Abstract
Bacterial promoters are recognized by RNA polymerase (RNAP) σ subunit, which specifically interacts with the −10 and −35 promoter elements. Here, we provide evidence that the β′ zipper, an evolutionarily conserved loop of the largest subunit of RNAP core, interacts with promoter spacer, a DNA segment that separates the −10 and −35 promoter elements, and facilitates the formation of stable closed promoter complex. Depending on the spacer sequence, the proposed interaction of the β′ zipper with the spacer can also facilitate open promoter complex formation and even substitute for interactions of the σ subunit with the −35 element. These results suggest that there exists a novel class of promoters that rely on interaction of the β′ zipper with promoter spacer, along with or instead of interactions of σ subunit with the −35 element, for their activity. Finally, our data suggest that sequence-dependent interactions of the β′ zipper with DNA can contribute to promoter-proximal σ-dependent RNAP pausing, a recently recognized important step of transcription control.
Keywords: promoter, RNA polymerase core, sigma subunit, transcription pausing, zipper domain
Introduction
The formation of promoter complex, a step that commits bacterial RNA polymerase (RNAP) to transcribe a gene, requires the RNAP specificity (σ) subunit (Burgess and Anthony, 2001; Borukhov and Severinov, 2002). Within the context of the RNAP holoenzyme, σ regions 2 and 4 (σR2 and σR4) make specific interactions with promoter elements located, correspondingly, around positions −10 and −35 relative to the transcription start site (Helmann and deHaseth, 1999; Burgess and Anthony, 2001; Borukhov and Severinov, 2002). A minor class of extended −10 promoters, instead of the −35 motif, rely on a TG motif located immediately upstream of the −10 element (Barne et al, 1997). The TG motif is also recognized by the σ subunit. σ Region 1.2 was also shown to make sequence-specific interactions with non-template nucleotide downstream of the −10 element (region referred to as ‘discriminator’ in stable RNA promoters; Haugen et al, 2006).
The available medium-resolution structure of Thermus aquaticus RNAP σA holoenzyme (EσA) complex with fork-junction DNA (Murakami et al, 2002), a synthetic nucleic acid substrate mimicking promoter DNA, as well as structural modelling results (Naryshkin et al, 2000; Murakami and Darst, 2003) show that in addition to the expected contacts with σ, promoter DNA is close to residues from various regions of large (β′ and β) RNAP core subunits throughout the complex. At the downstream ‘end’ of the open complex, the β′ jaw, a structural element that is part of a trough where the double-stranded DNA downstream of the catalytic centre binds, contributes to open complex stability by wrapping the downstream DNA and firmly securing it in the trough (Ederth et al, 2002). These interactions were proposed to contribute to species specificity of promoter utilization (Artsimovitch et al, 2000). At the upstream end, both specific and non-specific interactions of the α subunit C-terminal domains (αCTDs) with DNA upstream of the −35 promoter element have a strong stimulatory effect on promoter complex formation. Non-specific interactions of RNAP core with a region between positions −30 to −40 were also proposed (Nechaev and Geiduschek, 2006). DNA around the transcription initiation start point makes intimate contacts with β and β′ residues that form the RNAP catalytic centre (Murakami and Darst, 2003). Several structural elements of RNAP core also come in close proximity with promoter spacer located between the −10 and −35 promoter elements (Murakami et al, 2002). These structural elements are depicted in Figure 1A and include the β′ zipper, the β′ zinc-binding domain, and the β flap. We previously showed the critical role of the β flap for promoter selectivity (Kuznedelov et al, 2002b). The β flap contributes to promoter recognition indirectly, by enabling σR4 interaction with the −35 promoter element. The functional role, if any, of other RNAP core elements proximal to the spacer in promoter recognition is presently unclear.
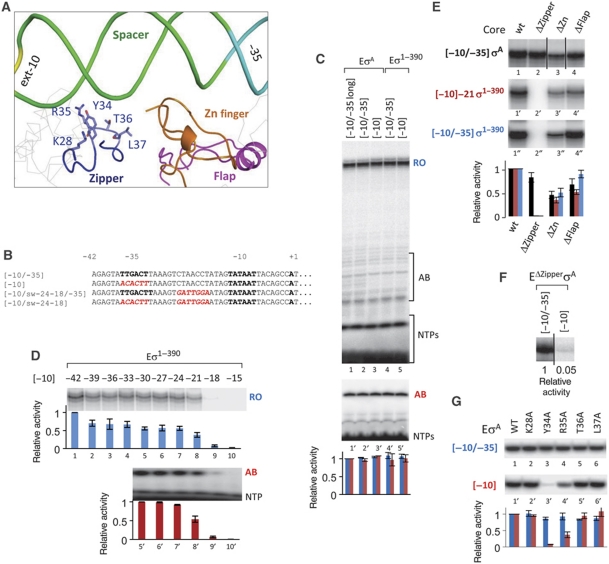
Figure 1.
The β′ zipper contributes to promoter utilization in the absence of −35–σR4 interactions. (A) T. aquaticus RNAP core domains that are close to the DNA in the promoter complex (Murakami et al, 2002) are shown as ribbons. β′ zipper amino acids analysed in the study are shown. σA Domains are in grey. (B) T7A1 derivatives used in our study (also see Supplementary Figure S1A). Functional elements are in bold and mutations are in red. (C) Run-off (lanes 1–5) and abortive (lanes 1′–5′) transcriptions by EσA and Eσ1–390 on promoters depicted above the gels. Promoter [−10/−35]long has 84 bp upstream of the transcription start site, other promoters-42 bp (B). RO—run-off product; AB—abortive product; NTP—unincorporated NTP. Here and after, histogram below the gels shows quantification of the corresponding lanes, error bars represent the standard deviation of at least three independent experiments. Blue bars—RO, red bars—AB. All activities were normalized to the activity of EσA on [−10/−35]long. (D) Transcription by Eσ1–390 on the truncated variants of [−10] promoter (Supplementary Figure S1A). Abbreviations as in (C). Activities were normalized to the activity of Eσ1–390 on [−10] (designated as −42) for run-off, and to the activity of Eσ1–390 on [−10]−30 for abortive transcription. (E) Transcription by wild-type (wt) and mutant RNAPs, lacking β′ zipper (ΔZipper), β′ zinc finger (ΔZn) or β flap (ΔFlap), with σ subunits and on promoter depicted to the left of each gel. Here and after, black vertical lines separate lanes originating from the same gel that were brought together. In the histogram: black bars—σA holoenzymes on [−10/−35]; red bars—σ1–390 holoenzymes on [−10]−21; blue bars—σ1–390 holoenzymes on [−10/−35]. Activities of mutant enzymes were normalized to the activity of wild-type RNAP on corresponding promoter (see text for details). (F) Activity of EΔZipperσA in the presence and in the absence of the −35 element. (G) Transcription by σA holoenzymes bearing single-alanine substitution in the β′ zipper, depicted above the gels on [−10/−35] and [−10] promoters. Quantification as in (E). Blue bars—transcription on [−10/−35] and red bars—transcription on [−10].
