Abstract
The presence of tumor-infiltrating lymphocytes (TILs) in epithelial ovarian cancer indicates a host antitumor response and is associated with improved survival. We wished to determine the extent to which TIL density differs from site to site within a given patient. We initially studied multiple paired metastases from serous ovarian carcinoma obtained at the time of primary debulking. The expression of genes in specific immune-related pathways was profiled on a pilot set of five patients. We then used immunohistochemistry and quantitative PCR to estimate the density of CD3+, CD8+ and Foxp3+ TILs in these same tumors. To extend the findings to a larger cohort, we semiquantitatively measured intraepithelial and stromal TILs in a tissue microarray (TMA) containing both primary tumors and metastases from 50 patients. In the pilot group, genes related to antimicrobial signaling and TGFβ signaling showed between-site heterogeneity, whereas cytokines and antigen presentation transcripts were more homogeneous in any given patient. IHC and qPCR for T-cell markers were concordant. In the TMA cohort, two-way ANOVA showed that TIL heterogeneity between sites was present in some but not all patients. The stroma of extra-ovarian metastases showed significantly greater TIL infiltration than ovarian sites. A simulation showed that at clinically meaningful levels of precision, up to 3% of patients will be misclassified for intraepithelial TILs by a single biopsy. In conclusion, between-site heterogeneity exists in some patients with metastatic serous ovarian cancer. The predictive value of biopsies should be considered in clinical trial design.
Key words: ovarian neoplasms, lymphocytes, tumor-infiltrating, adoptive immunotherapy, metastasis, microarray analysis
Introduction
Tumor immunotherapy is an emerging therapeutic modality that aims to augment the patient's host antitumor response through personalized cancer vaccines, adoptive transfer of lymphocytes and/or immunomodulatory therapy. Positive results have been reported in phase II and III trials for prostate cancer,1 low-grade non-Hodgkin lymphoma,2 melanoma,3–5 breast cancer6 and other tumors. Critical to the success of immunotherapy is the ability of effector T cells to infiltrate the tumor microenvironment. A variety of factors at the tumor microenvironment control T-cell homing, engraftment and function. Significant effort has therefore been directed to understanding the factors that contribute to this microenvironment, including cellular constituents and soluble factors that recruit T-cell subsets and modulate their activity.
Clinical studies testing novel tumor immune therapies have relied to date upon blood and serum assays to monitor the biological effects of immune therapy in vivo and interpret disease response. These assays include cytokine profiling, analysis of peripheral blood lymphocyte phenotype and specificity, and ex vivo activation assays on peripheral lymphocytes.7 However, these assays are poorly informative about events occurring in the patient's tumor, because antitumor immune response in periphery may be disconnected from that in tumors, due to factors acting specifically at the tumor microenvironment. Being able to monitor the microenvironment of tumors during immune therapy is a powerful tool to gain knowledge on the biological effects of immune therapies, understand the impact of immunomodulation therapy and interpret clinical results. Furthermore, tissue sampling of the tumor can be used as an inclusion criterion or as a means to identify biomarkers that predict response. In a companion article, we show evidence that CT-compatible core needle samples of ovarian tumor tissue are a suitable and reliable method for interrogating the tumor microenvironment. These studies have made the implicit assumption that a single tumor site is a valid proxy for the patient's overall disease. However, tumors with multiple metastatic deposits represent a unique challenge with respect to tissue based immune monitoring. It is in this case unclear whether the characteristics of one metastatic deposit will be representative of distant lesions.
Ovarian cancer is the most lethal gynecologic cancer, with 21,880 new cases and 13,850 deaths estimated to occur in the US in 2010, and is a well-established model system for the role of the immune system in cancer. Because of ovarian cancer's relatively nonspecific symptoms, it often presents at an advanced stage with disseminated peritoneal disease. Optimal resection is strongly predictive of survival, but even optimally debulked patients have some degree of residual peritoneal metastases. Complete response to chemotherapy is quite common, but recurrence is the norm. These pathologic features make immunotherapy especially appealing in ovarian cancer. The density of intraepithelial lymphocytic tumor infiltration is associated with survival in ovarian cancer,8–11 which was confirmed by multiple independent studies.12–17 Spontaneous antitumor immunity, in the form of tumor-reactive T cells and antibodies, has been detected in peripheral blood of patients with advanced stage disease at diagnosis.18,19 Furthermore, pilot clinical data indicate that ovarian cancer patients respond to interleukin-2 (IL-2),20,21 CTLA-4 antibody22,23 and adoptive transfer of ex vivo expanded TIL.12,13 Because of the multiple peritoneal nodules commonly seen, ovarian cancer is a suitable tumor to study the level of heterogeneity among metastatic deposits. In particular, it is important to understand whether multiple tumor sites within a given patient have different immunologic microenvironments, as assessed by lymphocytic infiltration. This question is especially important in case tumor biopsies must be used to monitor or interpret therapy. The magnitude of this between-site heterogeneity has not been well quantified in ovarian cancer. We report here a series of experiments addressing these questions. Using expression profiling we demonstrated that immune-related gene expression differed from site to site within a patient, then using quantitative PCR and immunohistochemistry we investigated between-site heterogeneity in lymphocytic infiltration in some patients and modeled the significance of this heterogeneity for clinical trial design.
Results
Between-site heterogeneity in immune gene expression profiles.
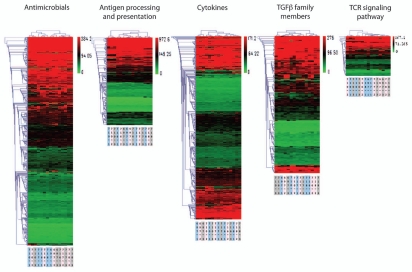
To determine whether the immune microenvironment of tumors is highly overlapping or rather heterogeneous among different metastatic sites within a given patient, we performed gene expression analysis using Affymetrix arrays and examined genes that are part of immune-related pathways. We focused on three tumor sites (the primary and two intraperitoneal metastases) sampled from each of five different patients as a pilot study. Genes pertaining to five immune-related pathways, as categorized by the ImmPort immunology bioinformatics project (25), were included, encompassing the T-cell receptor (TCR) signaling pathway (which captures T-cell-related signatures); antigen processing and presentation (which captures dendritic cell/monocyte-related signatures); TGFβ family members (which captures signatures of wound healing and immune tolerance pathways); antimicrobials (which represents a mixture of inflammation and innate immunity) and cytokines.
