Abstract
Venezuelan equine encephalitis virus (VEEV), a member of the membrane-containing Alphavirus genus, is a human and equine pathogen, and has been developed as a biological weapon. Using electron cryo-microscopy (cryo-EM), we determined the structure of an attenuated vaccine strain, TC-83, of VEEV to 4.4 Å resolution. Our density map clearly resolves regions (including E1, E2 transmembrane helices and cytoplasmic tails) that were missing in the crystal structures of domains of alphavirus subunits. These new features are implicated in the fusion, assembly and budding processes of alphaviruses. Furthermore, our map reveals the unexpected E3 protein, which is cleaved and generally thought to be absent in the mature VEEV. Our structural results suggest a mechanism for the initial stage of nucleocapsid core formation, and shed light on the virulence attenuation, host recognition and neutralizing activities of VEEV and other alphavirus pathogens.
Keywords: alphavirus, bioweapon, cryo-EM, modelling, VEEV
Introduction
Venezuelan equine encephalitis virus (VEEV) is a mosquito-borne viral pathogen that has caused periodic, extensive outbreaks of human and equine diseases throughout the Americas, including in Texas (Weaver et al, 2004). Epidemics emerge when enzootic VEEV strains, which circulate among rodents in sylvatic or swamp habitats, acquire mutations that alter their host range to mediate equine amplification and transmission by mosquitos which have more promiscuous host preferences. In several countries including the United States, VEEV has been developed as a biological weapon (Bronze et al, 2002). Consequently, VEEV is classified as an NIAID (National Institute of Allergy and Infectious Diseases) Category B priority pathogen. Despite its threat to the public, no human vaccines or antiviral drugs thus far have been licensed. The attenuated TC-83 strain (Berge et al, 1961) is one of the few experimental vaccines that have been used to protect laboratory workers and military personnel. It was derived by 83 passages of the wild-type Trinidad donkey strain in guinea pig heart cells, which resulted in 12 nucleotide substitutions, 8 of which are non-synonymous (Kinney et al, 1993).
VEEV is one of the species in the Alphavirus genus of Togaviridae (Strauss and Strauss, 1994), a family of membrane-containing single-stranded RNA viruses, which also includes Sindbis (SINV), Semliki Forest (SFV), Ross River (RRV) and Chikungunya (CHIKV) viruses. To date, crystal structures have been obtained for domains of the three major alphavirus structural proteins: the C-terminal protease domain of the capsid protein (CP; Choi et al, 1991, 1997) and the elongated ectodomains of the envelope glycoproteins E1 (Lescar et al, 2001; Gibbons et al, 2004; Roussel et al, 2006; Li et al, 2010; Voss et al, 2010) and E2 (Li et al, 2010; Voss et al, 2010). However, the crystal structures of E1 and E2 endodomains are still missing. (The term ‘ectodomain’ refers to the portion of the protein outside the membrane, while ‘endodomains’ refers to the regions within or inside the membrane.) In addition, electron cryo-microscopy (cryo-EM) has been used to determine the entire structures of various alphaviruses and their mutants, but limited to moderate resolution (Mancini et al, 2000; Pletnev et al, 2001; Zhang et al, 2002, 2005; Mukhopadhyay et al, 2006; Sherman and Weaver, 2010; Kostyuchenko et al, 2011). These structures have not only revealed two nested shells (an outer glycoprotein shell and an inner nucleocapsid shell) with the same T=4 icosahedral symmetry, but also have localized several glycosylation and antibody binding sites on their surfaces.
