Abstract
Evolution results from molecular-level changes in an organism, thereby producing novel phenotypes and, eventually novel species. However, changes in a single gene can lead to significant changes in biomolecular networks through the gain and loss of many molecular interactions. Thus, significant insights into microbial evolution have been gained through the analysis and comparison of reconstructed metabolic networks. However, challenges remain from reconstruction incompleteness and the inability to experiment with evolution on the timescale necessary for new species to arise. Despite these challenges, experimental laboratory evolution of microbes has provided some insights into the cellular objectives underlying evolution, under the constraints of nutrient availability and the use of mechanisms that protect from extreme conditions.
Introduction
While evolution involves the change of genetic sequence over time, it more importantly changes molecular interactions. Therefore, evolution often leads to modifications in molecular-network topology and ultimately in the systems-level functions of an organism (i.e., phenotype).
Metabolism, a complex network of chemical reactions, is an ideal system to study network-level changes in evolution. First, it is the best characterized biomolecular network. The underlying genes and proteins are well-characterized, and their interactions and biochemistry have been studied comprehensively. Second, because metabolism is critical to all cellular functions, many core reactions have ancient origins and are conserved across all kingdoms of life[1]. While small differences exist in metabolic pathway topology within different strains of one species (e.g., in Pseudomonas syringae[2]), examination of conserved reactions across species nonetheless facilitates clear comparisons for evolutionary insight.
Selective pressures resulting from cellular objectives help guide the expansion and pruning of metabolic pathways through evolution (Figure 1). The cellular objectives underlying the selective pressure can involve maximizing the nutritional potential of the environment and surviving environmental stress. Here we review recent insights into metabolic pathway evolution, which have been gained from both computational and experimental approaches.
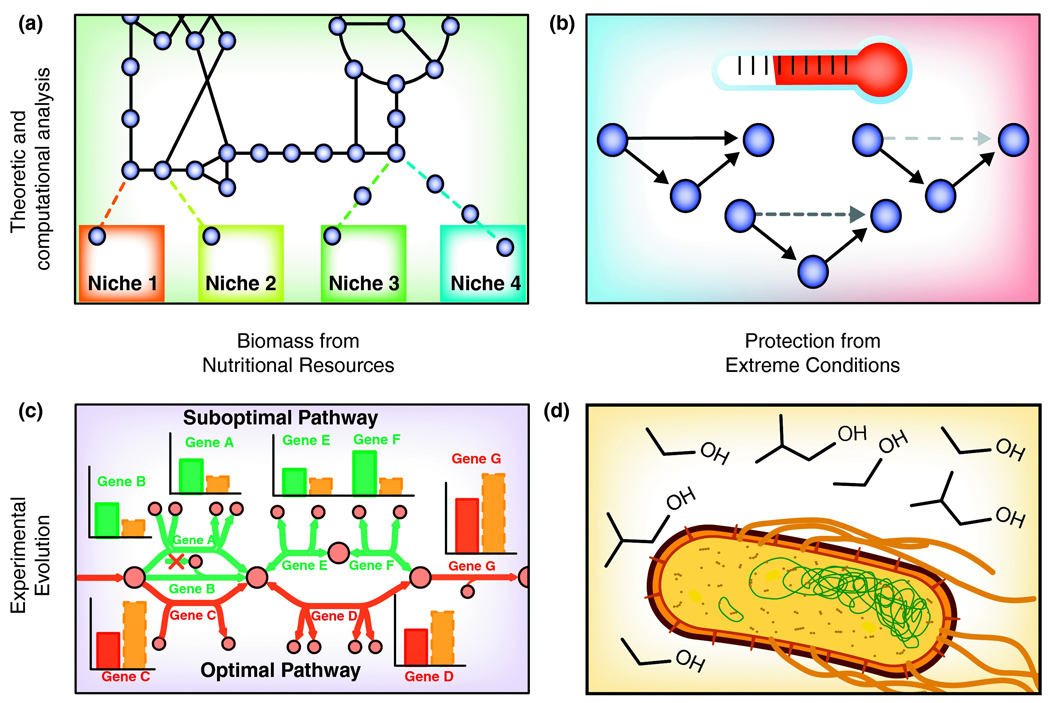
Figure 1. Cellular objectives and selective pressures guiding pathway evolution.
