Abstract
Chronic pain states are characterized by long-term sensitization of spinal cord neurons that relay nociceptive information to the brain. Among the mechanisms involved, up-regulation of Cav1.2-comprising L-type calcium channel (Cav1.2-LTC) in spinal dorsal horn have a crucial role in chronic neuropathic pain. Here, we address a mechanism of translational regulation of this calcium channel. Translational regulation by microRNAs is a key factor in the expression and function of eukaryotic genomes. Because perfect matching to target sequence is not required for inhibition, theoretically, microRNAs could regulate simultaneously multiple mRNAs. We show here that a single microRNA, miR-103, simultaneously regulates the expression of the three subunits forming Cav1.2-LTC in a novel integrative regulation. This regulation is bidirectional since knocking-down or over-expressing miR-103, respectively, up- or down-regulate the level of Cav1.2-LTC translation. Functionally, we show that miR-103 knockdown in naive rats results in hypersensitivity to pain. Moreover, we demonstrate that miR-103 is down-regulated in neuropathic animals and that miR-103 intrathecal applications successfully relieve pain, identifying miR-103 as a novel possible therapeutic target in neuropathic chronic pain.
Keywords: calcium channel, microRNA, neuropathic pain, spinal cord
Introduction
In chronic pain states, modifications of intrinsic amplification properties of dorsal horn spinal cord neurons lead to long-term sensitization (Derjean et al, 2003). Moreover, sensitization to pain involves the activation of voltage-gated calcium channels (VGCC) (Matthews and Dickenson, 2001; Bourinet et al, 2005; Altier et al, 2007), and in particular Cav1.2-LTC that regulates gene expression underlying long-term plastic changes (Dolmetsch et al, 2001; Fossat et al, 2010).
MiRNAs are short non-coding transcripts targeting mRNA 3′ untranslated region (3′UTR) for cleavage or translation inhibition (Lee et al, 1993; Kertesz et al, 2007). Recent findings have demonstrated a key role for miRNAs in synaptic plasticity through the regulation of NMDA receptor expression (Edbauer et al, 2010; Wibrand et al, 2010) or dendritic spine morphology (Schratt et al, 2006). Previous screening studies addressed the involvement of miRNA in the nociceptive system. Indeed, miRNA repertoires in trigeminal root ganglia, spinal cord and prefrontal cortex are modulated after inflammatory pain (Bai et al, 2007; Kusuda et al, 2011; Poh et al, in press). Similarly, peripheral nerve injury modulates miRNA expression in spinal cord and dorsal root ganglion (Kusuda et al, 2011; von Schack et al, 2011). Taken together, these data suggest a role in pain pathophysiology. In addition, two studies provided evidence for a causal role of miRNAs. First Zhao et al (2010) demonstrated that miRNAs regulate Nav1.8 sodium channel expression in dorsal root ganglion and therefore have a role in inflammatory pain. Second, Li et al (2011) suggested that miR-146a controls knee joint homeostasis and osteoarthritis-associated algesia.
The regulatory role of miRNAs is likely to involve multiple mRNAs since perfect matching to target sequence is not required for inhibition (Lim et al, 2005). We show here for the first time that such multi-target (or integrative) silencing regulates the different subunits of a macromolecular complex, namely Cav1.2-LTC. In an animal model for neuropathic pain, we demonstrate that miR-103 is down-regulated leading to an up-regulation of Cav1.2-LTC subunits. We further demonstrate a novel causal role of miRNA in the maintenance of pain sensitization, showing that miR-103 intrathecal applications repress Cav1.2-LTC up-regulation and consequently relieve spinal sensitization to pain, therefore identifying miR-103 as a possible therapeutic target.
Results
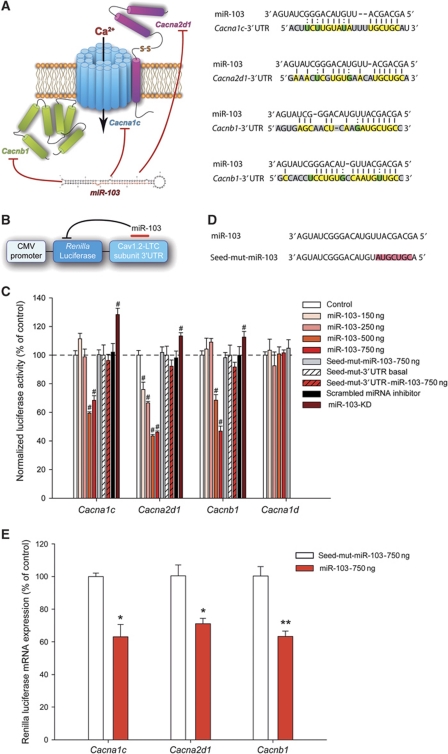
Neuronal Cav1.2-LTC comprises three subunits: Cav1.2-α1, α2δ1 and β1 encoded by Cacna1c, Cacna2d1 and Cacnb1 mRNAs, respectively. Bioinformatics analyses using miRanda and Targetscan algorithms (John et al, 2004; Lewis et al, 2005) revealed that all these mRNAs are putative targets of various miRNAs (Supplementary Table S1). They also suggested that a single miRNA (miR-103) possibly binds and inhibits all three subunits forming Cav1.2-LTC (Figure 1A). Indeed, Cacna1c and Cacna2d1 each contain one miR-103 target site while Cacnb1 has two miR-103 target sites. Moreover, the miR-103 target sequences in the Cacna1c, Cacna2d1 and Cacnb1 3′UTRs are well conserved across vertebrates and miR-103 does not target any other VGCC subunits (Supplementary Figure S1; Supplementary Table S2). In mammals, miRNAs are thought to regulate the expression of target mRNAs predominantly through the inhibition of productive translation (Bartel, 2004). Thus, to confirm these theoretical interactions, we generated reporter plasmids by fusing the 3′UTR of each Cav1.2-LTC subunit mRNA (containing miR-103 target sites) to the 3′ terminus of a Renilla luciferase coding sequence (Figure 1B). In COS-7 cells, co-expression of these Renilla luciferase constructs with a miR-103-encoding plasmid prevented luciferase expression in a dose-dependent manner (Figure 1C), whereby demonstrating miR-103 potency to inhibit translation of the three Cav1.2-LTC subunits. Interestingly, the amount of inhibition was different according to the Cav1.2-LTC subunit 3′UTR considered. Cacna2d1 3′UTR mediated significant inhibition at the lowest miR-103 dose tested whereas Cacna1c and Cacnb1 3′UTRs reporter activities remained unaffected. At higher miR-103 doses, the activity of all three Cav1.2-LTC subunit reporters was significantly repressed. Differences in the number and the location of miR-103 sites in 3′UTRs could explain these observations (Grimson et al, 2007; Kertesz et al, 2007). To demonstrate that inhibition was sequence specific, we cloned into the reporter plasmid, the 3′UTR of Cav1.3, another LTC-α1 subunit that contains no miR-103 target site. As expected, miR-103 had no effect on Cav1.3 reporter gene translation, even at the highest amounts tested (Figure 1C, Cacna1d). To illustrate the specificity of miR-103 targeting, we designed a miR-103 mutant (seed-mut-miR-103) differing from the original sequence in the ‘seed region’ of miR-103 (residues at positions 2–8), that controls hybridization with mRNAs (Lewis et al, 2003; Stark et al, 2003; Doench and Sharp, 2004). Nucleotide sequence of the ‘seed’ region was mutated to antisense sequence (Figure 1D) and using miRNA hybridization algorithm (Lewis et al, 2005), we confirmed that it theoretically results in a complete loss of targeting Cav1.2-LTC components. As expected, seed-mut-miR-103 was unable to inhibit any of the Cav1.2-LTC subunit reporters, even at the highest amount tested (Figure 1C). To further demonstrate the role of the seed regions of Cacna1c, Cacna2d1 and Cacnb1 3′UTRs in miR-103 specificity, we deleted the seed regions of all Cav1.2-LTC luciferase reporters. As a result, miR-103 was then unable to mediate translation inhibition of these modified reporters (Figure 1C, seed-mut-3′UTR). As miR-103 sequence is conserved between rat and COS-7 cells, we tested whether endogenous miR-103 expressed by COS-7 cells could regulate Cav1.2-LTC. Therefore, we knocked-down endogenous miR-103 by transfecting LNA-modified miR-103 inhibitor (miR-103-KD). As a result, miR-103-KD enhanced luciferase activity of all Cav1.2-LTC reporters (Figure 1C), demonstrating an endogenous and bidirectional regulation of Cav1.2-LTC by miR-103 in COS-7 cells. It is now well admitted that miRNAs can inhibit translation of target mRNAs either with or without mRNA degradation (Huntzinger and Izaurralde, 2011). To determine whether miR-103 induces target mRNA decay, we measured luciferase mRNA levels by qRT–polymerase chain reaction (PCR). Compared with seed-mut-miR-103 over-expression, miR-103 transfection induced a significant down-regulation of luciferase mRNA for all Cav1.2-LTC reporters (Figure 1E). This result indicates that the inhibition of Cav1.2-LTC subunit translation by miR-103 implies, at least, mRNA decay. Thus, we demonstrated, at the molecular level, the ability of miR-103 to silence simultaneously the three subunits of Cav1.2-LTC through binding their 3′UTR.