Alphaviruses enter the host cells by receptor-mediated endocytosis (Garoff et al, 1994). Acidification in the endosome triggers the structural rearrangement of the outer glycoprotein shell (Wahlberg and Garoff, 1992), which drives the fusion between the viral envelope and the endosomal membranes. The membrane fusion followed by the disassembly of nucleocapsid cores with ribosomal subunits (Wengler et al, 1992) allows the viral genome (∼11.5 kb) to be released into the host cytoplasm. Following the translation of the non-structural polyprotein from the genomic RNA and synthesis of negative strand RNA, a subgenomic 26S RNA is then transcribed and translated into a single structural polypeptide C-p62-6K-E1 (Rice and Strauss, 1981), which is subsequently cleaved into several structural proteins. The CP is autoproteolytically processed, and 240 CPs encapsidate one copy of viral RNA genome to form the nascent nucleocapsid core. The remaining polypeptide is translocated to the endoplasmic reticulum (ER) membrane (Liljestrom and Garoff, 1991), where it undergoes cotranslational cleavage to yield pE2, 6K and E1 proteins. The pE2 and E1 proteins form heterodimers (Barth et al, 1995; Andersson et al, 1997) in ER and are transported to the Golgi complex, where they form trimers of pE2-E1 heterodimers (Mulvey and Brown, 1996). In Golgi, the small E3 protein is cleaved from pE2 to yield the mature E2 protein (Jain et al, 1991; Salminen et al, 1992). Finally, the trimers of E1-E2 heterodimers are transported to the plasma membrane, where they interact with the nascent nucleocapsid cores in the cytoplasm to form the intact progeny viruses that bud out of the host cell for the next round of infection.
Here, we report an all-atom model of the entire VEEV derived from our 4.4 Å resolution cryo-EM density map. Our model covers the full-length E1 and E2 glycoproteins (including the ectodomain, stem region, transmembrane (TM) helix and C-terminal tail), the E3 protein, and all the structurally ordered portion of CP (including the protease domain and one additional helix). Our map and model provide new insights on how different components of an alphavirus interact to self-assemble into an infectious viral particle.
Results and discussion
3D reconstruction and averaging subunits within an asymmetric unit
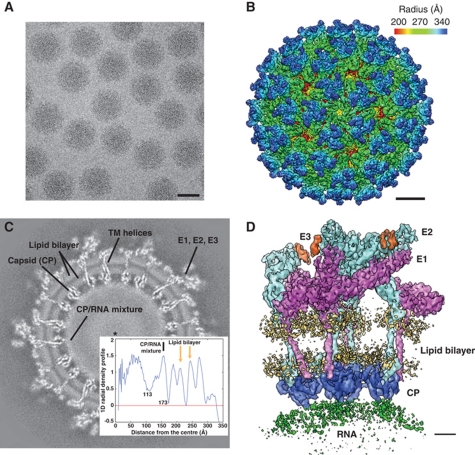
A cryo-EM structure of VEEV (TC-83 strain) was obtained initially at 4.8 Å resolution from ∼37 000 virus particle images recorded in a 300-keV electron cryo-microscope (Figure 1A). The reconstructed map (Figure 1B) shows 80 trimeric spikes on its surface, each containing three copies of E1 (48 kDa), E2 (47 kDa) and E3 (7 kDa) proteins. Three E1 molecules form the edges of a triangle and surround an E2 homotrimer, which protrudes outward forming the tip of the spike. A slice through the 3D density map (Figure 1C) provides a clear view of the viral membrane and E1/E2 TM helices, which extend from the outer glycoprotein shell to the inner nucleocapsid shell. Directly below the nucleocapsid shell is a layer of relatively disordered density that corresponds to a mixture of CP and genomic RNA. Interior to this CP/RNA shell, two additional shells are present: a low-density shell and a dense core.
Figure 1.
3D reconstruction of VEEV. (A) A typical CCD image of VEEV TC-83 strain embedded in vitreous ice. Scale bar: 50 nm. (B) Radially coloured 3D reconstruction of VEEV, showing the E1 basal triangle (green) and E2 central protrusion (blue) for each spike. Scale bar: 10 nm. (C) A slice through the 3D density map 20 pixels from the origin. The insert is the 1D radial density profile of the map and is aligned to the slice image. (D) One asymmetric unit of the virus containing four unique copies of E1 (magenta), E2 (cyan), E3 (orange) and CP (blue). The cryo-EM densities for the viral membrane (yellow) and genomic RNA (green) are also displayed at slightly lower isosurface threshold. Scale bar: 2 nm.