We used unsupervised clustering of gene expression profiles as a means of interrogating samples for molecular similarities. When profiles were clustered (Fig. 1), we found that the three tumor sites from each subject tended to resemble each other more than they resembled the profiles from other subjects' tumors, but this tendency varied among pathways. The degree of between-site similarity was quantitated for each subject's tumors by calculating the cophenetic distance between the subject's three tumor sites (Table S1). The mean between-site distance was smallest for genes related to cytokines (mean distance 2.0) and antigen processing and presentation (mean 2.4), indicating that the tumor sites from a given patient differed least in these profiles. In contrast, the genes related to antimicrobial responses (mean 3.6) and TGFβ signaling (mean 3.0) were most divergent between tumor sites within a given patient. The TCR signaling gene set showed an intermediate degree of variability between tumor sites (mean 2.8). The patients varied in their mean between-site cophenetic distance averaged across all gene sets (range 2.4–3.2), indicating that they differed in their overall tendency toward transcriptional diversity at different sites. Overall, the immune signatures of different tumor sites within a given patient were more similar to each other than to those from other patients, especially the cytokine and antigen processing and presentation signatures, but some heterogeneity was noted.
Figure 1.
Gene expression profiles within five functionally defined sets of genes, measured for three tumor samples harvested from each of five patients (15 samples total). Heat maps indicate above-average (green) or below-average (red) expression of specific genes. Profiles were subjected to hierarchical clustering, as captured by the dendrograms at top. Titles at bottom indicate the patient ID (three-digit code) and tumor site (single digit: 1, 2 or 3).
Between-site heterogeneity in T-lymphocyte infiltration in a pilot group.
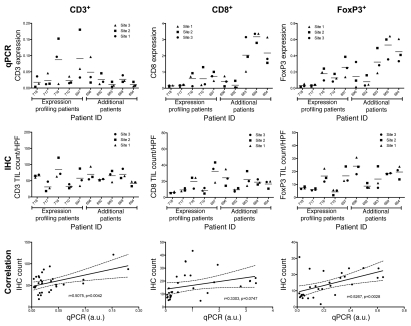
In the companion manuscript (Hagemann et al.) we show that there is little variation in T-cell infiltrate among different areas within the same tumor deposit. The results of the above gene expression analysis, however, suggest that in some patients there is variation in the immune milieu across different metastatic deposits within a given patient, especially those representing T-cell and tolerance pathways. We thus hypothesized that such molecular heterogeneity seen among tumor sites within subjects might be correlated with differences in T-cell infiltrates. Because in some patients tumor biopsies may yield tissue that is not suitable for IHC but could still be used for quantitative real-time PCR (qPCR), we first measured by qPCR T-cell markers differentiation markers CD3ε, CD8A and FoxP3 in the same RNA samples used to develop the gene expression data (Fig. 2, top). These markers are specifically expressed by and reflect the density of total CD3+ T cells, CD8+ T cells and FoxP3+ T regulatory cells, respectively. Data were normalized to β-actin as a loading control. In addition to the above 15 tumor samples from five subjects, we analyzed three tumor sites (primary and two intraperitoneal metastases) per subject from five additional subjects. Two-way ANOVA for qPCR measurement of lymphocyte markers showed that patients differed significantly from one another in their mean CD8A (p < 0.0001) and FoxP3 (p = 0.0001) expression, but did not differ significantly for CD3ε. There was no main effect of site in the ANOVA (p > 0.26 for all three markers). We noted that in some patients, the three sampled sites were very similar to one another, while in other patients the sites showed heterogeneous expression of lymphocytic markers. Importantly, there was a positive correlation between CD8A and FoxP3 genes (two-tailed Pearson r = 0.85, p < 0.0001), but not between CD3ε and CD8A, nor between CD3ε and FoxP3. Presumably due to the small sample size and the complexity of the underlying biology, there was no specific relationship between heterogeneity in gene expression profiles shown in Figure 1 and heterogeneity in lymphocytic markers.
Figure 2.
qPCR and immunohistochemical assessment of T-lymphocyte markers in a pilot set of patients. (Top) expression of T-lymphocyte markers CD3 (total T lymphocyte), CD8 (cytotoxic T lymphocyte) and FoxP3 (T regulatory lymphocyte) in tumor from three different metastatic sites within each of ten patients, as determined by qPCR. Data points represent mean of triplicate experiments. Lines indicate the grand mean for each patient. (Middle) Density of CD3+, CD8+ and Foxp3+ lymphocytes in tumor as determined by immunohistochemistry and visual scoring. (Bottom) Correlation between qPCR and IHC assessment of the immunologic microenvironment. r indicates the Pearson correlation coefficient. Dashed lines indicate the 95% confidence band of the best-fit line; a.u., arbitrary units.
Next, we examined whether qPCR analysis of T-cell subsets through differentiation markers accurately captured T-cell infiltration. We compared qPCR data with immunohistochemical assessment of CD3+, CD8+ and FoxP3+ cells (Fig. 2, middle and bottom). Tumor tissue from these ten subjects was subjected to immunoperoxidase staining for CD3, CD8 and FoxP3. CD3 was used as a pan-T-cell marker, CD8 was used as a marker for cytotoxic T cells and FoxP3 was used as a highly sensitive and specific marker for regulatory T cells.24 The average number of positive cells per high power field was determined by light microscopy (Fig. 2, middle). We found a strong correlation between qPCR and IHC quantification of total CD3+ T cells (two-tailed Pearson r = 0.51, p = 0.0042) and FoxP3 (r = 0.53, p = 0.0028), and a measurable but not statistically significant correlation between qPCR and IHC for CD8+ (r = 0.33, p = 0.0747) (Fig. 2, bottom). Similar to the qPCR data, there was no specific correlation between heterogeneity in histologically defined lymphocyte infiltration and heterogeneity in gene expression profiles. Finally, we found a positive correlation between IHC-quantified CD3+ and CD8+ cells (two-tailed Pearson r = 0.56, p = 0.0014); CD3+ and FoxP3+ cells (r = 0.51, p = 0.0036); and CD8+ and FoxP3+ cells (r = 0.75, p < 0.0001). Therefore, qPCR analysis or immunohistochemical assessment of T-cell subsets provide overlapping information and either one could be used to capture the adaptive cellular immune response in ovarian cancer. However, for the purpose of immune monitoring, these data collectively suggest that sampling of one single tumor deposit may not necessarily provide complete information on the cellular and transcriptional milieu of the overall tumor in some patients.
Between-site heterogeneity in T-lymphocyte infiltration in a larger patient cohort.
