Abstract
STAT proteins are obligate promoters of T helper cell differentiation and initial studies suggested that activation of a single STAT protein resulted in a particular phenotype. More recent work has supported a more complex paradigm wherein the activation of several STAT proteins is required for differentiation to a single effector lineage.
Key words: STAT, differentiation, CD4 T helper cell, cytokine, gene regulation
Signal Transducer and Activator of Transcription (STAT) proteins are phosphorylated by Janus (JAK) tyrosine kinases following stimulation of a cell with cytokines or growth factors. Phosphorylation of specific tyrosine residues converts latent cytoplasmic STATs into dimers that move to the nucleus, bind DNA and activate transcription. Cytokines or growth factors activate specific members of the seven member STAT family generating one component of specificity in responses to extracellular signals.
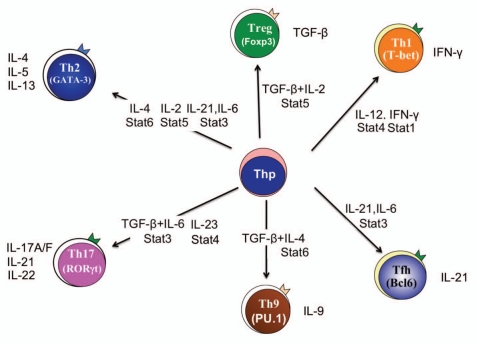
The differentiation of CD4+ T helper (Th) cells into effector subsets that secrete specific cytokines is largely dependent upon the cytokine environment present when naïve T cells are activated by antigen from pathogens, foreign proteins such as allergens, or self-antigens in the case of autoimmunity (Fig. 1). An environment containing IL-12 promotes the STAT4-dependent development of IFNγ-secreting Th1 cells.1,2 IL-4 induces the STAT6-dependent differentiation of Th2 cells.3–5 Similarly, several cytokines including IL-6, IL-21 and IL-23 promote the STAT3-dependent development of Th17 cells that secrete IL-17 and other cytokines.6–8 STAT5, activated in response to IL-2, contributes to inducible regulatory T cell development.9,10 Each of the STATs binds to hundreds of loci that contribute to the effector phenotype.11–14 All of these data supported a paradigm wherein activation of a particular STAT protein in T cells would result in the activation of a specific effector phenotype. However, continuing investigations revealed that a one STAT-one phenotype paradigm was too simple to define the complex responses of differentiating T cells to a diverse cytokine milieu.
Figure 1.
T helper (Th) cell phenotypes. Cytokine environments, and the downstream STAT proteins that are subsequently activated (adjacent to arrows from naïve T helper precursors (Thp) to each effector subset) promote the development of various Th cell effector phenotypes. The hallmark transcription factor for each phenotype is indicated within brackets in each cell type, and the cytokines produced by each subset are shown. Th, T helper; Treg, regulatory T cell; Tfh, T follicular helper cell.
The first evidence that more than one STAT was involved in a particular differentiation pathway was observed following the identification of T-bet, a T-box transcription factor that promotes the Th1 phenotype.15 Although both STAT4 and T-bet promote Th1 development, efforts to place these factors in a linear pathway failed because IFNγ-activated STAT1, rather than IL-12-activated STAT4, resulted in increased T-bet expression.16,17 More recent work suggested that IFNγ activates T-bet expression early, and IL-12 activates it later in differentiation, supporting parallel pathways where both STAT4 and T-bet are required for separate and overlapping aspects of the Th1 genetic program.18,19
A similar parallel requirement was observed for STAT5 in Th2 development (Fig. 2). Based on the observations that IL-2 increased the development of Th2 cells, more detailed studies demonstrated that STAT5 promoted Th2 cytokine production through a parallel pathway that involved accessibility of Th2 cytokine loci.20 STAT5 also contributed to allergic inflammation in vivo, demonstrating that both STAT5 and STAT6 proteins collaborated on the induction of Th2-mediated inflammation.21 These examples added to the complexity of the role of STATs in Th development. They demonstrated that although one cytokine might be the predominant initiator of a differentiation pathway, the developing effector T cell responds to additional cytokines in the environment, and those cytokines activate additional STAT proteins. Thus, the T cell is able to integrate multiple signals in the process of differentiation. Our recent work adds to the more complex paradigm by demonstrating that STAT3 is required for Th2 development.