Theoretical (a,b) and experimental (c,d) approaches have been used to gain insight into how the interactions between cellular objectives and selective pressures guide the evolution of metabolic pathways. Specifically, these efforts have shown how (a) microbes gain and lose pathways with an objective to gain biomass under the pressure of limited nutritional resources, and that (c) the metabolic pathways used are consistent with the in silico-predicted optimal pathways. In addition, cells also have an indirect objective of self-protection against a selective pressure of stressful environmental conditions. Consistent with this, (b) adaptation to different temperatures correlates with topological changes in metabolic pathways, and (d) microbes evolve mechanisms to metabolize or protect against toxic environmental conditions.
Theoretical advances in cellular objectives and pathway evolution
Over the past decade, numerous studies have identified topological properties of metabolic networks that are shaped through evolution, such as their robustness, modularity, and scale-free organization[3]. Recently, computational studies of metabolic network evolution have extended beyond topology to assess the interaction of cellular objectives and selective pressures in pathway evolution. The selective pressures affecting bacterial populations can be divided into two categories: 1) nutrient availability and 2) environmental stress.
Nutrient availability guides the evolution of metabolic pathways, since the central role of metabolism is the conversion of small molecules to biomass and energy (Figure 1.a). For instance, catabolism is poorly conserved relative to anabolism because the presence or absence of specific pathways is governed by an organism’s habitat and the access to different nutrients[4]. Also, Escherichia coli central metabolism is structured as a minimal number of enzymatic steps between nutrients and biomass precursors[5], thereby achieving metabolic functionality with a minimal set of enzymes[6]. Since there seems to be a selection for minimal metabolic pathways given the environmental conditions, the accessible nutrients for a species may be inferred by analyzing the network topologies. The essential sets of compounds for 478 species have been computed through a phylogenic analysis of network topologies[7]. To account for the evolution of metabolic pathways according to environmental conditions, the toolbox model of evolution was proposed. This model suggests that prokaryotic genomes are shaped by horizontal gene transfer of co-regulated metabolic pathways and prompt removal of redundant genes based on gain and loss of environmental nutrients[8].
Living organisms can adapt to environments that impose extreme stresses, such as high temperatures and oxidative conditions (Figure 1.b). Adaptation to different temperatures has accompanied evolution since ancient forms of life endured high temperatures[9,10]. Interestingly, temperature changes correlate with structural differences in metabolic networks[11]. As the optimal growth temperature increases amongst 113 prokaryotes, the network edge density, clustering coefficient, and subgraph concentration all decrease. Also, a strong selective constraint at high temperature tends to inhibit the emergence of shortcut pathways[12]. Therefore, metabolic networks tend to transit from heterogeneous and highly-modular structures at lower temperatures to homogeneous structures with low modularity and fewer shortcuts in higher temperatures.
Experimental evolution in studying pathway evolution
Metabolic network analysis provides insight into microbial evolution and suggestions for the evolutionary objectives. However, all of these studies are retrospective analyses, in which traits within current species are compared to get insights into evolution. Since the species analyzed in retrospective studies often diverged millions of years ago, it is difficult to rigorously test these evolutionary theories. Fortunately, some theories may be experimentally tested through adaptive laboratory evolution (ALE). ALE has successfully demonstrated changes to the transcriptional regulatory network and the metabolic network topology. These changes provide further support that pathway evolution is shaped in part by the interaction between cellular objectives and selective pressures, such as biomass production subject to nutrient availability and cellular protection under harsh environmental conditions.
Pathways adapt to optimize metabolism subject to nutrient availability
Exponentially growing bacteria often have growth phenotypes (e.g., substrate uptake rate, secretion products, etc.) consistent with the predictions of flux balance analysis using genome-scale models of metabolism[13]. Strains that fail to grow according to model predictions can evolve toward the model-predicted optimal phenotypes, as demonstrated on sub-optimal media conditions with E. coli[14] and Lactobacillus plantarum[15] and with several E. coli mutants[16]. The improved growth rates and biomass yields for exponentially growing E. coli are accompanied by improved use of the model-predicted optimal metabolic pathways[17]. Through ALE, many essential genes and the optimal pathways are up-regulated, while genes and proteins associated with reactions that could not carry flux were all significantly down-regulated (Figure 1.c). Thus, in short-term evolution, we see a tendency to tune protein expression levels[18], to prune out pathways that are not necessary, and to enhance expression of essential metabolic pathways[17]. This optimization of pathway usage in ALE is often driven by regulatory genes mutations that improve fitness for that specific environment[19–22].