To more systematically determine the prevalence, extent and significance of TIL infiltration and between-site TIL heterogeneity, we constructed a tissue microarray containing multiple primary and metastatic samples from 50 serous ovarian cancer cases. Paraffin sections of the microarray were stained for T cells by immunohistochemistry using CD3, CD8 and FoxP3 antibodies.
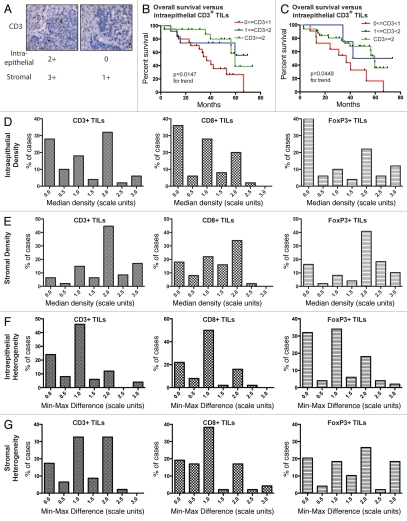
The density of intraepithelial and stromal tumor-infiltrating lymphocytes (TILs) was visually scored on a semiquantitative scale (Fig. 3A). As expected, we found that some tumors were densely infiltrated by TILs while others showed sparse TILs (Fig. 3A), and this was true across all three lymphocytic subsets.
Figure 3.
Immunohistochemical assessment of T-lymphocyte density and heterogeneity in a set of 50 patients. (A) Representative CD3-stained histocores with corresponding scores for intraepithelial and stromal lymphocytes. (B) Frequency distribution of the 50 cases according to the median density of intraepithelial lymphocytes in each of the three subsets examined (CD3+, CD8+, Foxp3+), averaged across all tumor sites. (C) Frequency distribution of the 50 cases according to median density of stromal lymphocytes. (D) Frequency distribution of the 50 cases according to the heterogeneity of their intraepithelial lymphocyte infiltration, assessed as the difference between the maximal and minimal TIL score for each patient's tumors. (E) Frequency distribution of the 50 cases according to the heterogeneity of their stromal lymphocyte infiltration. (F) Overall survival of patients stratified according to median intraepithelial CD3+ TIL density. (G) Overall survival of patients stratified according to stromal CD3+ TIL density.
Lymphocytic infiltration has been well documented to be a prognostic factor for survival in serous ovarian cancer.8 To test whether the patient cohort under examination was representative of ovarian cancer patients with respect to TIL density and clinical outcome, we performed survival analysis on the TMA population stratified into three groups according to median CD3+ TIL density (Fig. 3B and C). Intraepithelial TILs were positively associated with survival in this population (p = 0.0147) by log-rank test for trend. Thus, this population is representative of ovarian cancer patients as presented by numerous prior publications.8–17 In addition, stroma TILs were positively associated with survival in this population (p = 0.0448 by log-rank test).
Considering first only intraepithelial TILs (Fig. 3D) and averaging together all tumor sites for each patient, we found that very low TIL density (median score of 0) was relatively common, occurring in 28–40% of patients for all three subsets studied (CD3+, CD8+ and FoxP3+). In contrast, for stromal TILs, again averaged on a per-patient basis, absence of TILs in any given subset (CD3+, CD8+ and FoxP3+) was relatively uncommon, occurring in 6–18% of patients (Fig. 3E).
We next asked whether the multiple tumor sites examined for each patient were similar in TIL density or, on the contrary, showed between-site heterogeneity. As a measure of between-site heterogeneity, we used the numerical difference between the patient's highest and lowest TIL score (min-max difference). The majority of patients did show some between-site heterogeneity, as evidenced by a nonzero min-max difference (Fig. 3F and G); this was seen for all three lymphocyte subsets and for both intraepithelial and stromal TILs. For intraepithelial TILs, the degree of heterogeneity observed was relatively small: a min-max difference exceeding 1.0 scale unit was seen in only 22–30% of patients (Fig. 3F). On the whole, stromal TILs were more heterogeneous than intraepithelial TILs (Fig. 3G): a minmax distance exceeding 1.0 scale unit was still seen in 25–58% of patients.
Having demonstrated that variability exists in TIL density, we used two-way ANOVA to test whether this variability reflected consistent differences between patients, differences between tumor sites, or some combination of these factors, examining separately intraepithelial and stromal T cells (Table 1). The interaction term of the ANOVA was highly significant (p < 0.0001), indicating that some patients showed significant site-to-site intraepithelial TIL heterogeneity and some did not. These results confirmed the previous qPCR and IHC findings in the smaller cohort of patients. Thus, it appears that serous ovarian cancer varies from patient to patient in its propensity to show between-site heterogeneity in lymphocytic infiltration.
Table 1.
Two-way ANOVA analyses of intraepithelial and stromal CD3+ TIL density, performed in a cohort of 50 patients to test for heterogeneity among patients and between tumor sites
| CD3 | CD8 | FoxP3 | ||||
| Source of variation | % of total variation | p | % of total variation | p | % of total variation | p |
| Factors associated with density of intraepithelial TILs | ||||||
| Site | 0.17 | 0.3802 | 0.16 | 0.3407 | 0.49 | <0.0001 |
| Patient | 52.90 | <0.0001 | 54.86 | <0.0001 | 55.27 | 0.0892 |
| Interaction | 27.62 | <0.0001 | 28.67 | <0.0001 | 19.24 | <0.0001 |
| (Residual) | 19.31 | 16.31 | 25.00 | |||
| Factors associated with density of stromal TILs | ||||||
| Site | 1.84 | <0.0001 | 2.88 | <0.0001 | 2.30 | <0.0001 |
| Patient | 49.61 | <0.0001 | 42.50 | <0.0001 | 43.57 | <0.0001 |
| Interaction | 29.61 | <0.0001 | 33.89 | <0.0001 | 27.61 | <0.0001 |
| (Residual) | 18.94 | 23.61 | 26.52 | |||
Six separate analyses were run: one for each of CD3, CD8 and FoxP3, for both intraepithelial and stromal TILs.
Stromal TILs showed similar behavior to intraepithelial TILs in that there was a significant interaction between patient and site (both p < 0.0001; Table 1). Unlike intraepithelial TILs, stromal TILs appeared to be significantly associated with the tumor site (primary vs. metastasis). As shown in Table 2, there was no systematic difference between intraepithelial TILs in primary and metastatic tumors. However, CD3+, CD8+ and FoxP3+ cell density in the stroma of metastases was on average 0.41, 0.29 and 0.52 scale units higher than in primary ovarian tumor stroma (p = 0.0003, 0.0001 and 0.0003 respectively). Together, these data confirm that besides the previously described patient-to-patient variability in tumor lymphocytic infiltration, some degree of variability also exists between sites (primary vs. metastases) within a given patient. These findings are quite important for interpreting clinical experiments with translational endpoints based on tumor assessment.
