
- CTD
carboxy-terminal domain
- AR
acidic repeat
- EC
elongation complex
- ChIP
chromatin immunoprecipitation
References
- 1.Friedman DI, Baron LS. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology. 1974;58:141–148. doi: 10.1016/0042-6822(74)90149-4. [DOI] [PubMed] [Google Scholar]
- 2.Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- 3.Ha KS, Toulokhonov I, Vassylyev DG, Landick R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J Mol Bio. 2010 doi: 10.1016/j.jmb.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Worbs M, Bourenkov GP, Bartunik HD, Huber R, Wahl MC. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell. 2001;7:1177–1189. doi: 10.1016/s1097-2765(01)00262-3. [DOI] [PubMed] [Google Scholar]
- 5.Prasch S, Jurk M, Washburn RS, Gottesman ME, Wöhrl BM, Rösch P. RNA-binding specificity of E. coli NusA. Nucl Acids Res. 2009;37:4736–4742. doi: 10.1093/nar/gkp452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenmann A, Schwarz S, Prasch S, Schweimer K, Rösch P. The E. coli NusA carboxy-terminal domains are structurally similar and show specific RNAP- and λ N interaction. Protein Sci. 2005;14:2018–2029. doi: 10.1110/ps.051372205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasch S, Schwarz S, Eisenmann A, Wöhrl BM, Schweimer K, Rösch P. Interaction of the intrinsically unstructured phage lambda N protein with E. coli NusA. Biochemistry. 2006;45:4542–4549. doi: 10.1021/bi0523411. [DOI] [PubMed] [Google Scholar]
- 8.Schweimer K, Prasch S, Santhanam SP, Bubunenko M, Gottesman ME, Rösch P. NusA interaction with the α-subunit of E. coli RNA polymerase is via the UP-element site and releases autoinhibition. Structure. 2011 doi: 10.1016/j.str.2011.03.024. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mah TF, Kuznedelov K, Mushegian A, Severinov K, Greenblatt J. The alpha subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev. 2000;14:2664–2675. doi: 10.1101/gad.822900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 11.Jin DJ, Cashel M, Friedman DI, Nakamura Y, Walter WA, Gross CA. Effects of rifampicin resistant rpoB mutations on antitermination and interaction with nusA in Escherichia coli. J Mol Biol. 1988;204:247–261. doi: 10.1016/0022-2836(88)90573-6. [DOI] [PubMed] [Google Scholar]
- 12.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: The basic mechanisms. Cell. 2001;107:437–449. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 14.Altieri AS, Mazzulla MJ, Horita DA, Coats RH, Wingfield PT, Das A, et al. The structure of the transcriptional antiterminator NusB from Escherichia coli. Nat Struct Biol. 2000;7:470–474. doi: 10.1038/75869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason SW, Li J, Greenblatt J. Direct interaction between two Escherichia coli transcription antitermination factors, NusB and ribosomal protein S10. J Mol Biol. 1992;223:55–66. doi: 10.1016/0022-2836(92)90715-v. [DOI] [PubMed] [Google Scholar]
- 16.Burmann BM, Luo X, Wahl MC, Rösch P, Gottesman ME. Fine tuning of the E. coli NusB:NusE complex affinity to BoxA RNA is required for processive antitermination. Nucleic Acids Res. 2010;38:314–326. doi: 10.1093/nar/gkp736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo X, Hsiao HH, Bubunenko M, Weber G, Court DL, Gottesman ME, et al. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol Cell. 2008;32:791–802. doi: 10.1016/j.molcel.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greive SJ, Lins AF, von Hippel PH. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. J Biol Chem. 2005;280:36397–36408. doi: 10.1074/jbc.M507146200. [DOI] [PubMed] [Google Scholar]
- 19.Noller HF, Nomura M. Neidhardt FC, Curtiss R, Ingraham J, Lin ECC, Low KB, editors. Ribosomes. Escherichia Coli and Salmonella: Cellular and Molecular Biology. Am Soc Microbiol. 1996;1:167–186. [Google Scholar]
- 20.Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rösch P. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 21.Kyrpides NC, Ouzounis CA. Transcription in archaea. Proc Natl Acad Sci USA. 1999;96:8545–8550. doi: 10.1073/pnas.96.15.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5 and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burmann BM, Schweimer K, Scheckenhofer U, Rösch P. Domain interactions of the transcription-translation coupling factor E. coli NusG are intermolecular and transient. Biochem J. 2011;435:783–789. doi: 10.1042/BJ20101679. [DOI] [PubMed] [Google Scholar]
- 24.Mooney RA, Schweimer K, Rösch P, Gottesman ME, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 2009;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belogurov GA, Mooney RA, Svetlov V, Landick R, Artsimovitch I. Functional specialization of transcription elongation factors. EMBO J. 2009;28:112–122. doi: 10.1038/emboj.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbert KM, Zhou J, Mooney RA, Porta AL, Landick R, Block SM. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399:17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Severinov K. RNA polymerase structure-function: Insights into points of transcriptional regulation. Curr Opin Microbiol. 2000;3:118–125. doi: 10.1016/s1369-5274(00)00062-x. [DOI] [PubMed] [Google Scholar]
- 29.Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: An on-again, off-again relationship? Mol Cell. 2005;20:335–345. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 30.Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci USA. 2009;106:15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldman SR, Ebright RH, Nickels BE. Direct detection of abortive RNA transcripts in vivo. Science. 2009;324:927–928. doi: 10.1126/science.1169237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 36.Opalka N, Brown J, Lane WJ, Twist KF, Landick R, Asturias FJ, Darst SA. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol. 2010;8:1000483. doi: 10.1371/journal.pbio.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts JW. Molecular biology. Syntheses that stay together. Science. 2010;328:436–437. doi: 10.1126/science.1189971. [DOI] [PubMed] [Google Scholar]