To further improve the resolvability of various structural features, we performed an additional averaging (Zhou et al, 2000; Zhang et al, 2008, 2010; Wolf et al, 2010) of the four unique sets of E1-E2-E3-CP molecules within one asymmetric unit of the virus (Figure 1D), resulting in an averaged map at 4.4 Å resolution (Supplementary Figure S1). This averaged map substantially improves the density connectivity for the loop regions and the TM helices (Figures 2A and 5A). In addition, the β-strands are better resolved, for example, in the E2 ectodomain (Figure 2B). It is noteworthy that the averaged density appears smoother than the original map, with a significant reduction of noise and the disappearance of some putative side-chain densities. However, density bulges corresponding to many of the bulky side chains (F, W and Y) remain visible in the averaged map. As a result, we primarily used this averaged density map for the subsequent structural modelling and interpretation.
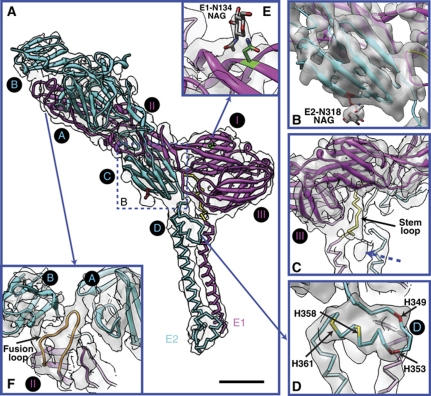
Figure 2.
E1 and E2 ectodomains. (A) Our models for VEEV E1 (magenta) and E2 (cyan). The homology model parts and de novo model parts are shown as ribbon and stick, respectively. The asymmetric unit averaged map is shown in transparent grey. The de novo modelled part of E1 stem region (residues 390–402) is coloured in yellow. Subdomains of E1 (I, II and III) and E2 (A, B, C and D) are labelled in black circle, following the previous definition (Lescar et al, 2001; Voss et al, 2010). Scale bar: 2 nm. (B) The separation of β-strands at E2 subdomain C, displayed at slight higher isosurface threshold. It also shows the protrusion density for glycan at E2-318 and the annotated atomic structure of N-acetylglucosamine (NAG). (C) A 180° rotation of (A) shows the E1 stem region wraps around E1 subdomain III. The blue dashed arrow points to a small, unidentified density. (D) The E2 subdomain D. (E) The protrusion density for the glycan at E1-N134 and the annotated NAG. (F) The E1 fusion loop (orange) which sits between E2 A and B subdomains.
E1 and E2 ectodomains
We built the full-length VEEV E1 and E2 models using a combination of homology modelling and de novo modelling. A majority of the E1, E2 ectodomains (E1: subdomains DI, DII, DIII; E2: subdomains A, B, C) were constructed by homology modelling; while part of the E1 stem loop (residues 390–402, Figure 2A and C), a previously unidentified E2 subdomain D (residues 342–367, Figure 2D), and the entire E1, E2 endodomains were built de novo. Subsequently, the homology part and the de novo part were stitched together and refined against cryo-EM density to produce our final E1 and E2 atomic models (see Materials and methods). Since the side-chain conformations (rotamers) except those bulky ones with evident protruding densities in our final model are not fully restrained by the cryo-EM densities, their accuracies are limited, as with any crystal structure determined at a similar resolution.
To compare the differences between our final model of VEEV and the CHIKV crystal structure, we fit the CHIKV E1 and E2 ectodomains separately into our asymmetric unit averaged map of VEEV as a rigid body and then calculated the individual root-mean-square-deviation (RMSD) per Cα atom for E1 and E2 between the fitted CHIKV model and our VEEV model. The results (Supplementary Figure S2) show that the E2 ectodomain (residues 1–341) has more variations (RMSD 4.2 Å) than E1 ectodomain (residues 1–391) (RMSD 1.8 Å), with the largest deviations mapped to not only the loop regions but also some of the β-strands. Similarly, we also computed the RMSD between our VEEV model and the fitted SINV E1-E2 crystal structure at low pH (PDB ID: 3MUU, chain A) (Li et al, 2010), in which subdomain B at E2 ectodomain is not resolved. Our results show an RMSD of 2.4 Å for E2 ectodomain and 2.9 Å for E1 ectodomain, with the most variations mapped to the E1 fusion loop region (Supplementary Figure S3).