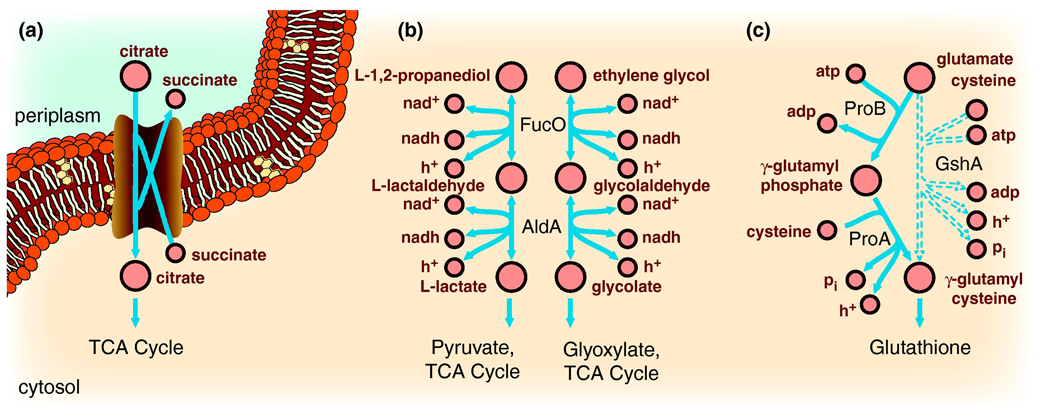
Moreover, metabolic pathways can be extended by to utilize novel nutrients. After ~33,127 generations of growth on glucose-poor/citrate-rich minimal media, E. coli evolved the capability to aerobically use citrate as its sole carbon source[23] (Figure 2.a). This capability does not solely result from the activation of a latent citrate transporter, since it depends on a genetic background containing additional mutations, obtained over thousands of generations. In another example, wild-type E. coli K12 evolved the ability to grow on L-1,2-propanediol (L-1,2-PDO) and 1,2-ethanediol (ethylene glycol). It evolved the ability to metabolize L-1,2-PDO when glycerol minimal media was slowly replaced with L-1,2-PDO minimal media[24]. Further mutagenesis allowed for the isolation of E. coli K12 strains that metabolize ethylene glycol[25]. These functions require the constitutive expression and mutation of fucO, and ethylene glycol metabolism required a mutation in lactaldehyde dehydrogenase (aldA) (Figure 2.b). Thus, the acquisition of a new metabolic function required mutations that induced expression of a latent pathway, and mutations that changed substrate specificity to catabolize the metabolites. Therefore, prokaryotes will evolve to a metabolic phenotype consistent with an efficient use of the metabolic network as constrained by nutrient availability.
Figure 2. Extension of metabolic pathways through laboratory evolution.
Laboratory evolution under a defined selective pressure has identified a few cases in which a new metabolic function arose. Three of these examples include the ability that E. coli gained to (a) transport citrate after 33,000 generations, (b) metabolize L-1,2-propanediol and ethylene glycol, and (c) synthesize glutathione when a key enzyme in its synthesis was deleted.
Pathways adapt to protect the microbe
The optimization of metabolic efficiency is seen in exponentially-growing cells. However, some enzymes that do not contribute to biomass are used to protect cells from harmful metabolites. For example, the product of the E. coli dut gene, which dephosphorylates dUTP, is predicted in silico to waste resources; however, in exponentially growing E. coli, the dut mRNA and protein are up-regulated following ALE[17]. This is because its metabolic role in vivo maintains genome integrity by precluding dUTP from integrating into the genome during replication, and its absence decreases the growth rate in E. coli[26].