Table 2.
Linear mixed effects modeling of TIL infiltration, performed on a cohort of 50 patients to estimate the magnitude of the difference between intraepithelial and stromal TIL infiltration at primary (ovarian) and metastatic sites
| Intraepithelial TILs | ||||||
| CD3 | CD8 | FoxP3 | ||||
| Site | Avg. Diff. (95% CI) | p value | Avg. diff. (95% CI) | p value | Avg. diff. (95% CI) | p value |
| Primaries (ovaries) | (ref) | (ref) | (ref) | (ref) | (ref) | (ref) |
| Metastases | −0.07 (−0.30, 0.17) | 0.57 | −0.09 (−0.29, 0.12) | 0.40 | 0.12 (−0.14, 0.17) | 0.37 |
| Significant global effects? | No, p = 0.30 | No, p = 0.35 | No, p = 0.32 | |||
| Stromal TILs | ||||||
| CD3 | CD8 | FoxP3 | ||||
| Site | Avg. Diff. (95% CI) | p value | Avg. diff. (95% CI) | p value | Avg. diff. (95% CI) | p value |
| Primaries (ovaries) | (ref) | (ref) | (ref) | (ref) | (ref) | (ref) |
| Metastases | 0.41 (0.21, 0.62) | 0.020 | 0.39 (0.20, 0.57) | 0.00002 | 0.22 (0.04, 0.47) | 0.086 |
| Significant global effects? | Yes, p = 0.0003 | Yes, p = 0.0001 | Yes, p = 0.0003 | |||
Next, we tested whether between-site heterogeneity in TIL density was associated with survival. The OS of patients with homogeneously high (n = 18) and with heterogeneous CD3+ TIL density (n = 11) was significantly longer than that of patients with homogeneously low CD3+ density (n = 21) (p = 0.0069; Fig. S1). Median survival was 59.6 mo for patients with homogeneously high CD3+ TILs and 59.5 mo for those with heterogeneous CD3+ TIL density, but only 34.1 mo for those with homogeneously low CD3+ TILs. Thus, having low CD3+ TILs at all sites was an adverse prognostic marker in this population. A similar analysis was performed for CD8+ and FoxP3+ TIL subsets, but no significant effect was found, most likely due to small sample size.
Estimating the predictive value of limited tumor sampling.
Accurately estimating T-cell subset infiltration through limited tumor sampling is an important task for clinical translational studies. The current paradigm in tumor vaccine therapy is that biopsy of a single site provides useful information about the patient's tumor as a whole. We tested this hypothesis by computing the predictive value of a small number of tumor biopsies for estimating the patient's overall TIL density as averaged across all sites. Tumor sampling was modeled using an “urn” model in which k biopsies are used to sample a patient with a total of n tumor deposits. Parameters of the model included k, the number of biopsies and ε, the maximum error that one is willing to make in estimating the median TIL density of the patient's tumor.
The simulation showed that for the purpose of predicting a patient's median intraepithelial CD3+ TIL density with an error not exceeding 0.5 scale units (ε = 0.5), a single biopsy (k = 1) is only 73% accurate (Table 3). Predictably, the performance characteristics of single biopsy improved as the value of ε was increased; if the median TIL density is only to be estimated within 1.0 scale units, then a single biopsy is 97% accurate. This value is adequate for the design of a clinical trial. One way to more accurately estimate the patient's median TIL density is to biopsy more than one site and average these estimates. Simulation showed that by biopsying two separate tumor deposits, the procedure becomes 92% accurate for predicting intraepithelial TIL density within ε = 0.5 and 98% for ε = 1.0. Similar findings arose when the simulation was repeated for stromal CD3+ TILs (Table 3) or for other lymphocyte subsets (data not shown).
Table 3.
Predictive value of single or multiple biopsies for estimating a patient's overall median TIL density
| Intraepithelial CD3+ TILs | ||
| Error tolerance of the estimate (ε) (scale units) | Accuracy of 1 biopsy | Accuracy of 2 biopsies |
| 0.5 | 73% | 92% |
| 1 | 97% | 98% |
| 1.5 | 398% | 100% |
| Mean error (absolute value, in scale units) ± SD | 0.34 ± 0.43 | 0.21 ± 0.32 |
| Stromal CD3+ TILs | ||
| Error tolerance of the estimate (ε) (scale units) | Accuracy of 1 biopsy | Accuracy of 2 biopsies |
| 0.5 | 69% | 84% |
| 1 | 91% | 99% |
| 1.5 | 96% | 100% |
| Mean error (absolute value, in scale units) ± SD | 0.47 ± 0.55 | 0.28 ± 0.38 |
The error tolerance (referred to as ε in the text) indicates the precision with which one wishes to estimate the actual TIL density, measured in scale units.
To present this information in another way, we computed the mean error that one would make if using a limited number of biopsies as a surrogate for a patient's overall TIL density, as determined by the urn model described above. As shown in Table 3, using a single biopsy to estimate intraepithelial CD3+ TILs results in an estimate that is on average different by 0.34 scale units from the patient's true TIL density as averaged across all tumor deposits. By averaging biopsies taken from any two tumor deposits, the mean error is lowered to 0.21 scale units. The results are similar for stromal TILs, except that the magnitude of the error is larger (0.47 and 0.28 scale units for one or two biopsies, respectively). Overall, the predictive value modeling indicates that a biopsy of a single tumor site will not always accurately describe the patient's overall tumor microenvironment, and the accuracy can be improved by sampling additional tumor sites. These are important findings that can guide the design of clinical trials requiring enrollment biopsy or longitudinal immune monitoring, as discussed below.
Discussion
During immune monitoring studies, a key question relates to the heterogeneity of therapeutic targets among multiple metastatic tumor sites in any given patient. This question has not been asked in ovarian cancer before, and studies of the immunologic response to ovarian cancer have generally examined a single disease site. We thus undertook a series of studies to assess whether tumor from different disease sites within a given patient showed different immune milieu.