The number and positions of the N-linked glycosylation sites among the glycoproteins in alphaviruses are not absolutely conserved (Strauss and Strauss, 1994), and the locations and types of glycans in VEEV have not been well characterized. The N-linked glycosylation sites are generally identified by an Asn-X-Thr/Ser motif, where X is any amino acid except Proline (Gavel and von Heijne, 1990). Based on this criterion, only two residues (E1-N134 and E2-N318) are likely glycosylated in VEEV. In our cryo-EM map, we indeed observe prominent protruding densities at both sites, either of which can accommodate only a monosaccharide (N-acetylglucosamine) rather than a disaccharide (Figure 2B and E). It is noteworthy that the E1 glycan is surface exposed, while the E2 glycan is buried near the outer lipid membrane.
Our map and models also reveal several features that are implicated in the alphavirus fusion process in the endosome. In our density map of mature VEEV at neutral pH condition, the E1 fusion loop is clearly visualized at the cleft between E2 subdomains A and B (Figure 2F), consistent with the crystal structure of CHIKV (Voss et al, 2010). Additionally, several histidines, specifically two completely conserved (H349 and H353) and two partially conserved (H358 and H361) (Supplementary Figure S4), are located in the previously unidentified E2 subdomain D, which consists of a loop and a helix (Figure 2D). The location suggests that upon low pH exposure in the endosome, the protonation of these histidines may promote their interactions with the neighbouring negative charged lipid head groups and anchor E2 to the lipid membrane, thus facilitating the separation of E1-E2 heterodimers along with the E1 homotrimerization.
The structure of VEEV TC-83 vaccine strain is different from its parental Trinidad donkey strain by five residues (K7N, H85Y, T120R, V192D and T296I) in E2 and one residue in E1 (L161I) (Kinney et al, 1993). The E2 T120R mutation has been found to be the major structural determinant of attenuation (Kinney et al, 1993). In our structure, this residue R120 is located at the E2 trimeric interface and at the top surface of the spike complex (Figure 3). This location suggests that the in vitro adaption of residue T120 from neutral to positive charged residue may result from binding to the negative charged heparan sulphate, which is the putative receptor on the surface of cell cultures; while the attenuation in vivo is likely a result of less efficient spreading of the virus due to binding to some negatively charged molecules (Klimstra et al, 1998; Byrnes and Griffin, 2000).
Figure 3.
Mapping of specific residues on VEEV E1 and E2. (A) Model of an E1-E2 heterodimer. Two N-linked glycosylation sites (E1-N134 and E2-N318) are labelled in green and red, respectively. The major determinant of virulence attenuation (residue E2-T120) is labelled in dark blue. Two sets of residues (E2-193/213 and E2-218) whose mutations strongly affects the equine and mosquito host range are labelled in grey and black, respectively. The previously identified VEEV epitopes for murine monoclonal antibodies mMAbs (residues 182–207) and human hMAbs (residues 115–119) are coloured in orange and yellow, respectively. (B) Model of an E2 homotrimer of VEEV in one asymmetric unit. The residue labelling is the same as (A).
Our VEEV E2 model also provides the structural basis to interpret the previous genetic studies on the host range and neutralizing activities of VEEV. An S218N mutation is believed to have mediated the recent emergence of VEE in southern Mexico by adapting subtype IE enzootic strains to more efficiently infect the epidemic vector, Aedes (Ochlerotatus) taeniorhynchus (Brault et al, 2004). The G193R and T213R mutations are associated with the 1992 subtype IC epidemic emergence in Venezuela (Rico-Hesse et al, 1995), and experimental equine infections demonstrated that the T213R mutation mediates enhanced equine viraemia, the critical driver of epidemic VEEV spread (Anishchenko et al, 2006). The close clustering of all of these host range mutations on the tip of the spikes (Figure 3) provides strong circumstantial evidence that this location is directly involved in receptor binding. Interestingly, the VEEV epitopes for murine monoclonal antibodies (mMAbs) and human hMAbs map to E2 residues 182–207 (Johnson et al, 1990) and 115–119 (Hunt et al, 2010), respectively, which are also located at the tip of the spikes (Figure 3).