Figure 1.
Integrative regulation of Cav1.2-LTC by miR-103. (A) Bioinformatics analyses revealed that all Cav1.2-LTC subunit mRNA contain a target site for miR-103. Cacna1c and Cacna2d1 each contain one miR-103 site while Cacnb1 has two miR-103 sites. Hybridization to Cacna1c, Cacna2d1 and Cacnb1 3′UTR, yellow indicates perfect base pairing, green indicates wobble base pairing and grey indicates no match. (B) Luciferase reporters consisted of a Renilla luciferase coding sequence under CMV promoter control that was fused on its 3′ end to the 3′UTR of the different Cav1.2-LTC subunits. (C) Compared with control conditions, Cacna2d1 3′UTR mediated significant inhibition of Renilla luciferase activity at the lowest miR-103 dose (150 ng, −24%, P<0.001) whereas Cacna1c and Cacnb1 3′UTRs reporter activities were still unaffected. At higher miR-103 dose (500 ng), all Cav1.2-LTC subunit reporter activities were significantly repressed: −40.51%, P<0.001, −56.45%, P<0.001 and −31.51%, P<0.001 for Cacna1c, Cacna2d1 and Cacnb1, respectively. At the highest miR-103 dose (750 ng), inhibition reached a plateau for Cacna1c and Cacna2d1 (−31.59%, P<0.001 and −53.94%, P<0.001, respectively) whereas Cacnb1 reporter was more efficiently inhibited (−53.07%, P<0.001). Mutation of miR-103 seed region (seed-mut-miR-103) or reporter 3′UTR seed region (seed-mut-3′UTR) resulted in a complete abolition of miR-103 inhibition. Endogenous miR-103 knockdown resulted in a moderate but significant increase of all Cav1.2-LTC subunit reporter activities (miR-103-KD). #P<0.001, ANOVA, Holm–Sidak post hoc test. Data are expressed as mean values±s.e.m. n=6. (D) Mutation of miR-103 (seed-mut-miR-103) at nucleotide positions 2–8 (antisense of original sequence, highlighted in red). (E) qRT–PCR analysis of luciferase mRNA levels showed that miR-103 over-expression induced reporter mRNA decay for all Cav1.2-LTC subunits (Cacna1c −36.88%, P=0.021; Cacna2d1 −28.94 %, P=0.013; Cacnb1 −36.74%, P=0.004). *P<0.05, **P<0.01. Data are expressed as mean values±s.e.m. n=3.
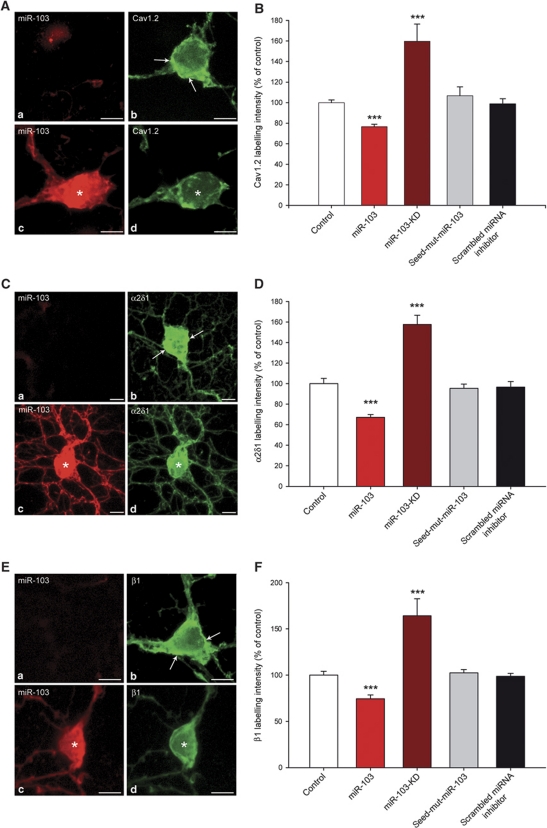
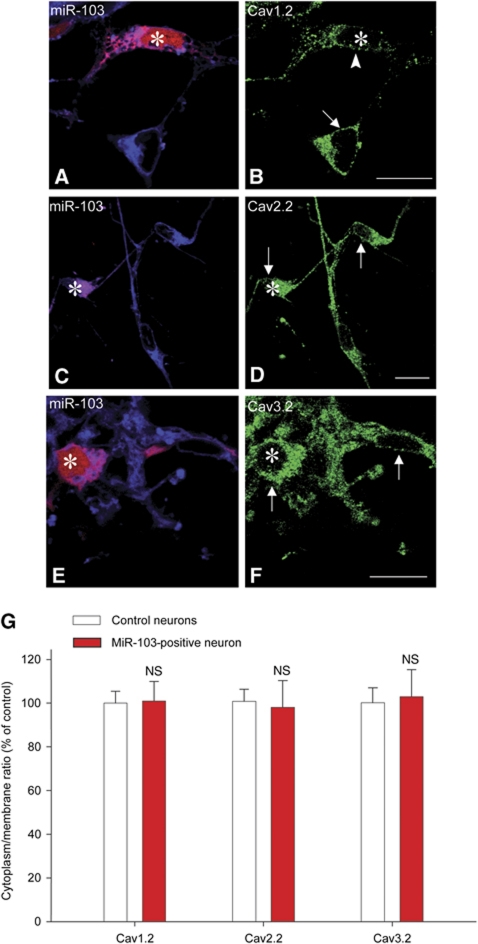
Next, we assessed the effect of miR-103 regulation on Cav1.2-LTC expression in neurons. We performed immunolabelling for Cav1.2-α1, α2δ1 and β1 subunit (Figure 2A, C and E, respectively) on primary spinal cord neuron cultures and we quantified immunostaining intensity on confocal images. Compared with control non-transfected conditions, over-expression of miR-103 induced a significant decrease in the intensity of all Cav1.2-LTC subunit staining, namely Cav1.2-α1, α2δ1 and β1 subunits (Figure 2B, D and F, respectively). On the contrary, miR-103 knockdown with miR-103-KD caused an increase in all Cav1.2-LTC subunit labelling intensity (Figure 2B, D and F), demonstrating a role for endogenous miR-103 to limit Cav1.2-LTC expression in neurons. As a control, we used either seed-mut-miR-103 or scrambled miRNA inhibitor that both had no effect on Cav1.2-LTC expression (Figure 2B, D and F). These results confirmed in neurons the luciferase assay results, indicating an integrative regulation of the three Cav1.2-LTC subunits by miR-103. Moreover, these experiments indicate that the regulation of Cav1.2-LTC by miR-103 is bidirectional since knocking-down or over-expressing miR-103, respectively, up- or down-regulate the level of Cav1.2-LTC translation. Among the three subunits forming Cav1.2-LTC, Cav1.2-α1 is specific of this calcium channel subtype whereas α2δ1 and β1 subunits could be associated with other VGCC. Combinatorial subunit associations are still poorly understood (Davies et al, 2007); however, α1 subunits of other calcium channels implicated in pain such as N-type Cav2.2 (Chaplan et al, 1994; Altier et al, 2007) or T-type Cav3.2 (Matthews and Dickenson, 2001; Bourinet et al, 2005) could be associated with α2δ1 and β1 subunits. To evaluate a possible impact of α2δ1 and β1 regulation by miR-103 on other pain-related calcium channel expression, we quantified immunolabelling of Cav2.2 and Cav3.2. Neither miR-103 over-expression, nor miR-103 knockdown had an effect on Cav2.2 and Cav3.2 expression (Supplementary Figures S2 and S3). We further tested for changes in Cav1.2, Cav2.2 and Cav3.2 trafficking by measuring the ratio between membrane and cytoplasm immunostaining. We first validated this method to quantify membrane trafficking by monitoring the GABAB receptor, which is known to be trafficked to the membrane only when associated as a heterodimer (Supplementary Figure S4). Then, we quantified the membrane to cytoplasm ratio for Cav1.2, Cav2.2 and Cav3.2 with and without miR-103 application. This ratio remained constant in both conditions for Cav1.2, Cav2.2 and Cav3.2 proteins, thus strongly suggesting that miR-103 does not induce changes in their trafficking (Figure 3).