Our companion manuscript (Hagemann et al.) shows that global gene expression profiles are highly overlapping among different sites within the same metastatic tumor deposit in any given patient, with correlation coefficients for within-tumor comparisons close to one (range 0.970–0.995), whereas between-patient correlations were weaker (range 0.888–0.922). Here we have used expression profiling and lymphocyte density counting to evaluate serous ovarian carcinoma deposits for between-site heterogeneity. A pilot investigation of genes related to immune pathways revealed some degree of within-patient heterogeneity across tumor sites, especially in genes related to TGFβ signaling and antimicrobial response, while expression of genes related to cytokines and antigen processing/presentation pathways were relatively constant. In addition, in a pilot set of patients analyzed by qPCR and IHC, and a larger TMA-based cohort examined by IHC, we found that some patients were indeed heterogeneous with regard to TIL density (especially stroma TILs), while the majority (over 70–80%) showed between-site homogeneity, especially with regard to intraepithelial TILs. This is important, as intraepithelial TILs have been correlated with improved survival and are considered the most important immune biomarker, while stromal TILs have not in most studies.
The TMA cohort we analyzed in this study was representative of previous studies, as we found a clear correlation of increased intraepithelial TILs with improved survival as previously reported by others and us. Importantly, although there was between-site heterogeneity in some patients, the TMA cohort did not show any systematic difference in intraepithelial lymphocyte infiltration between primary and metastatic sites. In other words, although there was variability among sites, the variability between primary and metastatic sites did not reproducibly contribute to this overall variability. Heterogeneity was seen in all three lymphocytic subsets (CD3+, CD8+, FoxP3+) studied. This study of between-site heterogeneity in ovarian cancer raises the question of whether heterogeneity itself may be a prognostic factor. In survival analyses (not shown here), we did not find this to be the case. While homogeneously low intraepithelial TILs appeared to be an adverse marker as compared with homogeneously high or heterogeneous intraepithelial TILs, the homogeneously low state is highly confounded with the low-median intraepithelial TIL state. Thus, overall, there is some degree of heterogeneity of TILs across tumor sites within patients with ovarian cancer, but intraepithelial TILs are less affected than stroma TILs, and this heterogeneity does not impact significantly on disease outcome, at least in this cohort.
Between-site immunologic heterogeneity is likely to reflect factors that are both intrinsic and extrinsic to the tumor. Intrinsic differences between tumor deposits may arise due to incremental acquisition of mutations in the course of tumor evolution. Tumors of unifocal and probably monoclonal origin25 spread as polyclonal collections of related tumor deposits,26,27 and these genetically heterogeneous tumors may present distinct tumor antigens and thus support a different immunologic microenvironment. Additionally, it appears that multiple lesions sometimes represent multiple independent tumors as predicted by the “field carcinogenesis” model,28,29 with similar implications for the tumor immune microenvironment. Factors extrinsic to the tumor, but intrinsic to the site of involvement, may also play a role in shaping the specific immunologic microenvironment. These factors could include resident stroma cell populations, local vascular factors, the molecular composition of the local extracellular matrix, or the influence of nearby secondary lymphoid tissues, all of which could affect quality, trafficking and function of T cells in the local microenvironment of metastatic deposits. This molecular heterogeneity was quite evident in the expression profiling data presented here. If the immunologic milieu were homogeneous across deposits of a single tumor, one would expect expression profiles to cluster closely together, whereas our data show that expression profiles of a given tumor may sometimes be more closely related to deposits of another patient's tumor. It was interesting to observe in this small cohort of tumors that genes related to TGFβ family members and antimicrobial response showed the highest degree of within-site heterogeneity, while expression of genes related to cytokines and antigen processing and presentation pathways were relatively constant. TGFβ pathways capture signatures of immune tolerance as well as wound healing and stroma, while antimicrobials pathways represent a mixture of inflammation and innate immunity signatures. Increased heterogeneity in these pathways may reflect underlying differences in stroma biology and inflammatory response intrinsic to the various sites. A similar difference for example was observed for stromal TILs between primary and metastatic sites, again possibly reflecting different composition of stroma. It appears that ovarian stroma is relatively resistant to infiltration by TILs, as compared with the stroma at extra-ovarian sites of metastasis. In contrast, the malignant epithelial component of serous ovarian cancer was not more densely infiltrated by lymphocytes at extra-ovarian sites than in the ovary, suggesting that this density is an autonomous property of the tumor that is not significantly conditioned by the surrounding stroma. It is interesting to note that the antigen processing and presentation pathways, which capture dendritic cell and monocyte-related signatures; the cytokine signatures, which capture both tumor cell and immune cell signatures; and the T-cell receptor (TCR) signaling pathways, which capture T-cell signatures, were more homogeneous, which may reflect the relative homogeneity of intraepithelial T cells and/or tumor cells. Given that our study was only exploratory, this interesting molecular variability among different metastatic deposits warrants further investigation through systems biology approaches using larger cohorts.
Our data indicate that some patients demonstrate heterogeneity in TIL density between tumor deposits. This observation has implications for clinical trial design. First, in trials whose design involves longitudinal tissue sampling, an attempt should be made to biopsy the same tumor nodule at each time point. In our companion manuscript (Hagemann et al.) we show that multiple biopsies of a single tumor deposit are in fact quite comparable to one another. Thus longitudinal sampling of the same tumor deposit in the course of therapy should accurately reflect microenvironment changes within that specific deposit. Vice versa, sampling different tumor deposits could possibly lead to erroneous conclusions, given the underlying within-patient heterogeneity. We estimated the level of error based on our TMA data; for CD3+ TILs the average magnitude of error was estimated to be 0.34 scale units, as computed in Table 3. Our study also indicates that the relative density of T-cell infiltration (along with other genes of tumor microenvironment signatures) could be captured adequately by qPCR, if tissue is not sufficient or adequate for IHC. In fact, qPCR and IHC data correlated reasonably well and this could be sufficient particularly for longitudinal studies, for example requiring pre-, intra- and/or post-therapy tumor sampling.
Using a single biopsy may potentially be misleading when used for the purpose of stratifying patients or as an inclusion/exclusion criterion. The likelihood of a misleading event will depend on the specific criterion being assessed (e.g., intraepithelial CD3+ TIL density), the scoring rubric used to classify it (e.g., a semi-quantitative scale as used here), and the number of total sites to biopsy. For example, we observed several tumors for which one deposit was markedly different from the others (e.g., 3+ intraepithelial CD3+ TILs in one ovary and one metastatic site, but absent TILs in another site). Multiple biopsies will more accurately capture the patient's mean value, and will also more accurately depict the range for that patient. It is not entirely clear how large an error one should tolerate in determining TIL density or any other clinicopathologic variable. Usually, the assumption in clinical research is that such variables can be measured accurately without error. Based upon survival data (Fig. 3B and C), a difference in TIL density of 1 unit on the scale used here appears to be prognostically significant, particularly for tumors that are overall low in lymphocytic infiltration. Therefore a clinically useful value of ε may be somewhere between 0.5 and 1 scale units. At this level of ε, a protocol using a single biopsy to assess TILs will only misclassify 3% of patients for intraepithelial TILs and 9% for stromal TILs.