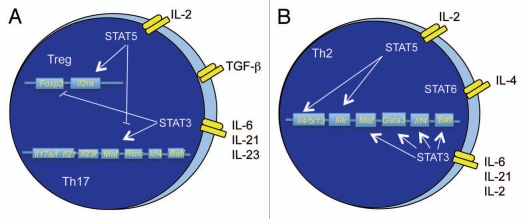
Figure 2.
Opposing and cooperative STAT pathways. (A) During Treg development, STAT5 promotes the expression of Foxp3 and Il2ra, while repressing Th17 development by binding to the Il17a/f locus, and potentially other loci. Conversely, STAT3 binds to multiple gene loci that comprise a Th17 genetic program, while inhibiting the expression of Foxp3. (B) In developing Th2 cells, STAT6 binds to multiple loci associated with the Th2 genetic program. STAT5 works in concert by binding to the Il4r locus, and the Il4 locus. STAT3 binds to loci expressing transcription factors that contribute to the Th2 phenotype. The loci indicated are only a subset of potential binding sites for each factor.
As mentioned above, a number of cytokines that activate STAT3 are present in the developing Th2 environment from autocrine or paracrine sources, including IL-6, IL-21 and IL-2, all of which might contribute to the activation of specific components of the Th2 genetic program.22–24 We demonstrated that STAT3 is activated throughout Th2 development, and that neutralization of all of the cytokines listed above was required to decrease phosphorylated STAT3 within developing Th2 cells, suggesting that there is some redundancy in their function.25 Moreover, STAT3-deficient T cells had diminished production of Th2 cytokines, reduced expression of transcription factors required for Th2 development, and altered histone modification patterns at the loci encoding those transcription factors. STAT3 is bound to the promoters of loci encoding transcription factors expressed in Th2 cells in both naïve and differentiated Th2 cells (Fig. 2). Importantly, STAT3 was not required for normal activation of STAT6, but was required for the development of Th2 cells in vivo, as well as the development of Th2-mediated allergic inflammation.
How the cell integrates signals from multiple STATs is still unclear, and several mechanisms might be involved. Data suggest that the STAT1-STAT4 interplay has a temporal basis, each STAT acting on common loci at different times,18 likely due to changes in the expression of receptors for the requisite activating cytokines. STAT5 also works with STAT4 by binding to gene loci important for Th1 development.26,27 In Th2 development, STAT5 and STAT6 appear to work separately, although STAT5 regulates receptors for the STAT6-activating cytokine IL-4.20,28 The theme of regulating the receptor for another cytokine and altering the activation of downstream STATs recurs in several instances during T helper cell differentiation, including regulation of IL-2R, IL-4R and IL-12R.27,29
However, we observed normal expression of IL-4R and normal activation of STAT6 in the absence of STAT3. Despite this, STAT6 was not bound appropriately to target loci.25 The lack of STAT6 binding suggests either that STAT3 and STAT6 cooperate in binding, or that STAT3 mediates accessibility of the loci to facilitate binding of STAT6. Indeed, we observed decreased accessibility at several Th2 transcription factor loci in the absence of STAT3. It is still possible that STAT3 and STAT6 might physically interact to mediate binding to target loci. Several STAT proteins can form hetero- as well as homodimers. STAT6 is thought to function mostly as a homodimer, but it is possible these factors might heterodimerize. STAT proteins can also “tetramerize” through interactions of N-terminal domains that facilitate cooperative binding to non-consensus DNA elements, and though these interactions are thought to be homotypic, occurring only among dimers of like STATs, it is conceivable that there could be higher-order interactions in an enhanceosome complex. STAT3 might also separately contribute to recruitment of factors to a STAT6-dependent enhanceosome.30 All of these functions could occur in parallel to the direct effect of STAT3 on additional documented targets including Maf and Socs1.31,32