In this work, we show that the β′ zipper, which is part of the evolutionarily conserved segment B present in the largest subunits of all multi-subunit RNAPs, is directly involved in promoter recognition. Moreover, we show that, depending on the spacer sequence, interactions of the β′ zipper with the spacer can substitute for σR4 interactions with the −35 element during the open complex formation. These results highlight the unexpected complexity of the process of bacterial promoter recognition and suggest novel ways in which promoter specificity of bacterial RNAP can be regulated.
Results
Interaction of RNAP with DNA around position −21 of the spacer
The strategy of our study was to search for previously unrecognized interactions of RNAP with DNA by sequentially removing known interactions. We analysed transcription by T. aquaticus holoenzyme, EσA, on derivatives of strong −10/−35 class T7A1 promoter bearing the consensus −10 element (here this promoter is referred to as [−10/−35]; Figure 1B; Supplementary Figure S1A). We analysed both abortive and run-off transcription and obtained essentially the same results, indicating that promoter escape by T. aquaticus EσA is independent on the strength of promoter interactions, at least on promoters studied here (see Supplementary data for details). Transcription was performed under conditions of excess (five-fold) of promoter fragments over the enzyme. Given that T. aquaticus RNAP does not form stable open promoter complexes even on strong promoters (Kuznedelov et al, 2003; Kulbachinskiy et al, 2004; Schroeder and deHaseth, 2005), transcription in such conditions reported on the overall efficiency of closed and/or open complexes formation. To further distinguish between closed and open complexes formation, we used DNase I footprinting and KMnO4 probing.
Interactions of αCTDs with DNA 45–80 bp upstream of the transcription initiation start point strongly influence promoter utilization in E. coli (Estrem et al, 1999). In the case of T. aquaticus EσA, αCTDs do not have such a role (Wada et al, 2000). Consistently, the upstream truncation of the promoter fragment from position −85 to position −42 did not influence promoter activity (Figure 1C, lanes 1 and 2). Next, we removed the −35 promoter element by changing it to a non-functional sequence and generating the [−10] promoter fragment (Figure 1B). Curiously, removal of the −35 element had no effect on promoter activity (Figure 1C, lanes 2 and 3; see Supplementary data for details). Since the TG motif characteristic of extended −10 promoters (Barne et al, 1997) is absent from the [−10/−35] promoter and its derivatives, the result may be explained by postulating that non-specific favourable interactions of σR4 with DNA suffice for [−10] promoter activity. To test this idea, experiments were repeated with RNAP holoenzyme reconstituted from the wild-type core and σA mutant lacking region 4.2 (σ1–390). Unexpectedly, as seen from Figure 1C (lanes 4 and 5), the mutant holoenzyme was active on both [−10/−35] and [−10] promoters (see Supplementary data for details). Therefore, we conclude that interaction(s) other than the interaction of σR4 with DNA is responsible for [−10] promoter activity.
To identify DNA regions important for [−10] promoter utilization, derivatives progressively truncated from the upstream end (Supplementary Figure S1A) were tested in transcription with Eσ1–390 (Figure 1D). As can be seen, the mutant enzyme was still able to transcribe from promoters containing as little as 21 bp of DNA upstream of the transcription start point ([−10]−21 promoter; Figure 1D, lane 8). Further deletion (to position −18) abolished promoter activity ([−10]−18 promoter; Figure 1D, lane 9). This effect was not due to altered promoter escape since the abortive initiation assay gave essentially the same result (Figure 1D, lanes 5′–10′). Therefore, the experiment indicates that RNAP interactions with DNA at/or immediately downstream of position −21 contribute to promoter utilization. The slight decrease in promoter activity upon the truncation to position −39 (Figure 1D, compare lanes 1 and 2) can be explained by the elimination of the proposed non-specific interaction of RNAP core with this region (Nechaev and Geiduschek, 2006).
The β′ zipper is required for promoter utilization in the absence of specific interactions of σR4 with the −35 element
To explain our data, we hypothesized that there exists a domain of RNAP that contacts DNA on deeply truncated promoter fragments. This interaction appears to involve promoter spacer at/or around position −21, since truncation beyond this point destroys promoter activity, presumably by preventing the interaction. On the basis of structural considerations, the β flap, the β′ zinc-binding domain, or the β′ zipper could interact with spacer DNA (Figure 1A). We tested transcription by RNAPs lacking these domains: EΔFlap, EΔZn, and EΔZipper, respectively (Figure 1E). As above, the experiment was done in the five-fold molar excess of the promoter over RNAP. Differences in transcription by various RNAPs can, therefore, reflect both the efficiency of promoter utilization and/or RNAP-specific activity. In order to compare transcription by mutant enzymes on different templates between each other, their activities were normalized to the activity of EσA (or Eσ1–390) on each template (Figure 1E, histogram). The activities of mutant holoenzymes reconstituted with σA on the [−10/−35] promoter are shown in Figure 1E (lanes 1–4). Next, wild-type and mutant RNAP cores were combined with σ1–390, and the resultant holoenzymes were tested using the shortest active truncated promoter [−10]−21 as a template (Figure 1E, lanes 1′–4′). As can be seen, deletion of the β flap or the β′ zinc-binding domain had no significant effect on transcription from [−10]−21 (lanes 3′ and 4′). In contrast, removal of the β′ zipper abolished transcription (lane 2′), indicating that the β′ zipper is essential for utilization of the truncated promoter. This result suggests that the β′ zipper is also responsible for transcription activity of Eσ1–390 on full-length [−10/−35] promoter (Figure 1C, lane 4). Indeed, EΔZipperσ1–390 but not other double mutant RNAP holoenzymes was inactive on [−10/−35] (Figure 1E, compare lanes 1″–4″). The result further suggests that activity of EσA on promoter lacking the −35 element was also determined by the β′ zipper (Figure 1C, lane 3). Indeed, EΔZipper reconstituted with full-length σA was inactive on the [−10] promoter (Figure 1F). Note that, as tested in elongation complexes, kinetics of nucleotide addition and specific activities of wild-type and EΔZipper core enzymes were similar (Supplementary Figure S2A). This indicates that the observed differences in activities of EσA and EΔZipperσA indeed reflect differences in promoter utilization.