As immune or immunomodulatory therapies become progressively more effective, there is an increasing need to monitor events in the tumor microenvironment in addition to what can be measured in peripheral blood. For clinical trials that require measurement of TILs as an inclusion criterion or as one of the translational end-points, sample size calculations must take into account the above methodological sampling errors. Intraepithelial TILs are the most meaningful biomarker based on most prior studies; if one agreed that an error of ε ≤ 1.0 scale unit was tolerable, then only 3% of patients would be misclassified for TILs in a clinical study involving a single biopsy, which is probably acceptable.
In summary, in this study we examined the issue of within-patient between-site heterogeneity of antitumor immune response. Our companion manuscript shows that multiple biopsies of the same tumor nodule yield highly reproducible results, while here we show that for some patients, between-site heterogeneity could introduce some error in the estimation of TILs. Understanding the magnitude of this error is critical in trial design and interpretation of data related to the tumor immune microenvironment.
Materials and Methods
Prospective tissue collection.
Institutional Review Board approval was obtained. A biopsy was obtained intraoperatively from each of three metastatic EOC lesions in ten consecutive patients undergoing debulking surgery between October 2005 and May 2006 from a total of ten patients ranging in age from 39–73, median age of 58. Nine patients were undergoing primary surgery while one was undergoing secondary debulking for recurrent disease. On clinicopathologic grounds, two patients were considered to have primary peritoneal carcinoma involving the ovary; the others had serous ovarian cancer. All patients were stage IIIC. Only metastatic tumor was sampled, comprising omental and peritoneal metastases. Just prior to tumor collection, plastic cryomolds were filled half to three-quarters full with Optimal Cutting Temperature (OCT) Compound (Sakura Tissue-Tek, Torrance, CA) and placed on dry ice. Immediately upon removal from the patient, fresh tumor nodules 2–5 cm in diameter were subjected to open-air needle biopsy in the operating room. Each tumor was biopsied in up to three different areas at least 0.5 cm apart using a 16-gauge Quick-Core Biopsy Needle (Cook Medical, Bloomington, IN); for each area we obtained up to three needle cores. The tumor tissue cores were immediately transferred into the frozen cryomold prefilled with OCT, covered in OCT and stored at −80°C. A total of three specimens were collected for each patient.
Gene expression profiling.
Gene expression studies were performed on samples from five patients at the Cancer Biology Branch, National Cancer Institute, as previously described in reference 30. Two rounds of amplification were used as previously described. Briefly, during first round cDNA synthesis, 100 ng of total RNA was reverse transcribed using the Two-Cycle cDNA Synthesis Kit (Affymetrix, Santa Clara, CA) and oligo-dT24-T7 (5′-GGC CAG TGA ATT GTA ATA CGA CTC ACT ATA GGG AGG CGG-3′) primer according to the manufacturer's instructions. First round amplification used the T7 promoter coupled double stranded cDNA as template and the MEGAscript T7 Kit (Ambion, Inc., Austin, TX). Following cleanup of the cRNA with a GeneChip Sample Cleanup Module IVT column (Affymetrix), second round double stranded cDNA was generated and purified. After amplification and biotinylation with the IVT Labeling Kit (Affymetrix), a 15.0 mg aliquot of labeled product was fragmented by heat and ion-mediated hydrolysis and hybridized to human U133A 2.0 oligonucleotide GeneChip arrays (Affymetrix), which comprise over 500,000 unique oligonucleotide features covering more than 18,400 transcripts and variants on a single chip. The microarrays were subsequently stained for visualization in a Fluidics Station 450 and scanned using the laser confocal GeneChip Scanner 3000 (Affymetrix). Affymetrix array CEL files were processed in R using an algorithm that uses genes in the least variant set (LVS) to normalize the expression data.31
Quantitative PCR.
RNA was isolated from tumor tissue of ten patients using the TRIzol Reagent (Invitrogen) and reconstituted in RNase-free water. RNA quantity and quality were measured using the Agilent 2100 Bioanalyzer. RNA samples were treated with DNase I (Invitrogen) and cDNA was transcribed for each sample using Superscript II First Strand Synthesis Kit for RT-PCR (Invitrogen). Experiments with real-time quantitative (q)PCR were performed with the use of the ABI Prism 7900 Analyzer and SYBR Green PCR kits (Applied Biosystems). cDNA levels were normalized against β-actin.
Tissue microarray construction and staining.
We identified 50 patients with metastatic ovarian cancer (FIGO stage IIC and above) undergoing primary resection at our institution between 2005 and 2008 (Table 1). All cases were invasive carcinomas; borderline tumors were excluded. Inclusion criteria included papillary serous histology or poorly differentiated histology with some serous features, clinicopathologic findings consistent with ovarian primary and availability of paraffin blocks representing at least three tumor sites including one ovarian primary site. Mean age was 59.5 y (SD, 11.7); FIGO stage was IIC for three patients, IIIB for two patients, IIIC for 40 patients and IV for six patients. Three patients had moderately differentiated histology, the remainder being poorly differentiated. Of the patients, 33/50 = 66% were optimally debulked and 17/50 = 34% suboptimally debulked.
H&E-stained slides were reviewed and annotated and paraffin-embedded tissue blocks were used to construct a tissue micro-array of primary and metastatic tumors. Including primary sites and metastases, a total of 206 tumor sites were represented on the array. A mean of 3.8 sites were included per patient, including one to two primary (ovarian) sites and one to seven metastatic sites. Eighty-nine ovarian deposits were included. The most common metastatic sites included omentum (41 deposits), peritoneum (e.g., cul-de-sac) (21 deposits), uterine serosa (21 deposits) and bowel wall (18 deposits). Previously frozen samples (frozen section remnants) were permitted, but avoided when possible. Lymph node metastases represented a small number (seven) of the deposits. For each tumor site represented on the array, triplicate 0.6 mm cores of tumor were taken from a single archival paraffin block and placed on a tissue microarray using a manual arrayer. The resulting tissue microarray spanned three paraffin blocks, each containing up to 20 × 14 tissue cores including cases and controls.