Figure 2.
MiR-103 bidirectionally regulates Cav1.2-LTC expression in spinal cord neurons. To assess all Cav1.2-LTC subunit expressions, we performed a specific labelling for Cav1.2-α1, α2δ1 and β1 subunits (A, C, E, respectively). In non-transfected neurons (a, b), labelling appears more intense, particularly at the plasma membrane (arrows), than in miR-103 over-expressing cells (star in c, d). Quantification of immunostaining intensity on confocal images demonstrated that miR-103 over-expression induced a significant decrease in Cav1.2-α1, α2δ1 and β1 labelling intensity compared with control conditions (−23.47% (B), −32.75% (D) and −25.66% (F), respectively, ***P<0.001). In miR-103 knockdown experiment, Cav1.2-α1, α2δ1 and β1 expressions were significantly increased (+59.64% (B), +57.62% (D) and +64.20% (F), respectively, P<0.001). Transfection with seed-mut-miR-103 or scrambled miRNA inhibitor had no effect on Cav1.2-LTC subunit labelling intensities. Data are expressed as mean values±s.e.m. of four experiments with a number of quantified neurons ranging from 11 to 19 for each condition, bar=10 μm.
Figure 3.
MiR-103 does not affect Cav1.2, Cav2.2 and Cav3.2 trafficking. We assessed Cav1.2, Cav2.2 and Cav3.2 trafficking by measuring the ratio between membrane and cytoplasm immunostaining with and without miR-103 application. Membrane is identified with a lipophilic dye, DiD (blue in A, C, E). (A, B) miR-103 over-expressing neurons (red, A, star) showed an overall decrease in Cav1.2 labelling (green, B, arrow and arrowhead) but trafficking was not affected. In contrast, miR-103 over-expression (red, star in C, E) did not alter Cav2.2 nor Cav3.2 labelling (green; D, F). (G) Statistical analysis showed no difference in Cav1.2, Cav2.2 and Cav3.2 trafficking, data are expressed as mean values±s.e.m. n=10 for each condition, bar=20 μm.
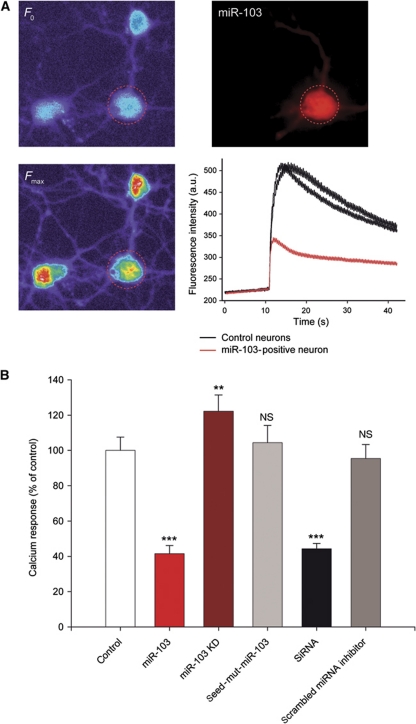
To assess the physiological relevance of such an integrative regulation of Cav1.2-LTC by miR-103, we imaged calcium transients elicited by KCl-induced depolarization in spinal cord neurons. Upon stimulation, neurons exhibited a large increase in intracellular calcium concentration due in part to ion influx through several types of voltage-dependent calcium channels including Cav1.2-LTC (Figure 4A). To demonstrate that miR-103 levels can dynamically regulate calcium influx through Cav1.2-LTC, we used either a miR-103 over-expression plasmid or miR-103-KD. MiR-103 plasmid transfection significantly decreased evoked calcium signals (−58.44%, P<0.001; Figure 4A and B), whereas miR-103-KD significantly increased calcium influx (+22.23%, P<0.01; Figure 4B). To assess the specificity of the miR-103 regulation, we used seed-mut-miR-103 and scrambled miRNA inhibitor, which had no effect on calcium transients. To evaluate the calcium signal mediated by Cav1.2-LTC in the global calcium response, we compared these data with the results obtained with anti-Cav1.2-α1 siRNAs. As shown in Figure 4B, in the presence of anti-Cav1.2-α1 siRNA, the decrease in calcium transients was equivalent to that induced by miR-103 (−55.69%, P<0.001, compared with control). This last result confirms that Cav1.2-LTC has an important role in calcium transients induced by KCl depolarization. Taken together, these calcium imaging experiments confirm that the integrative regulation of Cav1.2-LTC by miR-103 is bidirectional. Expression of Cav1.2-LTC is determinant for the dorsal horn neuron's firing properties (Morisset and Nagy, 1999; Fossat et al, 2010). Hence, our results strongly suggest that Cav1.2-LTC regulation by miR-103 controls neuron excitability in vitro.
Figure 4.