Further mapping has revealed that highly conserved Y34 and R35 (Supplementary Figure S3A) of the β′ zipper are responsible for its function (Figure 1G). In the crystal structure of the promoter complex, Y34 and R35 are in contact distance from spacer DNA (Figure 1A), suggesting that the β′ zipper may indeed function through direct interactions with promoter DNA around position −21.
The β′ zipper stabilizes promoter complexes
We were interested to determine which step of transcription initiation is affected by the deletion of the β′ zipper. We examined closed and open complexes formation using DNAse I footprinting (Figure 2A) and probing with KMnO4 (Figure 2B), respectively. KMnO4 probing was done at equilibrium conditions, in the absence of heparin (recall that T. aquaticus RNAP open promoter complexes are relatively unstable). As can be seen, EσA formed closed and open promoter complexes on both [−10/−35] (Figure 2A, lane 3 and Figure 2B, lane 3, respectively) and [−10] (Figure 2A, lane 6 and Figure 2B, lane 7, respectively) promoters. EΔZipperσA was able to form open (Figure 2A, lane 4) and closed (Figure 2B, lane 4) complexes on the [−10/−35] promoter. However, in the absence of the −35 element, EΔZipperσA failed to form either open (Figure 2B, lane 8) or closed (Figure 2A, lane 7) promoter complex. These results are consistent with the results of transcription assays with EσA and EΔZipperσA on [−10/−35] and [−10] promoters (Figure 2C), and indicate that the lack of activity of EΔZipperσA in the absence of specific interactions of σR4 with the −35 element is due to the inability of mutant enzyme to form closed promoter complex.
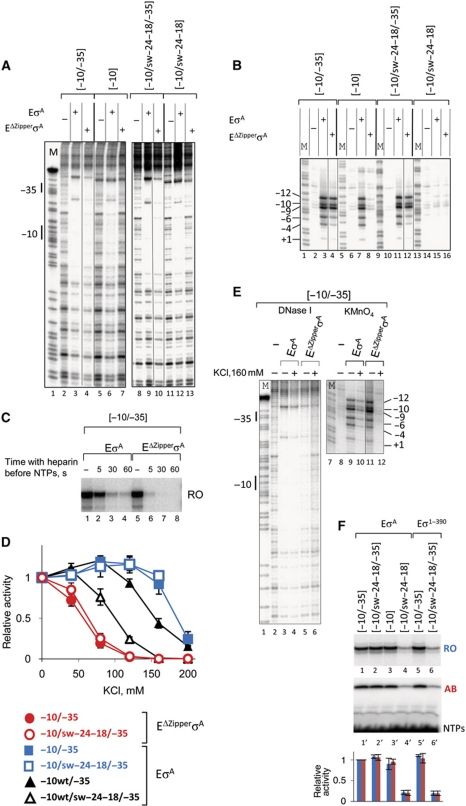
Figure 2.
Roles of the β′ zipper and the Z-element of promoters in promoter utilization. (A) The closed complexes formed at 30°C on templates (depicted above the gels), labelled at the 5′-end of template strand, by EσA and EΔZipperσA, were digested by DNase I. As a control, the corresponding DNA was digested without addition of RNAP. As a marker (M) one of the A+G reactions from (B) was used. Positions of −10 and −35 elements are shown to the left of the gel. (B) The open complexes formed by EσA and EΔZipperσA at 60°C on templates (depicted above the gel) labelled at the 5′-end of template strand were probed with KMnO4. As a control, the corresponding DNA was KMnO4 treated without addition of RNAP. As a marker, the A+G reaction on the corresponding templates was used. Modified positions are shown to the left of the gel. (C) Run-off transcription by EσA and EΔZipperσA on [−10/−35]. Promoter complexes were challenged with 0.1 mg/ml heparin for various times before the addition of NTPs. Note that transcription elongation is not affected by heparin (Supplementary Figure S2B). (D) Transcription by EσA and EΔZipperσA on promoters depicted below the plots. Promoter complexes were formed at various concentrations of KCl before NTPs were added. Transcription elongation is not affected by KCl concentrations used (Supplementary Figure S2B). For each RNAP–promoter pair, activities were normalized to the activity without KCl, which was taken as 1. (E) Closed and open promoter complexes formed by EσA and EΔZipperσA on [−10/−35] in different KCl concentrations were analysed as in (A) and (B). (F) Transcription by EσA and Eσ1–390 on the promoters depicted above the gels. Abbreviations as in Figure 1C. Histogram below the gels shows quantification of the corresponding lanes (blue bars—RO (lanes 1–6) and red bars—AB (lanes 1′–6′)). All activities were normalized to the activity of EσA on [−10/−35].
We next tested whether the β′ zipper contributes to promoter complexes formation in the presence of specific interactions of σR4 with the −35 element. We challenged transcription by EσA and EΔZipperσA on the [−10/−35] promoter with DNA competitor heparin and increased ionic strength. Given that transcription elongation was not affected by the concentrations of heparin and KCl used (Supplementary Figure S2B), the assay directly addresses the stability of promoter complexes. As shown in Figure 2C and D, transcription by EΔZipperσA was much more sensitive to heparin and high ionic strength treatment than transcription by the wild-type EσA. DNase I and KMnO4 analysis of promoter complexes also revealed that both closed and open promoter complexes formed by EΔZipperσA on [−10/−35] were far less resistant to salt challenge than those formed by EσA (Figure 2E). However, due to instability of promoter open complexes formed by T. aquaticus RNAP we cannot distinguish whether the effect of increased salt concentration on open complexes was direct, or was caused by destabilization of closed complexes. The results indicate that the β′ zipper contributes to stabilization of closed (and possibly open) promoter complexes both in the presence and in the absence of specific interactions of σR4 with the −35 element.