The role of protection is further apparent when E. coli is evolved to grow with toxic levels of isobutanol or ethanol. In isobutanol, the evolved strain increases the expression of an enzyme needed for the synthesis of glucosamine-6-phosphate, which enhances cell membrane synthesis[27]. In ethanol, expression increases for genes related to glycine and betaine synthesis, which both serve as protective osmolites[28]. In another study, ALE on ethanol resulted in strains that concurrently metabolize ethanol, even when ample glucose is provided, thereby potentially reducing the ethanol concentration in the cellular microenvironment[29]. All of these metabolic changes improve fitness under toxic conditions, presumably by protecting the integrity of the genome, macromolecules, and membrane.
Furthermore, microbes can evolve novel metabolic functions to protect the cell through the mutation of extant enzymes[30]. Glutathione (GSH) is essential under stress-inducing conditions such as oxidizing conditions[31]. When gshA, a gene in the GSH synthesis pathway, was deleted from E. coli, the mutant strain was unable to produce GSH, thereby reducing its tolerance to AsO43−. The mutant strain was evolved, and mutations in the proline synthesis genes proB and proA restored AsO43− tolerance and GSH production. The proB mutation affected its metabolic regulation, while the proA mutation changed the regulatory and catalytic capabilities of the enzyme. Wild type ProA normally catalyzes the conversion of γ-glutamyl phosphate to glutamic acid 5-semialdehyde. However, ProA in the evolved strain instead metabolizes γ-glutamyl phosphate to form γ-glutamyl cysteine, a precursor to GSH. Together these mutations changed the functions of extant enzymes from proline biosynthesis to GSH synthesis (Figure 2.c).
Future directions
Computational and experimental approaches have increased insight into how cellular objectives and selective pressure guide pathway evolution. Two models by which these pathways are believed to evolve are the retrograde and patchwork hypotheses[32,33] (Figure 3). The arrival of novel metabolic functions from enzyme mutations in ALE (Figure 2) provides support for the patchwork evolution hypothesis, as novel pathways are built with enzymes recruited from other pathways (Figure 3.b). However, many more analyses are needed to gain further understanding into pathway evolution. Two areas of research that may provide additional insight into how pathways change include: 1) the investigation into plasticity of promiscuous enzymatic activities and 2) the evaluation of how protein structures fit in the pathway context.
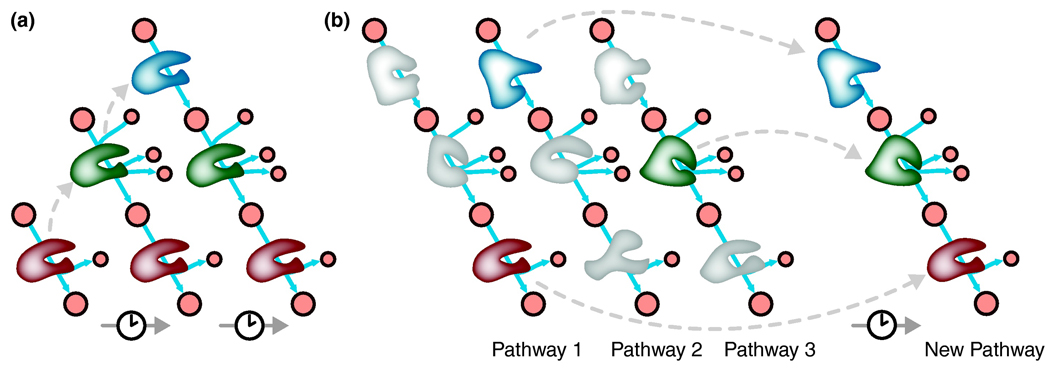
Figure 3. Two models of pathway evolution.
(a) The retrograde model suggests that duplication events of neighboring genes extends pathways from a key metabolite. (b) The patchwork model assumes novel pathways arise as broad-specificity enzymes are acquired or duplicated and mutated, thus forming a new pathway. Dashed grey arrows represent duplication events and mutation.
In case of promiscuous enzymatic activities, recent studies support the idea that novel metabolic functions may arise in a patchwork manner through refinement of promiscuous enzymatic functions[34,35]. Moreover, another recent study demonstrated the ability of artificial protein sequences to rescue 27 E. coli auxotrophs[36]. Thus, there is a wide range of potential metabolic functions that can be gained through the refinement of promiscuous activities of proteins within a cell or gained through horizontal gene transfer[37].