MiR-103 regulation modulates neuronal calcium transients in vitro. (A) Spinal cord neurons were loaded with Fluo4-AM calcium indicator and depolarization was induced by KCl bath application (25 mM). (B) MiR-103 over-expression strongly reduced calcium responses compared with control (−58.44%, P<0.001) while miR-103 knockdown increased calcium transients (+22.23%, P<0.01). Like miR-103, anti-Cav1.2 siRNA application significantly reduced calcium signals compared with control, (−55.69%, P<0.001). Seed-mut-miR-103 and scrambled miRNA inhibitor did not change calcium transients. **P<0.01, ***P<0.001, ANOVA, Holm–Sidak post hoc test. Data are expressed as mean values±s.e.m. n=16/7, 21/7, 30/8, 46/8, 44/9 and 18/7 (first and second numbers indicate the number of tested cells and cultures, respectively) for control, miR-103, miR-103-KD, seed-mut-miR-103, siRNAs and scrambled miRNA inhibitor experiments, respectively.
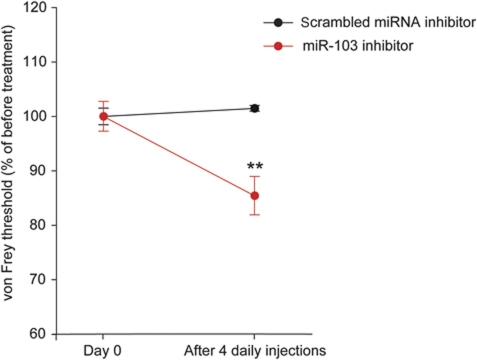
We previously demonstrated that up-regulation of Cav1.2-LTC in spinal dorsal horn is a key mechanism involved in chronic neuropathic pain. Indeed, specific knockdown of Cav1.2-α1 in the spinal dorsal horn reversed the neuropathy-associated mechanical hypersensitivity and hyperexcitability, and increased responsiveness of dorsal horn neurons (Fossat et al, 2010). Therefore, we investigated the role of miR-103 in neuronal sensitization in vivo. In the present study, we demonstrated that miR-103 regulates Cav1.2-LTC expression in vitro. Thus, modulating miR-103 in vivo should also modify Cav1.2-LTC expression and should therefore modify neuron excitability and pain sensitivity. To test this hypothesis, we performed four daily intrathecal injections of miR-103-KD in naive animals and measured their sensitivity to mechanical stimulations. Compared with control animals that received scrambled miRNA inhibitor, miR-103-KD-injected rats demonstrated a moderate but significant hypersensitivity to pain (−14.57% in mechanical threshold after treatment, P<0.01; Figure 5), suggesting a basal anti-nociceptive control by endogenous miR-103.
Figure 5.
MiR-103 knockdown in naive animals induces hypersensitivity to pain. Naive animals were subjected to four daily intrathecal injections of miR-103-KD and their sensitivity to pain was measured with mechanical stimulations. Compared with control animals who received scrambled miRNA inhibitor, miR-103-KD-injected rats demonstrated a slight but significant hypersensitivity to pain (−14.57% in mechanical threshold after treatment, P<0.01). Data are expressed as mean values±s.e.m. n=6 for miR-103-KD and scrambled miRNA inhibitor injected rats, **P<0.01, Wilcoxon signed rank test.
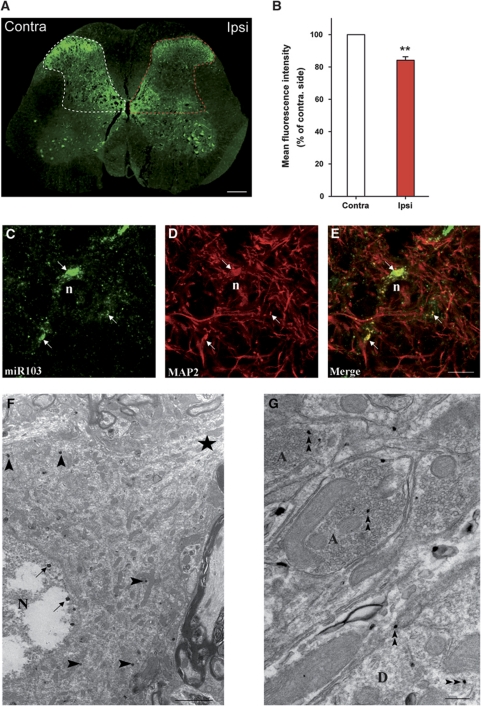
To uncover further the role of Cav1.2-LTC regulation by miR-103 in pain mechanisms, we used a rat model of chronic neuropathic pain. The neuropathy was induced by spinal nerve ligation (SNL) (Kim and Chung, 1992) and rats were tested for pain-related sensitization of mechanical sensitivity. We first investigated miR-103 expression in the spinal cord with in situ hybridization. Endogenous miR-103 was widely expressed throughout the grey matter, with the most intense signal in motoneurons and superficial dorsal horn (Figure 6A). In neuropathic animals, labelling intensity in spinal cord dorsal horn was clearly decreased ipsilaterally to the lesion (−15.92% compared with contralateral side, P=0.006; Figure 6B). Labelling was seen as clusters in cell bodies and processes (Figure 6C) and often co-localized with the somato-dendritic marker MAP2, indicating that miR-103 was expressed in neurons (Figure 6D and E). Electron microscopy confirmed this neuronal localization, in both soma and processes (Figure 6F and G). MiR-103 was found in the cytosol, but also in the vicinity of the plasma membrane, and in the nucleus, near the nuclear envelope. The localization of miR-103 in processes and its possible role in local translation remain to be studied.
Figure 6.
MiR-103 expression in the spinal cord is modulated in neuropathic pain conditions. (A) MiR-103 was detected in SNL rats by in situ hybridization with a double-digoxigenin-labelled probe. MiR-103 expression is enriched in superficial laminae of the dorsal horn and in ventral motoneurons. The signal intensity is weaker in the dorsal horn ipsilateral to the peripheral nerve injury (red mask). Bar: 500 μm. (B) Quantification of miR-103 labelling intensity showed a significant decrease in miR-103 expression in the ipsilateral dorsal horn of SNL animals (−15.92% compared with controlateral side, P=0.006). Data are expressed as mean values±s.e.m. n=5. (C–E) Double detection of miR-103 (green) and MAP2 (red) shows neuronal expression of the miRNA. MiR-103 is expressed as discrete clusters throughout cell bodies and dendritic processes (arrows). n=nucleus; bar=10 μm. (F, G) Ultrastructural localization of miR-103 in dorsal horn neurons by in situ hybridization, double-digoxigenin-labelled probe was visualized with silver-enhanced ultra-small gold particles. The expression of miR-103 is restricted to subdomains of the soma (arrowheads in F). It is also present in the nucleus, near the nuclear envelope (arrows in F). In processes, miR-103 is detected in both axons (A) and dendrites (D) (double arrowheads in F and G). N=nucleus, star indicates dentritic process; bar=2 μm in (F) and bar=200 nm in (G).