Taken together, the fact that the function of the β′ zipper is determined by two amino acids that face the promoter spacer (Figure 1G), the results on promoter shortening (Figure 1D and E), and the data on the role of the β′ zipper in stabilization of promoter complexes lend strong support to idea that the β′ zipper acts through direct contacts with promoter spacer. The β′ zipper may either provide additional favourable contacts with promoter spacer, or it may change the structure of promoter DNA, and thus optimize the recognition of the −10 promoter element by σ subunit and/or improve downstream RNAP–promoter interactions. In this regard, it is noteworthy that the DNase I footprint of closed complexes formed by EσA, but not by EΔZipperσA, contains a DNase I hypersensitive band upstream of the −10 element both in the presence and absence of the −35 element (Figure 2A, lanes 3 and 6), suggesting that, upon the interaction with the β′ zipper, promoter spacer indeed undergoes a conformational change.
The role of promoter spacer sequence in the β′ zipper function
Though our results suggest that the β′ zipper interacts with promoter spacer and participates in stabilization of promoter complexes, it is unclear if the contribution of the β′ zipper to promoter complex formation depends on the sequence of the spacer or is sequence independent. To address this issue, we changed (‘switched’) the sequence between positions −24 and −18 of [−10/−35] and [−10] promoters to a complementary sequence, and tested the activity of EσA and Eσ1–390 on resulting [−10/sw-24–18/−35] and [−10/sw-24–18] promoters (Figure 1B). As can be seen from Figure 2F, switching of the spacer sequence in the absence of either the −35 element (lanes 4′ and 4′) or σR4 domain (lanes 6 and 6′) decreased the level of transcription 4–5-fold. In the presence of the −35 element, transcription was not affected by the switch in the spacer sequence (Figure 2F, lanes 2 and 2′).
We examined what stage of transcription initiation is affected by a change in the spacer sequence in the absence of the −35 element. Permanganate probing and DNase I footprinting showed that EσA failed to open promoter DNA on [−10/sw-24–18] promoter (Figure 2B, lane 15), while the formation of closed complex was not affected (Figure 2A, lane 12). Note that closed complex on [−10/sw-24–18] persists in the conditions of permanganate probing (Supplementary Figure S4), indicating that the ‘switch’ of the spacer sequence affects only promoter opening. Though the switch in the spacer sequence did not affect the level of transcription from the [−10/−35] promoter, we found that in a background of non-consensus −10 element the spacer sequence between positions −24 and −18 contributed to promoter strength even in the presence of specific −35–σR4 interactions (Figure 2D, triangles). These results indicate that the sequence between positions −24 and −18 is important for open complex formation when RNAP interactions with promoter elements are weak. In the background of a strong −10 element, this sequence can substitute for the −35 element.
Switching of the spacer sequence could affect promoter strength by changing the curvature of promoter DNA, which, in turn, may influence the recognition of the −10 element or RNAP contacts with downstream DNA (Hook-Barnard and Hinton, 2009). However, in silico curvature prediction showed only small deviations in curvature upon the change of spacer sequence (Supplementary Figure S5A). Importantly, though EΔZipperσA formed very unstable promoter complexes (Figure 2C–E), its activity was the same on [−10/−35] and [−10/sw-24–18/−35] promoters, that is, was not affected by the switch of the spacer sequence (Figure 2A, lanes 4 and 10; Figure 2B, lanes 4 and 12; Figure 2D, compare filled and empty circles; see also experiments with E. coli RNAP below). These results suggest that the change of the spacer sequence, on its own, does not seem to significantly influence the strength of promoter.
The Z-element of promoters
Taking into account the proposed interaction of the β′ zipper with DNA around the position −21, our results suggest that the contribution of the sequence between positions −24 and −18 to promoter opening is mediated by the β′ zipper. The β′ zipper may either directly recognize the T7A1 spacer sequence around position −21 or ‘sense’ specific local structure of spacer DNA in this region. We name the spacer region that interacts with the β′ zipper ‘Z-element’ (for zipper). To determine if the β′ zipper and the Z-element contribute to promoter utilization by RNAPs other than T. aquaticus, we tested E. coli RNAP (EcEσ70) on the [−10/−35] promoter and its derivatives. Only abortive initiation was analysed, since EcEσ70 forms much more stable promoter open complexes than T. aquaticus enzyme, and biases arising at the stage of promoter escape were, therefore, expected. Transcription by EcEσ70 was more sensitive to removal of the −35 element, which led to ∼3-fold decrease in the level of transcription (Figure 3A). Interestingly, a switch of the Z-element also decreased transcription (∼2-fold) even in the presence of the −35 element (Figure 3A). Removal of both elements had a cumulative effect, and EcEσ70 utilized [−10/sw-24–18] ∼6 times less efficiently than [−10/−35] (Figure 3A). Next, we tested if the Z-element affected EcEσ70 initiation through the β′ zipper. Given that E. coli holoenzyme lacking the β′ zipper was inactive in transcription initiation, we used RNAP bearing a double alanine substitution of conserved tyrosine and arginine, which determine the function of the β′ zipper in promoter utilization by T. aquaticus RNAP (Figure 1G; β′Y47A/R48A in E. coli numbering, corresponding to β′Y34A/R35A of T. aquaticus). As can be seen from Figure 3A, removal of the Z-element, either in the presence or in the absence of the −35 element, had no effect on EcEY47A/R48Aσ70 activity. Removal of the −35 element decreased the level of transcription ∼8-fold irrespective of the presence or the absence of the Z-element (Figure 3A). These results suggest that the Z-element participates in promoter utilization by E. coli RNAP and acts through the β′ zipper, supporting the results obtained with T. aquaticus RNAP. We also tested Bacillus subtilis holoenzyme, BsEσA. As can be seen from Figure 3B, BsEσA behaved similarly to EcEσ70 with respect to the Z and the −35 elements contributions to promoter activity. These results suggest that participation of the Z-element and the β′ zipper in promoter utilization may be a ubiquitous phenomenon among bacteria, though the extent of the actual contribution may be species specific.
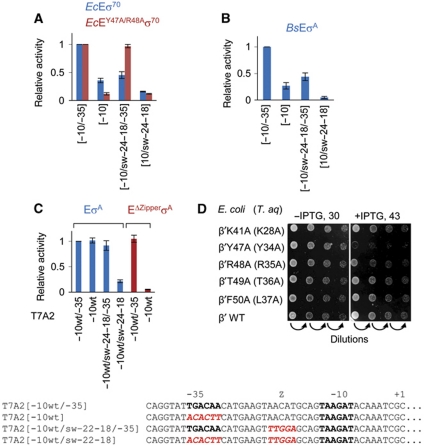
Figure 3.