The incorporation of protein structure into pathway analyses will allow an additional dimension of insight into pathway evolution. For example, an assessment of protein folds across a functional metabolic network model of Thermotoga maritima demonstrated that enzymes catalyzing similar reactions tended to share folds more frequently than enzymes that are connected in the same pathways, thereby providing stronger support for the patchwork model[38].
Thus, the integration of genome-scale science, experimental evolution, and network modeling will advance our understanding about the molecular mechanisms of evolution and how they are molded by cellular objectives and selective pressures. This will accelerate as novel experimental approaches develop and as systems biology evolves to incorporate more biochemical knowledge and to better reflect biological reality.
Acknowledgements
The authors would like to thank Daniel Zielinski and Joshua Lerman for insightful discussion concerning this work. This work was supported in part by US National Institutes of Health grants R01 GM062791 and R01 GM57089.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Whitaker JW, Letunic I, McConkey GA, Westhead DR. metaTIGER: a metabolic evolution resource. Nucleic Acids Res. 2009;37:D531–D538. doi: 10.1093/nar/gkn826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mithani A, Hein J, Preston GM. Comparative analysis of metabolic networks provides insight into the evolution of plant pathogenic and nonpathogenic lifestyles in pseudomonas. Mol Biol Evol. 2011;28:483–499. doi: 10.1093/molbev/msq213. [DOI] [PubMed] [Google Scholar]
- 3.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 4.Seshasayee AS, Fraser GM, Babu MM, Luscombe NM. Principles of transcriptional regulation and evolution of the metabolic system in E. coli. Genome Res. 2009;19:79–91. doi: 10.1101/gr.079715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noor E, Eden E, Milo R, Alon U. Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol Cell. 2010;39:809–820. doi: 10.1016/j.molcel.2010.08.031. The authors show that metabolites that act as biomass precursors are linked by the fewest enzymatic steps possible.
- 6.Pal C, Papp B, Lercher MJ, Csermely P, Oliver SG, Hurst LD. Chance and necessity in the evolution of minimal metabolic networks. Nature. 2006;440:667–670. doi: 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- 7.Borenstein E, Kupiec M, Feldman MW, Ruppin E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc Natl Acad Sci U S A. 2008;105:14482–14487. doi: 10.1073/pnas.0806162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maslov S, Krishna S, Pang TY, Sneppen K. Toolbox model of evolution of prokaryotic metabolic networks and their regulation. Proc Natl Acad Sci U S A. 2009;106:9743–9748. doi: 10.1073/pnas.0903206106. This work proposes a new model of evolution in which pathways are transferred to adapt to new nutritional environments.
- 9.Pace NR. Origin of life--facing up to the physical setting. Cell. 1991;65:531–533. doi: 10.1016/0092-8674(91)90082-a. [DOI] [PubMed] [Google Scholar]
- 10.Huang SL, Wu LC, Liang HK, Pan KT, Horng JT, Ko MT. PGTdb: a database providing growth temperatures of prokaryotes. Bioinformatics. 2004;20:276–278. doi: 10.1093/bioinformatics/btg403. [DOI] [PubMed] [Google Scholar]
- 11.Takemoto K, Nacher JC, Akutsu T. Correlation between structure and temperature in prokaryotic metabolic networks. BMC Bioinformatics. 2007;8:303. doi: 10.1186/1471-2105-8-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takemoto K, Akutsu T. Origin of structural difference in metabolic networks with respect to temperature. BMC Syst Biol. 2008;2:82. doi: 10.1186/1752-0509-2-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards JS, Ibarra RU, Palsson BO. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat Biotechnol. 2001;19:125–130. doi: 10.1038/84379. [DOI] [PubMed] [Google Scholar]
- 14.Ibarra RU, Edwards JS, Palsson BO. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- 15.Teusink B, Wiersma A, Jacobs L, Notebaart RA, Smid EJ. Understanding the adaptive growth strategy of Lactobacillus plantarum by in silico optimisation. PLoS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000410. e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong SS, Palsson BO. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat Genet. 2004;36:1056–1058. doi: 10.1038/ng1432. [DOI] [PubMed] [Google Scholar]