STAT3 and STAT5 are mutually antagonistic as they respectively promote Th17 and Treg generation (Fig. 2A). Th17 cells develop when STAT3 is activated in the presence of TGFβ or IL-1 and a combination of IL-6 and IL-23,33–36 and a constitutively active STAT3 can promote IL-17 production in several culture conditions.6,7 In the absence of STAT3, IL-6 does not inhibit iTreg development.7,10,37 However, STAT3 is required for the IL-10-induced function of Tregs in controlling Th17-mediated inflammatory disease.38,39 Conversely, an active STAT5 promotes expression of the Treg transcription factor Foxp3, and in the absence of STAT5, IL-2 neither promotes Treg development nor inhibits Th17 development.40 At the level of cytokine regulation, STAT3 and STAT5 bind to overlapping sites at the Il17a/f loci but have opposing effects on transcription from the loci.41
Despite opposing functions of STAT3 and STAT5 when they are activated in the presence of TGFβ-induced signals, in the absence of TGFβ, and in the presence of IL-4 and STAT6 activation, STAT3 and STAT5 cooperate with STAT6 in promoting Th2 development (Fig. 2B). When STAT6 is activated, STAT3 demonstrates reduced binding to Il17a/f genes, and induces the expression of Th2, rather than Th17, transcription factors.25 Moreover, multiple cytokine signals decrease binding of STAT5 to the Foxp3 locus as they decrease iTreg generation.37 Thus, signals from multiple STATs may integrate at a genome-wide level of resolution by altering the binding of one STAT when more than one STAT is activated.
Importantly, binding of STAT3 in Th2 cells is not entirely dissimilar to binding in Th17 cells. Although binding to Il17 genes is decreased, there is overlap in the binding of STAT3 to genes that are required for both Th17 and Th2 cells, including Irf4, Maf and Batf.25 In support of this concept, we have recently shown that STAT3 is required for IL-21 production in multiple T helper subsets.42 Similar targeting of STATs among commonly expressed genes might also provide a basis for some of the flexibility of programming in Th subsets.43 For example, we have shown that polarized Th17 cells will repress IL-17 production and induce IL-4 production when cultured under conditions that promote Th2 development.44 Thus, plasticity might result from an ability to integrate multiple signals and establish epigenetic modifications at the relevant loci that are poised to respond to additional changes in the cytokine environment.45
Conclusions
STAT protein activation is the first response of a differentiating T cell to the cytokine milieu and represents a necessary step in establishing an effector phenotype. Although one STAT protein might be the predominant factor required for the development of each phenotype, additional STAT proteins also impact differentiation. The requirement for multiple signal inputs likely reflects a necessity for a complex inflammatory cytokine environment to allow the development of Th subsets that potently enhance the inflammatory process. The balance of signals, not only pro- versus anti-inflammatory, but also in the integration of multiple potentially opposing pro-inflammatory signals, leads to changes in the phenotype of the differentiating cell. Changes in the balance could result in modest or significant shifts in the type of inflammation that develops to allow the fine-tuning of the immune response required to mediate pathogen immunity, but avoid damaging inflammatory disease.
Acknowledgments
We thank members of the Kaplan lab for comments. The preparation of this article was aided by PHS grant U19AI070448. G.L.S. was supported by T32 AI060519.