The Z-element of promoters. (A) Abortive transcription by E. coli EcEσ70 and EcEY47A/R48Aσ70 on promoters depicted under histogram was quantified and normalized to the activity of each enzyme on [−10/−35] promoter. (B) Abortive transcription by B. subtilis holoenzyme, BsEσA, on promoters depicted under histogram was quantified and normalized to the activity on [−10/−35] promoter. (C) Transcription by EσA and EΔZipperσA on T7A2 derivatives depicted below the histogram (promoter sequences are shown at the bottom of the panel). All activities by both enzymes were normalized to the activity of EσA on T7A2[−10wt/−35]. (D) Tyr47 and Arg48 (corresponding to Tyr34, Arg35 of T. aquaticus) of β′ zipper are important in vivo. E. coli cells with chromosomal rpoC that is inactive at 43°C were transformed with plasmids carrying wild-type or mutant rpoC genes under IPTG-inducible promoters. Mutations in the β′ zipper are depicted to the left of the photographs of petri dishes (corresponding mutations of T. aquaticus β′ are in brackets). The serial dilutions of cells were grown at permissive (30°C) and restrictive (43°C) temperatures.
Changes of small blocks of the spacer sequence (Supplementary Figure S5B) allowed us to localize the Z-element to a region extending from position −22 to position −18 of T7A1 promoter (AACCT). We noted that another well-characterized strong promoter T7A2, designated here as T7A2[−10wt/−35] (Figure 3C, Supplementary Figure S1B), while having no sequence similarity with T7A1, has an AACAT sequence in the −22 to −18 region, resembling the Z-element of T7A1. We tested the functionality of putative T7A2 Z-element with T. aquaticus RNAP. As can be seen from Figure 3C, derivatives of T7A2[−10wt/−35] acted in the same manner as [−10/−35] promoter derivatives with respect to Z-element function: (i) The Z-element was able to support transcription and was required on T7A2 derivative lacking the −35 element (T7A2[−10wt]); (ii) EΔZipperσA, while being as active as EσA on T7A2[−10wt/−35], failed to transcribe from T7A2[−10wt]. These results suggest the existence of a novel class of promoters that use interactions of the β′ zipper with the Z-element instead of and/or along with −35–σR4 interactions to facilitate promoter opening.
We analysed several single base-pair substitutions in the T7A1 Z-element of the [−10] promoter with T. aquaticus RNAP. Most of substitutions led to decreased level of transcription by EσA in the absence of the −35 element (Supplementary Figure S5C), suggesting that the sequence of the T7A1 Z-element may be close to consensus. However, the effects were modest (<2-fold decrease), suggesting that Z-element consensus may be ‘loose’. Though bioinformatics analysis of E. coli and B. subtilis promoters with known transcription start sites revealed that a significant proportion (∼10%) of them contain sequences that resemble the T7A1 Z-element (Supplementary data; Supplementary Tables S1 and S2), further analysis is required to determine functionality of these putative Z-elements.
Given that our results suggest that the β′ zipper is required for stabilization of promoter complexes, we expected these interactions to be important for cell viability. We, therefore, investigated the importance of proposed interaction of β′ zipper with promoter spacer in vivo. Mutations introducing single-alanine substitutions of β′ Lys41, Tyr47, Arg48, Thr49, Phe50 (corresponding to β′ Lys28, Tyr34, Arg35, Thr36, and Leu37 of T. aquaticus; Figure 1G), were created in plasmid-borne E. coli rpoC (codes for RNAP β′). Plasmids expressing mutant β′ subunits from an IPTG-inducible promoter were transformed into E. coli cells harbouring a chromosomal copy of rpoC coding for temperature-sensitive β′ (which is inactivated at 43°C). We analysed the ability of these plasmids to complement a temperature-sensitive phenotype of host cells (Figure 3D). As can be seen, growth of cells expressing β′ with Y47A substitution was strongly diminished at restrictive temperature. Complementation by β′ with R48A substitution was weaker than by the wild-type β′, though the effect was more moderate than that of the Y47A substitution. The β′ subunits with remaining substitutions complemented the temperature-sensitive phenotype as efficiently as wild-type β′ subunit (compare with the in vitro results in Figure 1G). Promoter escape, elongation, and termination by E. coli RNAP were not affected by Y47A and R48A substitutions in vitro (Supplementary Figure S6A and B), supporting the idea that in vivo effects of these mutations were caused by deficiencies in promoter utilization.
β′ Zipper–Z-element interactions affect σ-dependent promoter-proximal pausing
In addition to its essential role in transcription initiation, the RNAP σ subunit has a role in transcription elongation by causing transcriptional pausing in initially transcribed region of some promoters (Ring et al, 1996; Brodolin et al, 2004; Nickels et al, 2004; Hatoum and Roberts, 2008). The pause occurs when σ binds to elongating RNAP core and σR2 establishes specific contacts with the −10-like sequence in promoter-proximal transcribed DNA. Thus, the paused complex partially resembles the promoter open complex (Brodolin et al, 2004). We hypothesized that the β′ zipper may interact with DNA upstream of pause-inducing −10-like element during σ-dependent promoter-proximal pausing, and thus affect the pause efficiency.