- 17.Lewis NE, Hixson KK, Conrad TM, Lerman JA, Charusanti P, Polpitiya AD, Adkins JN, Schramm G, Purvine SO, Lopez-Ferrer D, et al. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol. 2010;6:390. doi: 10.1038/msb.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 19.Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, Palsson BO. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 20.Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, Gao Y, Palsson BO. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009;10:R118. doi: 10.1186/gb-2009-10-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charusanti P, Conrad TM, Knight EM, Venkataraman K, Fong NL, Xie B, Gao Y, Palsson BO. Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001186. e1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, Lewis NE, Landick R, Palsson BO. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee DH, Palsson BO. Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a Nonnative carbon source, L-1,2-propanediol. Appl Environ Microbiol. 2010;76:4158–4168. doi: 10.1128/AEM.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boronat A, Caballero E, Aguilar J. Experimental evolution of a metabolic pathway for ethylene glycol utilization by Escherichia coli. J Bacteriol. 1983;153:134–139. doi: 10.1128/jb.153.1.134-139.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hochhauser SJ, Weiss B. Escherichia coli mutants deficient in deoxyuridine triphosphatase. J Bacteriol. 1978;134:157–166. doi: 10.1128/jb.134.1.157-166.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atsumi S, Wu TY, Machado IM, Huang WC, Chen PY, Pellegrini M, Liao JC. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol. 2010;6:449. doi: 10.1038/msb.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez R, Tao H, Purvis JE, York SW, Shanmugam KT, Ingram LO. Gene array-based identification of changes that contribute to ethanol tolerance in ethanologenic Escherichia coli: comparison of KO11 (parent) to LY01 (resistant mutant) Biotechnol Prog. 2003;19:612–623. doi: 10.1021/bp025658q. [DOI] [PubMed] [Google Scholar]
- 29. Goodarzi H, Bennett BD, Amini S, Reaves ML, Hottes AK, Rabinowitz JD, Tavazoie S. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol Syst Biol. 2010;6:378. doi: 10.1038/msb.2010.33. E. coli is adapted to grow under toxic levels of ethanol, and evolves several mechanisms in the process.
- 30. Veeravalli K, Boyd D, Iverson BL, Beckwith J, Georgiou G. Laboratory evolution of glutathione biosynthesis reveals natural compensatory pathways. Nat Chem Biol. 2011;7:101–105. doi: 10.1038/nchembio.499. E. coli evolves a new pathway to synthesize glutathione when its native pathway is blocked. This new pathway results from the mutation of genes in proline synthesis.
- 31.Masip L, Veeravalli K, Georgiou G. The many faces of glutathione in bacteria. Antioxid Redox Signal. 2006;8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 32.Horowitz NH. On the Evolution of Biochemical Syntheses. Proc Natl Acad Sci U S A. 1945;31:153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 34. Kim J, Kershner JP, Novikov Y, Shoemaker RK, Copley SD. Three serendipitous pathways in E. coli can bypass a block in pyridoxal-5'-phosphate synthesis. Mol Syst Biol. 2010;6:436. doi: 10.1038/msb.2010.88. Authors amplify genes to show that promiscuous enzyme activities can build new metabolic pathways.
- 35. Soo VW, Hanson-Manful P, Patrick WM. From the Cover: Artificial gene amplification reveals an abundance of promiscuous resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:1484–1489. doi: 10.1073/pnas.1012108108. Authors amplify genes to identify proteins that can provide promiscuous antibiotic and toxin resistance.
- 36. Fisher MA, McKinley KL, Bradley LH, Viola SR, Hecht MH. De novo designed proteins from a library of artificial sequences function in Escherichia coli and enable cell growth. PLoS One. 2011;6:e15364. doi: 10.1371/journal.pone.0015364. Artificial proteins are screened to rescue auxotrophic strains of E. coli.
- 37.Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. Direct visualization of horizontal gene transfer. Science. 2008;319:1533–1536. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y, Thiele I, Weekes D, Li Z, Jaroszewski L, Ginalski K, Deacon AM, Wooley J, Lesley SA, Wilson IA, et al. Three-dimensional structural view of the central metabolic network of Thermotoga maritima. Science. 2009;325:1544–1549. doi: 10.1126/science.1174671. The authors use a metabolic network to analyze hundreds of protein structures.