Paraffin sections of the arrays were cut at 5 µm thickness and stained with hematoxylin-eosin or by immunohistochemistry using 3,3′-diaminobenzidine as chromogen and hematoxylin counterstain, according to standard protocols in our laboratory. Immunostains were performed for the immune markers CD3 (pan T lymphocyte), CD8 (cytotoxic T lymphocyte) and FoxP3 (regulatory T lymphocyte). CD3 (rabbit polyclonal, A0452, Dako Cytomation) and CD8 (clone C8/144B, M7103, Dako Cytomation) were used at 1:250 and 1:100 respectively after antigen retrieval by boiling in 1x citrate buffer for 15 min. FoxP3 (clone 206D, 320102, Biolegend) was used at 1:250 after antigen retrieval at pH 9–10 in a pressure cooker at 15 psi for 2 min.
Scoring of tumor-infiltrating lymphocytes in tissue microarray sections.
The immunostained microarrays were scored for tumor-infiltrating lymphocytes by a pathologist (I.S.H.) examining the tissue by light microscopy at 400x magnification. Intraepithelial lymphocytes (i.e., lymphocytes infiltrating the malignant epithelial compartment of the histospot) and stromal lymphocytes (all other lymphocytes in the histospot) were graded according to the following quantitative criteria: 0, absent TILs; 1, rare TILs [1–10/400x high-power field (hpf)]; 2, moderate TILs (11–20/hpf); 3, numerous TILs (>20/hpf). When multiple fields were available for review, the median of their scores was recorded for that core. The lymphocyte density in partial/incomplete fields was normalized based on a visual assessment of the percentage that consisted of tumor epithelium or stroma. Tumors for which fewer than two good-quality cores were present were considered unevaluable.
Biocomputational and statistical analyses.
Affymetrix data analysis. We analyzed expression of genes falling into five immune-related pathways as categorized by the ImmPort immunology bioinformatics project.32 Gene expression profiles were processed using unsupervised hierarchical clustering using genes in the least variant set (LVS) to normalize the expression data.31 The cophenetic spanning distance for any given patient and gene set was defined as the minimal dendrogram height at which the patient's three tumor sites are connected.33 Nearest neighbors have a distance of 1 and the smallest possible distance that can encompass a set of three samples is 2. These distances were manually computed for each for each subject and gene set.
Tissue microarray data analysis. For the heterogeneity analysis, patients with TIL counts for at least three tumor sites were included (40 patients for the CD3 analysis, 41 for CD8 and 42 for FoxP3) and a two-way ANOVA analysis (Prism 5, GraphPad Software) was performed on the first three tumor sites according to the order in which they were placed on the array, with site one always representing a primary site. To test between-site differences, linear regression with a mixed effects model was used (Stata 11, Stata Corp.), using the site (primary vs. metastasis) as a fixed effect and the patient as a random effect. This method controls for between-patient differences and allows the effect of tumor site to be independently determined. The average difference between sites was estimated. Between-patient variation and clustered effects between repeated measures from the same site were captured via random effects (random intercepts and/or random slopes). The need for a specific random effect was tested via likelihood ratio test.
Predictive value modeling. The predictive value of single or multiple tumor biopsies was modeled using an “urn model” simulation constructed in Microsoft Excel 2008. To simulate the act of biopsying a given patient, a specified number of biopsies were drawn at random from a simulated urn containing all of that patient's tumor sites and the resulting estimate of the patient's CD3+ TIL density was compared with the actual median of all of that patient's tumor sites. This simulation was iterated multiple times over the entire set of TMA patients to reach a total of 300 experiments.
Clinical outcomes. Overall survival (OS) was defined as the time from diagnosis to death from any cause, as determined from clinical and public records. Patients for whom death was not documented were censored at the last clinical encounter. The median follow-up period was 40 mo (range 3–74 mo). Kaplan-Meier curves were drawn and log-rank test was performed using Prism 5 (GraphPad Software).
Acknowledgments
We thank Li-Ping Wang and Emma Hung for outstanding technical support with TMA construction and immunostaining. This work was supported by Ovarian Cancer SPORE NIH P50-CA083638, the Ovarian Cancer Research Foundation and a core grant (NCI P30 CA016520-34) to the Abramson Cancer Center of the University of Pennsylvania.
Supplementary Material
References
- 1.Brill TH, Kubler HR, Pohla H, Buchner A, Fend F, Schuster T, et al. Therapeutic vaccination with an interleukin-2-interferon-gamma-secreting allogeneic tumor vaccine in patients with progressive castration-resistant prostate cancer: a phase I/II trial. Hum Gene Ther. 2009;20:1641–1651. doi: 10.1089/ hum.2009.101. [DOI] [PubMed] [Google Scholar]
- 2.Redfern CH, Guthrie TH, Bessudo A, Densmore JJ, Holman PR, Janakiraman N, et al. Phase II trial of idiotype vaccination in previously treated patients with indolent non-Hodgkin's lymphoma resulting in durable clinical responses. J Clin Oncol. 2006;24:3107–3112. doi: 10.1200/JCO.2005.04.4289. [DOI] [PubMed] [Google Scholar]
- 3.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat Rev Cancer. 2008;8:299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linette GP, Zhang D, Hodi FS, Jonasch EP, Longerich S, Stowell CP, et al. Immunization using autologous dendritic cells pulsed with the melanoma-associated antigen gp100-derived G280-9V peptide elicits CD8+ immunity. Clin Cancer Res. 2005;11:7692–7699. doi: 10.1158/1078-0432.CCR-051198. [DOI] [PubMed] [Google Scholar]
- 6.Czerniecki BJ, Koski GK, Koldovsky U, Xu S, Cohen PA, Mick R, et al. Targeting HER-2/neu in early breast cancer development using dendritic cells with staged interleukin-12 burst secretion. Cancer Res. 2007;67:1842–1852. doi: 10.1158/00085472.CAN-06-4038. [DOI] [PubMed] [Google Scholar]
- 7.Clay TM, Hobeika AC, Mosca PJ, Lyerly HK, Morse MA. Assays for monitoring cellular immune responses to active immunotherapy of cancer. Clin Cancer Res. 2001;7:1127–1135. [PubMed] [Google Scholar]