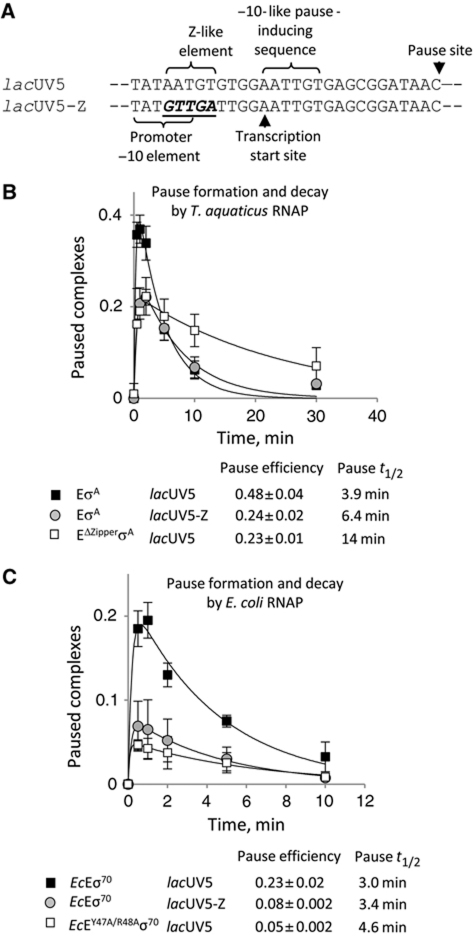
We used a transcription template carrying the lacUV5 promoter, which contains a −10-like σ-dependent pause-inducing sequence between positions +1 and +6 (Brodolin et al, 2004; Nickels et al, 2004; Figure 4A). Intriguingly, 4 bp upstream of the −10-like element, a sequence that resembles the Z-element (AATGT) is present. To test if the Z-like sequence is functional, we prepared a lacUV5 derivative that lacked this element (lacUV5-Z, Figure 4A) and measured efficiency of σ-dependent pausing on lacUV5 and lacUV5-Z by T. aquaticus and E. coli RNAPs. EσA paused two times (Figure 4B), while EcEσ70—three times less efficiently on lacUV5-Z than on lacUV5 (Figure 4C). To test if the action of the Z-like element is mediated by the β′ zipper, we examined pause formation by EΔZipperσA and EcEY47A/R48Aσ70. Pause efficiencies by mutant T. aquaticus (Figure 4B) and E. coli (Figure 4C) enzymes on lacUV5 were similar to those of corresponding wild-type RNAPs on lacUV5-Z, indicating that the effect of the Z-like element of pause-inducing sequence is abolished by removal of or mutations in the β′ zipper. We could not check pausing by mutant RNAPs on the lacUV5-Z template, because the mutant enzymes were inactive on this promoter due to downmutation in the −10 promoter element introduced by alterations in the Z-like sequence. The moderate effect of the Z-like element on pause efficiency may have been caused by the fact that this element lacked the equivalent of −20C, which could not be introduced into the sequence since the change destroyed the lacUV5 promoter activity (Supplementary Figure S7).
Figure 4.
β′ zipper and the Z-element contribute to the efficiency of σ-dependent elongation pausing. (A) The templates that were used in our study of promoter-proximal pausing. The functional elements are designated and the changes to the Z-like element are in bold and underlined. (B, C) Plots represent σ-dependent promoter-proximal pause formation and decay on the templates and by the enzymes indicated. Error bars show standard deviation between three independent experiments. The calculated efficiencies of pauses are presented (with standard errors of data fitting). Half-life times were calculated from the predicted rates of pause decay.
We cannot exclude a possibility that the Z-like element acts independently of the β′ zipper by influencing the recognition of the −10-like sequence by σ subunit. The results on promoter utilization, however, favour the possibility that the Z-element acts through interaction with the β′ zipper. As in the case of promoter utilization, during pausing, the proposed interaction of the β′ zipper with the Z-like element may increase the pause efficiency by either providing additional contacts of RNAP with the pause signal, or optimizing recognition of the −10-like element of the pause-inducing sequence by σR2 (via structural changes in DNA). Though the molecular details remain to be understood, our results suggest that interactions of the β′ zipper with DNA can influence transcription elongation rate by affecting the efficiency of σ-dependent pausing by bacterial RNAPs.
Note that the half-life of σ-dependent pausing by EΔZipperσA was longer than that of EσA (3.5 times, Figure 4B). This suggests that the β′ zipper may influence stabilization of the pause. The half-life of pausing by EcEY47A/R48Aσ70 increased only slightly, suggesting that deletion of β′ zipper amino acids other than conserved tyrosine and arginine is responsible for the observed effect. At present, we cannot explain the mechanism behind increased pause half-life of EΔZipperσA, but the finding that the σ-dependent pause on lacUV5 promoter is stabilized via backtracking (Brodolin et al, 2004), suggests that EΔZipperσA may be more prone to backtracking on the pause signal.
Discussion
Sequence-specific interactions with promoters are thought to be a prerogative of bacterial RNAP promoter specificity factor, the σ subunit, and the only known sequence-specific interaction of the core enzyme with promoters is that of CTDs of the α-subunits of RNAP with the Upstream Promoter element. RNAP core parts contacting downstream DNA of promoter were suggested to contribute to specificity in promoter utilization (Artsimovitch et al, 2000). Non-specific interactions were reported for various parts of RNAP core (see Introduction). The principal result of this work is the demonstration that a structural element of the RNAP core largest subunit, the β′ zipper, interacts with the promoter spacer and the outcome of this interaction can be modulated by the sequence of the spacer.
The β′ zipper contributes to promoter closed (and possibly open) complex stabilization. By interacting with the spacer around position −21, the β′ zipper either provides additional contacts between RNAP and promoter DNA, or optimizes interactions of σ and/or core with DNA downstream of the spacer. The presence of the Z-element in the region of spacer facilitates promoter opening and can even substitute for the canonical interaction of σ with the −35 element. The mechanism of the Z-element contribution to promoter opening remains to be understood. Since Z-element does not seem to have any significant effect on promoter structure or activity on its own but requires intact β′ zipper, direct favourable protein–DNA interaction is possible. Structural analysis supports this hypothesis. The β′ zipper may either directly recognize Z-element sequence or, instead, ‘sense’ local DNA conformation determined by this sequence. It is also possible that the Z-element is not recognized directly, but its sequence modulates the outcome of the β′ zipper interaction with the spacer on promoter opening. In the latter mechanism, the Z-element may determine the properties of the spacer (bendability, kinking, etc.) which, upon interaction with the β′ zipper, promote localized melting downstream of the Z-element. The effect of spacer sequence on the promoter strength was demonstrated for the lac promoter by selecting spacer sequences that made the activity of promoter activator independent (Liu et al, 2004). Our results open a possibility that differences in the spacer region may be either sensed by the β′ zipper, or may modulate the β′ zipper contribution to promoter activity.
In the presence of a strong −10 element, the proposed β′ zipper–Z-element interaction can substitute for the σR4 interaction with the −35 element during the promoter open complex formation. In the background of a weak −10 promoter element, both the −35 and the Z-elements contribute to promoter utilization. In this sense, the Z-element is similar to the TG motif of the ‘extended −10’ class promoters in that it facilitates open complex formation in the absence of or along with the −35 element. Therefore, the results suggest an existence of promoters of a new type that rely on the Z-element instead of and/or along with the −35 element (−10/Z and/or −10/Z/−35). Further investigations are required to determine the consensus sequence(s) of the Z-element and the mode of its recognition by the β′ zipper.