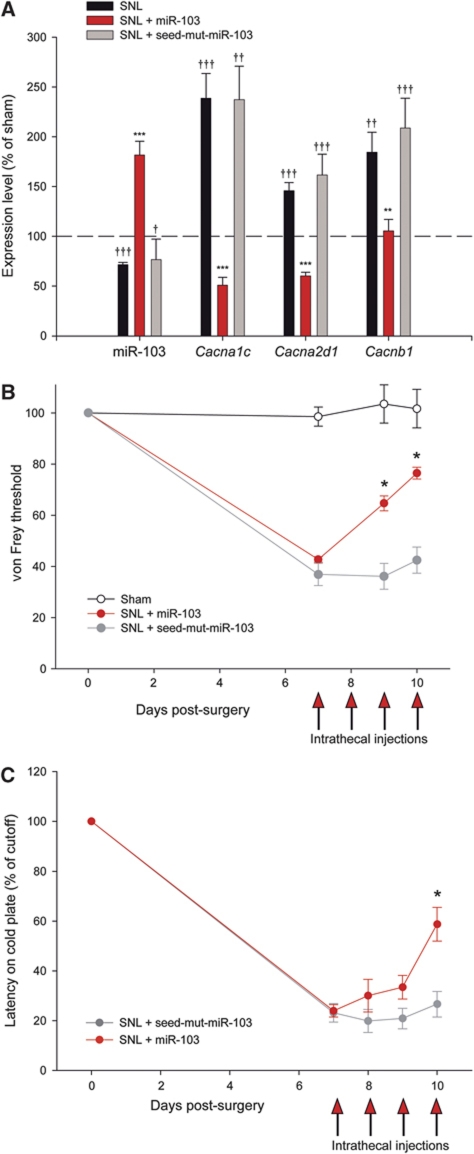
Using qRT–PCR, we compared the expression of Cacna1c, Cacna2d1, Cacnb1 mRNAs and miR-103 in the dorsal spinal cord of sham and SNL animals. In SNL rats, all three Cav1.2-LTC subunit mRNAs were over-expressed, while miR-103 was significantly down-regulated (Figure 7A). To test for a possible causal role of miR-103 down-regulation in Cav1.2-LTC subunits up-regulation, SNL rats received four daily intrathecal applications of miR-103 or seed-mut-miR-103. As determined by qRT–PCR, miR-103 injections significantly reduced the expression of all three Cav1.2-LTC subunits (Cacna1c −78.62%, P<0.001; Cacna2d1 −58.72%, P<0.001; Cacnb1 −42.93%, P=0.004, versus SNL). In contrast, seed-mut-miR-103 injections had no impact on Cav1.2-LTC subunits (Figure 7A). Finally, we tested for a control of pain sensitization by miR-103 in SNL neuropathic rats, which are known to exhibit both mechanical and cold allodynia (Choi et al, 1994). Seven days after the SNL, we monitored the mechanical threshold for ipsilateral paw withdrawal in response to stimulation with von Frey hairs and we compared it with the threshold measured in the same animal just before the surgery. Seven days after surgery, the threshold was lowered to 43.10% of the control in SNL animals, but remained unchanged in sham animals (98.51%; Figure 7B). SNL rats that were injected with miR-103 showed a significant recovery toward normal threshold sensitivity (76.14% of the pre-SNL threshold, P=0.031; Figure 7B). In contrary, seed-mut-miR-103-injected animals showed no response to treatment. Cold allodynia was evaluated with a cold-plate device where animals were placed on a thermostatic steel plate set to +4°C. Elevation of the injured hind limb latency is measured and compared with the latency of the same animal just before surgery. Likewise the response to mechanical stimulation, 7 days after surgery, SNL animals showed a much shorter latency, demonstrating hypersensitivity to cold (23.53% of pre-surgery latency; Figure 7C). Intrathecal miR-103 injections significantly reduced cold allodynia in SNL rats (58.75% of pre-surgery latency, P=0.018; Figure 7C), whereas seed-mut-miR-103-injected animals showed no response to treatment.
Figure 7.
MiR-103 dynamically regulates all Cav1.2-LTC subunits in vivo and is involved in pain sensitization. (A) In SNL animals, all three Cav1.2 subunit mRNAs were over-expressed as compared with sham (Cacna1c +138.57%, P<0.001; Cacna2d1 +47.09%, P<0.001; Cacnb1 +84.44%, P=0.005) while miR-103 was significantly decreased (−28.3%, P<0.001). ††P<0.01, †††P<0.001, ANOVA, Holm–Sidak post hoc test. MiR-103 intrathecal application significantly reduced Cacna1c, Cacna2d1 and Cacnb1 expressions (as compared with SNL level, −78.62%, P<0.001, −58.72%, P<0.001, −42.93%, P=0.004, respectively). In contrast, seed-mut-miR-103 did not change Cav1.2-LTC subunit expression. **P<0.01, ***P<0.001, ANOVA, Holm–Sidak post hoc test. Data are expressed as mean values±s.e.m. n=6, 6 and 3 for SNL, miR-103 and seed-mut-miR-103, respectively. (B) Rats were tested for pain sensitization using an electronic von Frey device: 7 days after surgery, they showed a decrease in paw withdrawal threshold in response to mechanical stimulation (40.70% of the pre-SNL threshold). Four daily intrathecal applications of miR-103 (days 7–10) alleviated mechanical allodynia (76.14% of pre-SNL threshold, P=0,031). Seed-mut-miR-103-injected animals showed no response to treatment (42.41% of pre-SNL threshold, P=0.219). *P<0.05, Wilcoxon signed rank test. Data are expressed as mean values±s.e.m. n=4, 4 and 6 for sham, miR-103 and seed-mut-miR-103-injected rats, respectively. (C) Cold allodynia was evaluated with a cold-plate device where animals are placed on a thermostatic steel plate set to +4°C. Elevation of the operated hind limb latency is measured and compared with the latency of the same animal just before surgery. Seven days after surgery (nerve ligation), SNL animals showed hypersensitivity to cold (23.53% of pre-surgery latency). Intrathecal miR-103 injections significantly reduced cold allodynia in SNL rats (58.75% of pre-surgery latency) whereas seed-mut-miR-103-injected animals showed no response to treatment. Data are expressed as mean values±s.e.m. n=6 and 4 for miR-103 and seed-mut-miR-103-injected rats, respectively.
Discussion
Recent studies uncovered the crucial role of miRNAs in the fine tuning of neuronal gene expression during development (Liu et al, 2004; Giraldez et al, 2005), but also in adult physiological functions such as circadian clock (Cheng et al, 2007). The implication of miRNAs in pathological mechanisms is increasingly documented and so are the demonstrations of miRNA involvement in central nervous system disorders. Recently, studies have suggested that deregulated miRNAs expression could contribute to Alzheimer's disease (for review see Satoh, 2010) or Parkinson's disease (Kim et al, 2007; Wang et al, 2008). The expression of miRNAs is modulated in the context of pain (Bai et al, 2007; Aldrich et al, 2009; Kusuda et al, 2011; Poh et al, in press; von Schack et al, 2011) but so far the mechanisms involved and the physiological impact remained scarcely explored. Here, we demonstrated that miR-103 regulates the translational level of the three subunits forming Cav1.2-LTC. Our in vitro experiments demonstrated that miR-103 hybridization to target mRNAs is sequence specific given that ‘seed’ mutation of miR-103 totally impairs inhibition of target translation. Moreover, this regulation is bidirectional since lower or higher miR-103 levels, respectively, up- or down-regulate the level of Cav1.2-LTC translation, thus confirming the role of miR-103 as an endogenous translational regulator of Cav1.2-LTC. Our calcium imaging experiments indicated that regulating Cav1.2 through miR-103 has an important impact on calcium influx through VGCC, suggesting a significant role in control of membrane potential and firing patterns of spinal neurons (Derjean et al, 2003). This control may constitute a dynamic way of modulating neuronal excitability without altering transcriptional level. It was previously demonstrated that Cav1.2-LTC is implicated in NMDA receptor-independent long-term potentiation (Moosmang et al, 2005). MiR-103 regulation of Cav1.2-LTC represents an upstream mechanism by which miRNA control intrinsic excitability of dorsal horn neurons, and may participate to neuronal plasticity mechanisms.