- 8.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 9.Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 10.Adams SF, Levine DA, Cadungog MG, Hammond R, Facciabene A, Olvera N, et al. Intraepithelial T cells and tumor proliferation: impact on the benefit from surgical cytoreduction in advanced serous ovarian cancer. Cancer. 2009;115:2891–2902. doi: 10.1002/cncr.24317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke B, Tinker AV, Lee CH, Subramanian S, van de Rijn M, Turbin D, et al. Intraepithelial T cells and prognosis in ovarian carcinoma: novel associations with stage, tumor type and BRCA1 loss. Mod Pathol. 2009;22:393–402. doi: 10.1038/modpathol.2008.191. [DOI] [PubMed] [Google Scholar]
- 12.Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci USA. 2007;104:3360–3365. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA. 2005;102:18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah CA, Allison KH, Garcia RL, Gray HJ, Goff BA, Swisher EM. Intratumoral T cells, tumor-associated macrophages and regulatory T cells: Association with p53 mutations, circulating tumor DNA and survival in women with ovarian cancer. Gynecol Oncol. 2008;109:215–219. doi: 10.1016/j.ygyno.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Tomsová M, Melichar B, Sedlakova I, Steiner I. Prognostic significance of CD3+ tumor-infiltrating lymphocytes in ovarian carcinoma. Gynecol Oncol. 2008;108:415–420. doi: 10.1016/j.ygyno.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Stumpf M, Hasenburg A, Riener MO, Jutting U, Wang C, Shen Y, et al. Intraepithelial CD8-positive T lymphocytes predict survival for patients with serous stage III ovarian carcinomas: relevance of clonal selection of T lymphocytes. Br J Cancer. 2009;101:1513–1521. doi: 10.1038/sj.bjc.6605274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milne K, Kobel M, Kalloger SE, Barnes RO, Gao D, Gilks CB, et al. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors. PLoS ONE. 2009;4:6412. doi: 10.1371/journal.pone.0006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlienger K, Chu CS, Woo EY, Rivers PM, Toll AJ, Hudson B, et al. TRANCE- and CD40 ligand-matured dendritic cells reveal MHC class I-restricted T cells specific for autologous tumor in late-stage ovarian cancer patients. Clin Cancer Res. 2003;9:1517–1527. [PubMed] [Google Scholar]
- 19.Goodell V, Salazar LG, Urban N, Drescher CW, Gray H, Swensen RE, et al. Antibody immunity to the p53 oncogenic protein is a prognostic indicator in ovarian cancer. J Clin Oncol. 2006;24:762–768. doi: 10.1200/JCO.2005.03.2813. [DOI] [PubMed] [Google Scholar]
- 20.Edwards RP, Gooding W, Lembersky BC, Colonello K, Hammond R, Paradise C, et al. Comparison of toxicity and survival following intraperitoneal recombinant interleukin-2 for persistent ovarian cancer after platinum: twenty-four-hour versus 7-day infusion. J Clin Oncol. 1997;15:3399–3407. doi: 10.1200/JCO.1997.15.11.3399. [DOI] [PubMed] [Google Scholar]
- 21.Vlad AM, Budiu RA, Lenzner DE, Wang Y, Thaller JA, Colonello K, et al. A phase II trial of intraperitoneal interleukin-2 in patients with platinum-resistant or platinum-refractory ovarian cancer. Cancer Immunol Immunother. 2010;59:293–301. doi: 10.1007/s00262-009-0750-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodi FS, Butler M, Oble DA, Seiden MV, Haluska FG, Kruse A, et al. Immunologic and clinical effects of antibody blockade of cytotoxic T lymphocyte-associated antigen 4 in previously vaccinated cancer patients. Proc Natl Acad Sci USA. 2008;105:3005–3010. doi: 10.1073/pnas.0712237105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodi FS, Mihm MC, Soiffer RJ, Haluska FG, Butler M, Seiden MV, et al. Biologic activity of cytotoxic T lymphocyte-associated antigen 4 antibody blockade in previously vaccinated metastatic melanoma and ovarian carcinoma patients. Proc Natl Acad Sci USA. 2003;100:4712–4717. doi: 10.1073/pnas.0830997100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramsdell F. FoxP3 and natural regulatory T cells: key to a cell lineage? Immunity. 2003;19:165–168. doi: 10.1016/S10747613(03)00207-3. [DOI] [PubMed] [Google Scholar]
- 25.Tsao SW, Mok CH, Knapp RC, Oike K, Muto MG, Welch WR, et al. Molecular genetic evidence of a unifocal origin for human serous ovarian carcinomas. Gynecol Oncol. 1993;48:5–10. doi: 10.1006/gyno.1993.1002. [DOI] [PubMed] [Google Scholar]
- 26.Khalique L, Ayhan A, Weale ME, Jacobs IJ, Ramus SJ, Gayther SA. Genetic intra-tumour heterogeneity in epithelial ovarian cancer and its implications for molecular diagnosis of tumours. J Pathol. 2007;211:286–295. doi: 10.1002/path.2112. [DOI] [PubMed] [Google Scholar]
- 27.Khalique L, Ayhan A, Whittaker JC, Singh N, Jacobs IJ, Gayther SA, et al. The clonal evolution of metastases from primary serous epithelial ovarian cancers. Int J Cancer. 2009;124:1579–1586. doi: 10.1002/ijc.24148. [DOI] [PubMed] [Google Scholar]
- 28.Buller RE, Skilling JS, Sood AK, Plaxe S, Baergen RN, Lager DJ. Field cancerization: why late “recurrent” ovarian cancer is not recurrent. Am J Obstet Gynecol. 1998;178:641–649. doi: 10.1016/S0002-9378(98)70473-9. [DOI] [PubMed] [Google Scholar]
- 29.Muto MG, Welch WR, Mok SC, Bandera CA, Fishbaugh PM, Tsao SW, et al. Evidence for a multifocal origin of papillary serous carcinoma of the peritoneum. Cancer Res. 1995;55:490–492. [PubMed] [Google Scholar]
- 30.Donninger H, Bonome T, Radonovich M, Pise-Masison CA, Brady J, Shih JH, et al. Whole genome expression profiling of advance stage papillary serous ovarian cancer reveals activated pathways. Oncogene. 2004;23:8065–8077. doi: 10.1038/ sj.onc.1207959. [DOI] [PubMed] [Google Scholar]
- 31.Calza S, Valentini D, Pawitan Y. Normalization of oligonucleotide arrays based on the least-variant set of genes. BMC Bioinformatics. 2008;9:140. doi: 10.1186/1471-2105-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.National Institute of Allergy and Infectious Diseases, author. ImmPort: Bioinformatics for the future of immunology. National Institute of Allergy and Infectious Diseases; Available at: www.immport.org. [Google Scholar]
- 33.Sokal RR, Rohlf FJ. The comparison of dendrograms by objective methods. Taxon. 1962;11:33–40. doi: 10.2307/1217208. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.