The interplay of specific interactions of the β′ zipper and σR4 with their respective elements may potentially be a target for genetic regulation. Curiously, in the structure of holoenzyme bound to a promoter DNA fragment containing a sequence similar to the Z-element, AAATT, 4 bp upstream of the −10 element, the σR4 recognition helix is shifted 6 Å upstream and does not interact with the −35 element specifically (Murakami et al, 2002).
RNAP stalling at promoter-proximal sequences is now believed to be a common way of transcription regulation in bacteria (Artsimovitch, 2008; Hatoum and Roberts, 2008). Though known σ-dependent pauses are close to promoters, the existence of distantly located σ-dependent pauses has also been proposed (Mooney and Landick, 2003; Deighan et al, 2011). Our results indicate that specific interactions of the β′ zipper with DNA can influence elongation rates by contributing to the efficiency of σ-dependent pauses.
The β′ zipper is highly conserved among bacteria (Supplementary Figure S3A), suggesting the conservation of the mechanisms of its interactions with DNA and its roles in transcription. In spite of a low level of sequence identity, the structural similarity of zipper domains of the largest subunits of bacterial, eukaryotic, and archaeal RNAPs (Hirata and Murakami, 2009; Supplementary Figure S3B) may indicate that interactions of zipper domain with promoter DNA may be utilized by RNAPs from all domains of life.
Materials and methods
Templates and proteins
Transcription templates were made by PCR and purified from agarose gel. Wild-type and mutant recombinant T. aquaticus core RNAPs and σA were purified as described (Kuznedelov et al, 2003). RNAPs lacking the β′ zipper (β′ residues 27–42) or the lid (β′ residues 526–539) domains were obtained as described (Zenkin et al, 2006). RNAP lacking the β flap domain (β residues from 757–786 were substituted for LeuGlu) was kindly provided by K Kuznedelov. RNAP lacking β′ zinc-binding domain was made by genetic substitution of β′ residues 54–82 for Gly-Gly linker. RNAPs with mutations in the β′ zipper were constructed by substitutions of single amino acids with alanines. T. aquaticus σA lacking domain 4.2 (σ1–390) was kindly provided by A Kulbachinsky. Wild-type and mutant E. coli RNAP core enzymes were isolated according to Kashlev et al (1996). E. coli σ70 was isolated as described (Borukhov and Goldfarb, 1993). To obtain E. coli RNAP cores bearing single-alanine substitutions of β′ Lys41, Tyr47, Arg48, Thr49, Phe50, double alanine substitution Y47A/R48A and deletion of the β′ zipper (positions β′ 40–55 replaced by GlyGly), the mutations were introduced in the β′ subunit coded under inducible promoter in plasmid pRL663 (Severinov et al, 1995). B. subtilis holoenzyme was isolated according to Polyakov et al (1995).
In vitro transcription
All reactions for E. coli and B. subtilis RNAP were done at 37°C and for T. aquaticus RNAP at 65°C. Reactions contained 20 nM holoenzyme in the case of B. subtilis, or 20 nM RNAP core and 100 nM σ in the case of T. aquaticus and E. coli, and 100 nM promoter fragment in transcription buffer (20 mM Tris–HCI, pH 7.9, 10 mM MgCl2). For salt stability experiments, KCl was added to the final concentrations specified in figures. For stability of complexes to heparin treatment, 0.1 mg/ml heparin was added for times specified in the figure legend before nucleotides addition. Transcription was initiated with 100 μM dinucleotide: CpA for T7A1 promoter derivatives and CpG for T7A2 derivatives. For abortive initiation 40 μM [α-32P] radiolabelled NTP (7.5 Ci/mmol) specified by the next position of the template was added. For run-off transcription all four NTPs, one of which was [α-32P] radiolabelled (7.5 Ci/mmol), were added to final concentration of 40–100 μM. Reactions were allowed to proceed for 2.5 min, and were terminated by the addition of an equal volume of loading buffer containing 8 M urea. Reaction products were resolved by electrophoresis in denaturing polyacrylamide gel, visualized by PhosphorImager (GE Healthcare), and quantified using the ImageQuant software (GE Healthcare). All experiments were repeated at least three times.
DNase I footprinting and KMnO4 probing
The reactions contained equimolar amounts of DNA template labelled at the 5′-end of the template strand and holoenzyme in the transcription buffer. Closed complexes footprinting was performed at 30°C (except for experiment in Supplementary Figure S4, see the legend for details) (given that the T. aquaticus RNAP opens promoter at temperatures above 45°C; Kuznedelov et al, 2002a) with one unit of DNase I (Roche) for 30 s. Reactions were stopped by addition of formamide, urea, and EDTA containing buffer. Permanganate probing was performed at 60°C for 10 s, without addition of heparin, as described (Kuznedelov et al, 2002a). Products were separated and analysed as described (Kuznedelov et al, 2002a).
In vivo experiments
pRL663 plasmids carrying wild-type and mutant rpoC genes (from above) were used to transform E. coli 397C cells (Christie et al, 1996), bearing genomic rpoC that codes for a temperature-sensitive β′ subunit. The transformants were grown in LB at 30°C (permissive temperature) in the presence of 100 μg/ml of ampicillin. Exponentially growing cells (A600 of ∼0.2) were diluted serially and plated by 2 μl spots on the minimal media (M9) agar, containing 100 μg/ml arginine, 0.5 μg/ml thiamine, 100 μg/ml ampicillin, with or without 1 mM IPTG, and were grown at 43°C (restrictive temperature) or 30°C overnight.
σ-Dependent pausing
Pausing on lacUV5 promoter was analysed as described in Brodolin et al (2004) except for +16 stalled elongation complex by T. aquaticus RNAP was formed at 65°C, and +16 complexes were chased by addition of 100 μM NTPs at 40°C. Products of reactions were analysed as above, and pause efficiency (fraction of transcribing RNAP that pause at +17) was determined by non-linear regression analysis using SigmaPlot software.
Supplementary Material
Acknowledgments
We thank Sergei Nechaev, Ann Hochschild, Peter E Geiduschek, and Kenn Gerdes for critical reading of the manuscript and for fruitful discussions, and Konstantin Kuznedelov and Andrey Kulbachinsky for proteins. This work was supported by research grants from the UK Biotechnology and Biological Sciences Research Council to NZ and NIH to KS.