MiRNAs are likely to regulate multiple mRNAs because perfect matching to the 3′UTR of target is not required to induce silencing (Lim et al, 2005). This feature is conserved from worms to mammals and therefore should benefit to cell function. One may speculate that if one molecule regulates the expression of multiple genes, it could lower the energy cost of translational regulation. Another possible advantage is that it regulates a function rather than only the expression of a single gene. Previous demonstrations of a concerted control by miRNAs were related to several components of a signalling pathway (Ferretti et al, 2008; Webster et al, 2009). Here, we show for the first time, to our knowledge, that a single miRNA may exert integrative regulation of all constitutive subunits of a given macromolecular complex. Thus, miR-103 simultaneously and bidirectionally regulates the expression of the three subunits forming Cav1.2-LTC: Cav1.2-α1, α2δ1 and β1. Whereas Cav1.2-α1 is specific of Cav1.2-LTC, combinatorial subunit associations involving α2δ1 and β1 are still only partially understood (Davies et al, 2007). Thus, other VGCC involved in pain such as N-type Cav2.2 (Chaplan et al, 1994; Altier et al, 2007) or T-type Cav3.2 (Matthews and Dickenson, 2001; Bourinet et al, 2005) can be associated with α2δ1 and β1 subunits. Our results indicate that miR-103 does regulate the translation of Cav1.2-α1, α2δ1 and β1, but that only Cav1.2-LTC channel expression is modulated, whereas neither Cav2.2- nor Cav3.2-comprising channels are affected. This result could be explained by the role of α2δ1 and β1 subunits as membrane trafficking auxiliary subunits, common to many VGCC. Expression levels of VGCC are not homogeneous, indicating that they should not be commonly regulated by a shared subunit. Therefore, one can hypothesize that α2δ1 and β1 subunits are not regulators of the global expression of VGCC but may be implicated in complex combinatorial subunit associations and region-specific membrane trafficking of VGCC (Bauer et al, 2009; Obermair et al, 2010). In line with this idea, our results suggest that the global expression level of functional Cav1.2-LTC is regulated by miR-103 through an integrative regulation of all its constitutive subunits. This novel mechanism could shed light on complex co-modulations of multiple mRNAs within a cell, in either normal or pathological conditions. Therefore, our results urge for combinatorial analyses of transcriptome and microRNAome.
We previously demonstrated that up-regulation of Cav1.2-LTC in spinal dorsal horn is a key mechanism involved in chronic neuropathic pain (Fossat et al, 2010). In this study, we demonstrated that miR-103 exerts a bidirectional and integrative regulation on Cav1.2-LTC and thus may be a major factor of chronic pain sensitization. Indeed, artificial down-regulation of miR-103 in naive animals induced hypersensitivity to pain. In addition, this regulation is impaired in an animal model for neuropathic pain and miR-103 intrathecal applications repressed Cav1.2-LTC up-regulation and subsequently relieved pain sensitization. Therefore, our study identifies miR-103 as a novel possible therapeutic target in neuropathic chronic pain.
Materials and methods
Ethics statement
All experimental procedures followed the ethical guidelines of the International Association for the Study of Pain and were approved by the local ethics committee (agreement no. 330110005-A).
Plasmids, siRNAs, knockdown LNA, RNA and DNA oligonucleotides
The rat Cacna1c, Cacna2d1, Cacnb1 and Cacna1c 3′UTR were amplified by PCR from rat brain cDNA and cloned into a modified phRL-CMV vector (Promega). For the miR-103 expression construct, a genomic sequence spanning 172 bp upstream and 166 bp downstream of the miR-103 sequence was PCR amplified and cloned into pcDNA3.1 (Invitrogen). Mutation of miR-103 and deletion of the seed region in Cav1.2-LTC subunits’ reporters were achieved using the Quick Change site-directed mutagenesis kit (Stratagene). To knockdown miR-103, we used specific miRCURY LNA™ microRNA Inhibitors and scrambled miRNA inhibitor (Exiqon). We used siRNA targeting several splice variants of Cav1.2 (‘Silencer Select Pre-designed and Validated siRNA’, Ambion). They consisted of a pool of two 21 nt duplex. siRNAs were selected to target two distinct CaV1.2mRNA regions to enhance silencing. The antisense sequences were as follows: 5′-UCUAUUGUCAUAUCGCAGG-3′ and 5′-UAUCCGAACAGGUAUAGAG-3′. For intrathecal miRNA applications, we used synthetic double-strand RNA (Eurogentec) designed as mature miR-103 sequence, seed-mutated miR-103 or mismatch miRNA. For in situ hybridization, we used LNA™ modified probes (Exiqon).
Cloning and mutation
- Cacna1c 3′UTR Fw:
5′-GCGCGCGGTACCTCGTTTCAATGTTCCTAATGGG-3′
- Cacna1c 3′UTR Bw:
5′-GCGCGCGGATCCCGACACCTTTGTTCACTTTGC-3′
- Cacna2d1 3′UTR Fw:
5′-GCGCGCGGTACCCCCTCTAAACCCATGATCTG-3′
- Cacna2d1 3′UTR Bw:
5′-GCGCGCGGATCCAAACCTCCCACAGAGAATGAG-3′
- Cacnb1 3′UTR Fw:
5′-GCGCGCGGTACCCACCATATCCATTCCATAACC-3′
- Cacnb1 3′UTR Bw:
5′-GCGCGCGGATCCCAGTGCTATCTTCTAACCTG-3′
- Cacna1d 3′UTR Fw:
5′-GCGCGCGGTACCGTACGAGTGATTTCACGCCT-3′
- Cacna1d 3′UTR Fw:
5′-GCGCGCGGATCCTGCAGTTCAAATACGGTACGA-3′
- miR-103 Fw:
5′-GCGCGCAAGCTTTTGAATGCACAGTCTAGATCTAAATGGG-3′
- miR-103 Bw:
5′-GCGCGCGGATCCAAAGGGTCCTCCTGTCTCCTC-3′
- miR-103-mut Fw:
5′-GCCTTGTTGCATATGGATCAAATAGCATTGTACAGGGCTATG-3′
- miR-103-mut Bw:
5′-CATCGCCCTGTACAATGCTATTTGATCCATATGCAACAAGGC-3′
- Cacna1c 3′UTR-mut Fw:
5′-GGATTCACTTCTTGTATATTTTGTAAAGTAAATCATTTTG-3′
- Cacna1c 3′UTR-mut Bw:
5′-CAAAATGATTTACTTTACAAAATATACAAGAAGTGAATCC-3′
- Cacna2d1 3′UTR-mut Fw:
5′-GGAAACTGAAACTCGTGTGAACTCATCTGTGTCCAACATC-3′
- Cacna2d1 3′UTR-mut Bw:
5′-GATGTTGGACACAGATGAGTTCACACGAGTTTCAGTTTCC-3′
- Cacnb1 3′UTR-mut1 Fw:
5′-GAGGAGTGAGCAACTCAAGCCCCCGCAACCCCTTCCAG-3′
- Cacnb1 3′UTR-mut1 Bw:
5′-CTGGAAGGGGTTGCGGGGGCTTGAGTTGCTCACTCCTC-3′
- Cacnb1 3′UTR-mut2 Fw:
5′-GTCTCGCCACCTCCTGTGCCCCCACCCCTTCCGGGGG-3′
- Cacnb1 3′UTR-mut2 Bw:
5′-CCCCCGGAAGGGGTGGGGGCACAGGAGGTGGCGAGAC-3′
In situ hybridization
- MiR-103:
5′-TCATAGCCCTGTACAATGCTGCT-3′
Real-time PCR
- SDHA Fw:
5′-TGCGGAAGCACGGAAGGAGT-3′
- SDHA Bw:
5′-CTTCTGCTGGCCCTCGATGG-3′
- Cacna1c Fw:
5′-AGACAGCCTTGCCCTTGCAT-3′
- Cacna1c Bw:
5′-GGCCTGGGGAACGTGGA-3′
- Cacna2d1 Fw:
5′-ATGTGCTGGAGGATTATACCG-3′
- Cacna2d1 Bw:
5′-CCAAAGATAGACCATAAGGAAGG-3′
- Cacnb1 Fw:
5′-AGGGCTATGAGGTAACTGAC-3′
- Cacnb1 Bw:
5′-GGAAATCCTGCCATCAAACC-3′
- miR-103 Fw:
5′-AGCAGCATTGTACAGGGCTATGA-3′
COS cell culture and luciferase assay
COS-7 cells were cultured in Dulbecco's Modified Eagle's Medium. We used a dual reporter system, with an empty Firefly luciferase vector (pGL3, Promega) as transfection control. Normalized Renilla luciferase activity was expressed as a ratio of reporter to control vector activities. Cells were co-transfected with Renilla reporter and Firefly control plasmids using FuGene transfection medium (Roche). We used 150, 250, 500 or 750 ng of miR-103 plasmid, together with 50 ng of Cav1.2-LTC subunit reporter plasmid and 10 ng of pGL3 control plasmid (Promega). Forty-eight hours later, luciferase activities were quantified with the Dual reporter system (Promega) and normalized to control experiment.