Abbreviations
- IL
interleukin
- STAT
signal transducer and activator of transcription
- Th
T helper
References
- 1.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 2.Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, et al. Requirement for Stat4 in interleukin-12 mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for the development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 4.Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, et al. Lack of IL-4 induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 5.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura SI, et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 6.Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, O'Malley JT, et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178:4901–4907. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- 7.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 8.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 9.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 10.Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elo LL, Jarvenpaa H, Tuomela S, Raghav S, Ahlfors H, Laurila K, et al. Genome-wide profiling of interleukin-4 and STAT6 transcription factor regulation of human Th2 cell programming. Immunity. 2010;32:852–862. doi: 10.1016/j.immuni.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Good SR, Thieu VT, Mathur AN, Yu Q, Stritesky GL, Yeh N, et al. Temporal induction pattern of STAT4 target genes defines potential for Th1 lineage-specific programming. J Immunol. 2009;183:3839–3847. doi: 10.4049/jimmunol.0901411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 16.Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 17.Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, et al. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci USA. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulz EG, Mariani L, Radbruch A, Hofer T. Sequential polarization and imprinting of type 1 T helper lymphocytes by interferon-gamma and interleukin-12. Immunity. 2009;30:673–683. doi: 10.1016/j.immuni.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, et al. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity. 2008;29:679–690. doi: 10.1016/j.immuni.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–748. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- 21.Takatori H, Nakajima H, Hirose K, Kagami S, Tamachi T, Suto A, et al. Indispensable role of Stat5a in Stat6-independent Th2 cell differentiation and allergic airway inflammation. J Immunol. 2005;174:3734–3740. doi: 10.4049/jimmunol.174.6.3734. [DOI] [PubMed] [Google Scholar]
- 22.Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, et al. Interleukin 2 plays a central role in Th2 differentiation. Proc Natl Acad Sci USA. 2004;101:3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rincon M, Anguita J, Nakamura T, Fikrig E, Flavell RA. Interleukin (IL)-6 directs the differentiation of IL-4-producing CD4+ T cells. J Exp Med. 1997;185:461–469. doi: 10.1084/jem.185.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wurster AL, Rodgers VL, Satoskar AR, Whitters MJ, Young DA, Collins M, et al. Interleukin 21 is a T helper (Th) cell 2 cytokine that specifically inhibits the differentiation of naive Th cells into interferon gamma-producing Th1 cells. J Exp Med. 2002;196:969–977. doi: 10.1084/jem.20020620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stritesky GL, Muthukrishnan R, Sehra S, Goswami R, Pham D, Travers J, et al. The transcription factor STAT3 is required for Th2 cell development. Immunity. 2011;34:39–49. doi: 10.1016/j.immuni.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bream JH, Hodge DL, Gonsky R, Spolski R, Leonard WJ, Krebs S, et al. A distal region in the interferon-gamma gene is a site of epigenetic remodeling and transcriptional regulation by interleukin-2. J Biol Chem. 2004;279:41249–41257. doi: 10.1074/jbc.M401168200. [DOI] [PubMed] [Google Scholar]
- 27.Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011;12:551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, et al. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat Immunol. 2008;9:1288–1296. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akaishi H, Takeda K, Kaisho T, Shineha R, Satomi S, Takeda J, et al. Defective IL-2-mediated IL-2 receptor alpha chain expression in Stat3-deficient T lymphocytes. Int Immunol. 1998;10:1747–1751. doi: 10.1093/intimm/10.11.1747. [DOI] [PubMed] [Google Scholar]
- 30.Goenka S, Kaplan MH. Transcriptional regulation by STAT6. Immunol Res. 2011;50:87–96. doi: 10.1007/s12026-011-8205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diehl S, Anguita J, Hoffmeyer A, Zapton T, Ihle JN, Fikrig E, et al. Inhibition of Th1 differentiation by IL-6 is mediated by SOCS1. Immunity. 2000;13:805–815. doi: 10.1016/s1074-7613(00)00078-9. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Ochando J, Yopp A, Bromberg JS, Ding Y. IL-6 plays a unique role in initiating c-Maf expression during early stage of CD4 T cell activation. J Immunol. 2005;174:2720–2729. doi: 10.4049/jimmunol.174.5.2720. [DOI] [PubMed] [Google Scholar]
- 33.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 34.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, et al. Generation of pathogenic T(H)17 cells in the absence of TGFbeta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 36.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 37.O'Malley JT, Sehra S, Thieu VT, Yu Q, Chang HC, Stritesky GL, et al. Signal transducer and activator of transcription 4 limits the development of adaptive regulatory T cells. Immunology. 2009;127:587–595. doi: 10.1111/j.1365-2567.2008.03037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich JM, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaplan MH, Glosson NL, Stritesky GL, Yeh N, Kinzfogl J, Rohrabaugh SL, et al. STAT3-dependent IL-21 production from T helper cells regulates hematopoietic progenitor cell homeostasis. Blood. 2011;117:6198–6201. doi: 10.1182/blood-2011-02-334367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Stritesky GL, Yeh N, Kaplan MH. IL-23 mediates stability but not commitment to the Th17 lineage. J Immunol. 2008;181:5948–5955. doi: 10.4049/jimmunol.181.9.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]