Author contributions: NZ and YY designed and performed the experiments and analysed the data, VT performed bioinformatics analysis; KS and NZ conceived the project and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Artsimovitch I (2008) Post-initiation control by the initiation factor sigma. Mol Microbiol 68: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Svetlov V, Anthony L, Burgess RR, Landick R (2000) RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J Bacteriol 182: 6027–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barne KA, Bown JA, Busby SJ, Minchin SD (1997) Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the ‘extended-10’ motif at promoters. EMBO J 16: 4034–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Goldfarb A (1993) Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Expr Purif 4: 503–511 [DOI] [PubMed] [Google Scholar]
- Borukhov S, Severinov K (2002) Role of the RNA polymerase sigma subunit in transcription initiation. Res Microbiol 153: 557–562 [DOI] [PubMed] [Google Scholar]
- Brodolin K, Zenkin N, Mustaev A, Mamaeva D, Heumann H (2004) The sigma 70 subunit of RNA polymerase induces lacUV5 promoter-proximal pausing of transcription. Nat Struct Mol Biol 11: 551–557 [DOI] [PubMed] [Google Scholar]
- Burgess RR, Anthony L (2001) How sigma docks to RNA polymerase and what sigma does. Curr Opin Microbiol 4: 126–131 [DOI] [PubMed] [Google Scholar]
- Christie GE, Cale SB, Isaksson LA, Jin DJ, Xu M, Sauer B, Calendar R (1996) Escherichia coli rpoC397 encodes a temperature-sensitive C-terminal frameshift in the beta′ subunit of RNA polymerase that blocks growth of bacteriophage P2. J Bacteriol 178: 6991–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighan P, Pukhrambam C, Nickels BE, Hochschild A (2011) Initial transcribed region sequences influence the composition and functional properties of the bacterial elongation complex. Genes Dev 25: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederth J, Artsimovitch I, Isaksson LA, Landick R (2002) The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J Biol Chem 277: 37456–37463 [DOI] [PubMed] [Google Scholar]
- Estrem ST, Ross W, Gaal T, Chen ZW, Niu W, Ebright RH, Gourse RL (1999) Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase alpha subunit. Genes Dev 13: 2134–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum A, Roberts J (2008) Prevalence of RNA polymerase stalling at Escherichia coli promoters after open complex formation. Mol Microbiol 68: 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL (2006) rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell 125: 1069–1082 [DOI] [PubMed] [Google Scholar]
- Helmann JD, deHaseth PL (1999) Protein-nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry 38: 5959–5967 [DOI] [PubMed] [Google Scholar]
- Hirata A, Murakami KS (2009) Archaeal RNA polymerase. Curr Opin Struct Biol 19: 724–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook-Barnard IG, Hinton DM (2009) The promoter spacer influences transcription initiation via sigma70 region 1.1 of Escherichia coli RNA polymerase. Proc Natl Acad Sci USA 106: 737–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlev M, Nudler E, Severinov K, Borukhov S, Komissarova N, Goldfarb A (1996) Histidine-tagged RNA polymerase of Escherichia coli and transcription in solid phase. Methods Enzymol 274: 326–334 [DOI] [PubMed] [Google Scholar]
- Kulbachinskiy A, Bass I, Bogdanova E, Goldfarb A, Nikiforov V (2004) Cold sensitivity of thermophilic and mesophilic RNA polymerases. J Bacteriol 186: 7818–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznedelov K, Korzheva N, Mustaev A, Severinov K (2002a) Structure-based analysis of RNA polymerase function: the largest subunit's rudder contributes critically to elongation complex stability and is not involved in the maintenance of RNA-DNA hybrid length. EMBO J 21: 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K (2002b) A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science 295: 855–857 [DOI] [PubMed] [Google Scholar]
- Kuznedelov K, Minakhin L, Severinov K (2003) Preparation and characterization of recombinant Thermus aquaticus RNA polymerase. Methods Enzymol 370: 94–108 [DOI] [PubMed] [Google Scholar]
- Liu M, Tolstorukov M, Zhurkin V, Garges S, Adhya S (2004) A mutant spacer sequence between −35 and −10 elements makes the Plac promoter hyperactive and cAMP receptor protein-independent. Proc Natl Acad Sci USA 101: 6911–6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Landick R (2003) Tethering sigma70 to RNA polymerase reveals high in vivo activity of sigma factors and sigma70-dependent pausing at promoter-distal locations. Genes Dev 17: 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Darst SA (2003) Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 13: 31–39 [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA (2002) Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296: 1285–1290 [DOI] [PubMed] [Google Scholar]
- Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright RH (2000) Structural organization of the RNA polymerase-promoter open complex. Cell 101: 601–611 [DOI] [PubMed] [Google Scholar]
- Nechaev S, Geiduschek EP (2006) The role of an upstream promoter interaction in initiation of bacterial transcription. EMBO J 25: 1700–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels BE, Mukhopadhyay J, Garrity SJ, Ebright RH, Hochschild A (2004) The sigma 70 subunit of RNA polymerase mediates a promoter-proximal pause at the lac promoter. Nat Struct Mol Biol 11: 544–550 [DOI] [PubMed] [Google Scholar]
- Polyakov A, Severinova E, Darst SA (1995) Three-dimensional structure of E. coli core RNA polymerase: promoter binding and elongation conformations of the enzyme. Cell 83: 365–373 [DOI] [PubMed] [Google Scholar]
- Ring BZ, Yarnell WS, Roberts JW (1996) Function of E. coli RNA polymerase sigma factor sigma 70 in promoter-proximal pausing. Cell 86: 485–493 [DOI] [PubMed] [Google Scholar]
- Schroeder LA, deHaseth PL (2005) Mechanistic differences in promoter DNA melting by Thermus aquaticus and Escherichia coli RNA polymerases. J Biol Chem 280: 17422–17429 [DOI] [PubMed] [Google Scholar]
- Severinov K, Markov D, Severinova E, Nikiforov V, Landick R, Darst SA, Goldfarb A (1995) Streptolydigin-resistant mutants in an evolutionarily conserved region of the beta′ subunit of Escherichia coli RNA polymerase. J Biol Chem 270: 23926–23929 [DOI] [PubMed] [Google Scholar]
- Wada T, Yamazaki T, Kyogoku Y (2000) The structure and the characteristic DNA binding property of the C-terminal domain of the RNA polymerase alpha subunit from Thermus thermophilus. J Biol Chem 275: 16057–16063 [DOI] [PubMed] [Google Scholar]
- Zenkin N, Naryshkina T, Kuznedelov K, Severinov K (2006) The mechanism of DNA replication primer synthesis by RNA polymerase. Nature 439: 617–620 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.