Neuron culture and calcium imaging
Dissociated cultures were made from the lumbar spinal cord of E18 rat embryos. The embryos were delivered by caesarean section from deeply anaesthetized pregnant females. Primary spinal neurons were cultured in Neurobasal medium+2% B-27 serum-free supplement+1% GlutaMAX-I supplement+1% Penicillin/Streptomycin (Gibco). One week after plating, neurons were transfected using Lipofectamine 2000 (Invitrogen) with MiR-103, seed-mut-miR-103 plasmids or anti-Cav1.2-siRNAs, together with a mCherry-encoding plasmid to visualize transfected cells. Two days later, neurons were incubated with Fluo4-AM (5 μM; Invitrogen) for 10 min. After three washes in Tyrode solution, cultures were mounted in an open chamber on a Nikon Ti inverted microscope equipped with appropriate filter sets. After selecting a field of view containing an mCherry-positive cell, 25 Hz image stacks of the calcium signal were taken with a HQ-CoolSNAP digital camera (Roper Scientific) driven by Metamorph (Universal Imaging). Neuron depolarization was induced by KCl bath application (25 mM).
Animal model, intrathecal injections and behavioural tests
The experiments were done on adult Wistar rats (250–300 g). The right L5 and L6 spinal nerves were isolated and tightly ligated with a 7.0 silk thread. After complete haemostasis, the incision was sutured. Surgery was performed on all rats under gaseous anaesthesia with a mixture of halothane (5% for induction and 2% for maintenance) and a 1:1 flow ratio of air/O2. The rats resumed normal activity within 30 min after termination of the gaseous anaesthesia. The rats were placed in the testing cage 1 h before the test for habituation. The withdrawal threshold of the leg on the operated side was determined in response to mechanical stimuli applied to the plantar surface of the foot. The limb withdrawal threshold was measured by an electronic device (Bioseb) derived from von Frey filaments. Synthetic miRNAs or siRNAs (2 μg) were solubilized in 10 μl of i-Fect reagent (Neuromics) following Neuromics instructions and published protocol (Luo et al, 2005), and applied intrathecally. Thermal allodynia to cold stimulus was assessed by using the hot/cold-plate analgesia metre (Bioseb). Briefly, rats were placed into compartment enclosures on the cold surface of the plate, which is maintained at a temperature of 4±0.5°C. Latency for paw withdrawal is the mean of three measurements with a cutoff time set to 30 s to avoid paw damages.
Immunohistochemistry, in situ hybridization and electron microscopy
Rats were perfused with 4% paraformaldehyde. For electron microscopy, glutaraldehyde (0.01 M) was added to the fixative solution. The lumbar spinal cord was rapidly dissected out, and post-fixed for 2 h in 4% paraformaldehyde. Tissue samples were then processed for in situ hybridization and immunohistochemistry with light or electron microscopy. For immunolabelling of Cav1.2, α2δ1, β1, Cav2.2 and Cav3.2 calcium channels, we used anti-Cav1.2 antibodies (NeuroMab 75-053, monoclonal, 1/200), anti-α2δ1 antibodies (Alomone ACC-015, polyclonal, 1/100), anti-β1 antibodies (NeuroMab 75-052, monoclonal 1/100), anti-Cav2.2 antibodies (Calbiochem 681505, polyclonal, 1/100) and anti-Cav3.2 antibodies (Alomone ACC-025, polyclonal, 1/100). For measuring Cav1.2-LTC subunits expression, confocal images were taken so that on each picture control, neurons were present together with miR-103 over-expressing or knockdown neurons. Thus, to obtain the most accurate data, quantification was performed by normalizing labelling intensity to adjacent control neurons.
For miR-103 in situ hybridization, a double-digoxigenin LNA-probe (Exiquon) was used at a final 20 nM concentration. Hybridization was performed according to the manufacturer's instructions; Digoxigenin was revealed with a tyramide-based method. For double labelling combining in situ hybridization and immunohistochemistry, sections were processed for MAP2 detection after hybridization. Then, the double-digoxigenin-labelled probe was visualized. For electron microscopy, in situ protocol was adapted and probes were visualized with a silver-enhanced ultra-small gold particle strategy (gold-conjugated anti-digoxigenin antibody (1/50; Roche) silver enhancement kit (Nanoprobes Inc.)). Sections were embedded in Epon resin and observed in a Hitachi H7650 electron microscope.
Quantitative RT–PCR
Total RNA was purified with the TRI Reagent (Sigma-Aldrich) according to the manufacturer's instructions. For mRNAs, cDNA was synthesized from 1 μg of total RNA using RevertAid H Minus first-strand synthesis kit (Fermentas) and primed with oligo-dT primers. For miRNAs, cDNA was synthesized using Ncode first-strand synthesis kit (Invitrogen). PCR amplification was performed on a DNA Engine Opticon2 fluorescence detection System (MJResearch/Bio-Rad) with primer pairs designed to span exon boundaries and to generate amplicons of ∼70 bp. Primer sets for Sdha, Cacna1c, Cacna2d1, Cacnb1 and miR-103 were tested by qRT–PCR and gel electrophoresis for the absence of primer–dimer artefacts and multiple products. Triplicate qRT–PCR reactions were done twice for each sample, using transcript-specific primers (600 nM) and cDNA (10 ng) in a final volume of 10 μl. The DyNAmo™ SYBR Green qPCR kit (Finnzymes) was used with the following PCR amplification cycles: initial denaturation, 95°C for 15 min; followed by 40 cycles with denaturation, 95°C for 20 s and annealing extension, 61°C for 35 s. A dissociation curve was generated at the end of the 40th cycle to verify that a single product was amplified. The Ct value of each gene was normalized against that of Sdha. The relative level of expression was calculated using the comparative (2ΔΔCt) method (Livak and Schmittgen, 2001).
Supplementary Material
Acknowledgments
We are grateful to Dr Etienne Gontier and Sabrina Lacomme, Bordeaux Imaging Center, for technical help in electron microscopy. This work was funded by ARC/INCA (07/3D1616/IABC-23-7/NC-NG), ANR (ANR-07NEURO-015-01), Conseil Régional d’Aquitaine (2008/30/023), Institut UPSA de la Douleur.
Author contributions: AF conceived and conducted the experiments, analysed the results and wrote the paper. OT conceived and conducted the experiments and analysed the results. RBB, VR, MAP, SAS, GD, CL and AC conducted the experiments. ML and FN analysed the results and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aldrich BT, Frakes EP, Kasuya J, Hammond DL, Kitamoto T (2009) Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience 164: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altier C, Dale CS, Kisilevsky AE, Chapman K, Castiglioni AJ, Matthews EA, Evans RM, Dickenson AH, Lipscombe D, Vergnolle N, Zamponi GW (2007) Differential role of N-type calcium channel splice isoforms in pain. J Neurosci 27: 6363–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai G, Ambalavanar R, Wei D, Dessem D (2007) Downregulation of selective microRNAs in trigeminal ganglion neurons following inflammatory muscle pain. Mol Pain 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bauer CS, Nieto-Rostro M, Rahman W, Tran-Van-Minh A, Ferron L, Douglas L, Kadurin I, Sri Ranjan Y, Fernandez-Alacid L, Millar NS, Dickenson AH, Lujan R, Dolphin AC (2009) The increased trafficking of the calcium channel subunit alpha2delta-1 to presynaptic terminals in neuropathic pain is inhibited by the alpha2delta ligand pregabalin. J Neurosci 29: 4076–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourinet E, Alloui A, Monteil A, Barrere C, Couette B, Poirot O, Pages A, McRory J, Snutch TP, Eschalier A, Nargeot J (2005) Silencing of the Cav3.2 T-type calcium channel gene in sensory neurons demonstrates its major role in nociception. EMBO J 24: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplan SR, Pogrel JW, Yaksh TL (1994) Role of voltage-dependent calcium channel subtypes in experimental tactile allodynia. J Pharmacol Exp Ther 269: 1117–1123 [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K (2007) microRNA modulation of circadian-clock period and entrainment. Neuron 54: 813–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Yoon YW, Na HS, Kim SH, Chung JM (1994) Behavioral signs of ongoing pain and cold allodynia in a rat model of neuropathic pain. Pain 59: 369–376 [DOI] [PubMed] [Google Scholar]
- Davies A, Hendrich J, Van Minh AT, Wratten J, Douglas L, Dolphin AC (2007) Functional biology of the alpha(2)delta subunits of voltage-gated calcium channels. Trends Pharmacol Sci 28: 220–228 [DOI] [PubMed] [Google Scholar]
- Derjean D, Bertrand S, Le Masson G, Landry M, Morisset V, Nagy F (2003) Dynamic balance of metabotropic inputs causes dorsal horn neurons to switch functional states. Nat Neurosci 6: 274–281 [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA (2004) Specificity of microRNA target selection in translational repression. Genes Dev 18: 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch RE, Pajvani U, Fife K, Spotts JM, Greenberg ME (2001) Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science 294: 333–339 [DOI] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M (2010) Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E, Screpanti I, Bozzoni I, Gulino A (2008) Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J 27: 2616–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat P, Dobremez E, Bouali-Benazzouz R, Favereaux A, Bertrand SS, Kilk K, Leger C, Cazalets JR, Langel U, Landry M, Nagy F (2010) Knockdown of L calcium channel subtypes: differential effects in neuropathic pain. J Neurosci 30: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF (2005) MicroRNAs regulate brain morphogenesis in zebrafish. Science 308: 833–838 [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004) Human MicroRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A (2007) A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317: 1220–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Chung JM (1992) An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain 50: 355–363 [DOI] [PubMed] [Google Scholar]
- Kusuda R, Cadetti F, Ravanelli MI, Sousa TA, Zanon S, De Lucca FL, Lucas G (2011) Differential expression of microRNAs in mouse pain models. Mol Pain 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115: 787–798 [DOI] [PubMed] [Google Scholar]
- Li X, Gibson G, Kim JS, Kroin J, Xu S, van Wijnen AJ, Im HJ (2011) MicroRNA-146a is linked to pain-related pathophysiology of osteoarthritis. Gene 480: 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luo MC, Zhang DQ, Ma SW, Huang YY, Shuster SJ, Porreca F, Lai J (2005) An efficient intrathecal delivery of small interfering RNA to the spinal cord and peripheral neurons. Mol Pain 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews EA, Dickenson AH (2001) Effects of ethosuximide, a T-type Ca(2+) channel blocker, on dorsal horn neuronal responses in rats. Eur J Pharmacol 415: 141–149 [DOI] [PubMed] [Google Scholar]
- Moosmang S, Haider N, Klugbauer N, Adelsberger H, Langwieser N, Muller J, Stiess M, Marais E, Schulla V, Lacinova L, Goebbels S, Nave KA, Storm DR, Hofmann F, Kleppisch T (2005) Role of hippocampal Cav1.2 Ca2+ channels in NMDA receptor-independent synaptic plasticity and spatial memory. J Neurosci 25: 9883–9892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisset V, Nagy F (1999) Ionic basis for plateau potentials in deep dorsal horn neurons of the rat spinal cord. J Neurosci 19: 7309–7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermair GJ, Schlick B, Di Biase V, Subramanyam P, Gebhart M, Baumgartner S, Flucher BE (2010) Reciprocal interactions regulate targeting of calcium channel beta subunits and membrane expression of alpha1 subunits in cultured hippocampal neurons. J Biol Chem 285: 5776–5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh KW, Yeo JF, Ong WY (2011) MicroRNA changes in the mouse prefrontal cortex after inflammatory pain. Eur J Pain (in press, doi:10.1016/j.ejpain.2011.02.002) [DOI] [PubMed] [Google Scholar]
- Satoh J-i (2010) MicroRNAs and their therapeutic potential for human diseases: aberrant microRNA expression in Alzheimer's disease brains. J Pharmacol Sci 114: 269–275 [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439: 283–289 [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM (2003) Identification of Drosophila MicroRNA targets. PLoS Biol 1: E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schack D, Agostino MJ, Murray BS, Li Y, Reddy PS, Chen J, Choe SE, Strassle BW, Li C, Bates B, Zhang L, Hu H, Kotnis S, Bingham B, Liu W, Whiteside GT, Samad TA, Kennedy JD, Ajit SK (2011) Dynamic changes in the microRNA expression profile reveal multiple regulatory mechanisms in the spinal nerve ligation model of neuropathic pain. PLoS One 6: e17670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM (2008) Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet 82: 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ (2009) Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 284: 5731–5741 [DOI] [PubMed] [Google Scholar]
- Wibrand K, Panja D, Tiron A, Ofte ML, Skaftnesmo KO, Lee CS, Pena JT, Tuschl T, Bramham CR (2010) Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur J Neurosci 31: 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Lee MC, Momin A, Cendan CM, Shepherd ST, Baker MD, Asante C, Bee L, Bethry A, Perkins JR, Nassar MA, Abrahamsen B, Dickenson A, Cobb BS, Merkenschlager M, Wood JN (2010) Small RNAs control sodium channel expression, nociceptor excitability, and pain thresholds. J Neurosci 30: 10860–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.