E3 protein
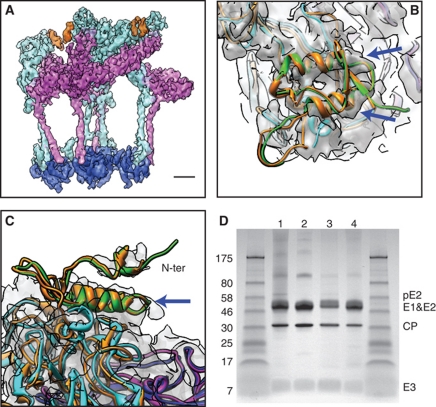
In both our original map (Figure 4A) and the asymmetric unit averaged map (Figure 4B and C), at a slightly lower isosurface threshold than used to visualize E1 and E2, we observed extra density decorating the outermost portion of E2 above subdomains A and B. The location of this density is consistent with the E3 densities observed in the previous cryo-EM structures of pE2 cleavage-impaired SINV and SFV mutants (Paredes et al, 1998; Wu et al, 2008).
Figure 4.
The presence of E3 protein in mature VEEV. (A) The densities for E3 (orange) in one asymmetric unit of the original 3D reconstruction. Note the E3 densities are displayed at slightly lower isosurface threshold than E1 (magenta) and E2 (cyan) densities. Scale bar: 2 nm. (B, C) Side and top views of the density for E3 in the asymmetric unit averaged map. The crystal structures of CHIKV pE2 (orange) and E1 (blue) (PDB code: 3N40) are fitted separately as a rigid body into the averaged density. Our models of VEEV E1, E2 and E3 are shown in magenta, cyan and green, respectively. The blue arrows point to the two rod-like features in the density. (D) SDS–PAGE result of VEEV TC-83 samples we used for imaging. The leftmost and rightmost lanes are molecular size markers. Lanes 1–4 are the four batches of VEEV TC-83 samples used for cryo-EM imaging.
The E3 protein, ∼60 residues in length, is cleaved from the pE2 protein in the Golgi complex to yield mature E2. Given the 52% sequence identity between VEEV E3 and CHIKV E3, we built a homology model for VEEV E3 (residues 1–59) using the CHIKV pE2 crystal structure (containing E3) (Voss et al, 2010) as the template and placed it in our asymmetric unit averaged map based on the fitting of CHIKV pE2 crystal structure (Figure 4B and C). There are two rod-like features in our E3 density that match the two α-helices of our E3 homology model (Figure 4B and C, blue arrows). Due to disconnected densities above these helices, presumably corresponding to the N-terminus of E3 (Figure 4C), our E3 homology model was not further refined against the cryo-EM density.
To determine whether E3 is present and cleaved from the precursor protein pE2 upon maturation in VEEV, we performed SDS–PAGE on the four batches of VEEV TC-83 samples that we used for cryo-EM imaging. A 4–20% polyacrylamide gel was used to resolve the small 7 kDa E3 protein, and the result confirmed the presence of E3 in the mature VEEV, although at lower stoichiometry (Figure 4D). Interestingly, a very faint band of pE2 (62 kDa) is seen in two of the four sample batches, indicating that in some cases the cleavage might not be 100% complete.
Based on our structural and biochemical analysis, it appears that the aforementioned density indeed represents the cleaved form of E3. While E3 has been found in another mature alphavirus SFV (Wu et al, 2008), this is the first time that E3 has been seen structurally in mature VEEV. It remains to be determined why E3 proteins stay associated with E2 in VEEV after cleavage. It is conceivable that E3 may function to maintain the relative orientation between E2 subdomains A and B, so as to protect the E1 fusion loop from premature exposure to the host membranes (Figure 2F) (Li et al, 2010; Voss et al, 2010).
E1 and E2 endodomains
Beyond the ectodomains, our cryo-EM structure of the entire virus provides us with the unique opportunity to explore the protein interactions in the E1 and E2 endodomains. In the asymmetric unit averaged map, almost all of the bulky side-chain densities along the TM helices remain visible after averaging, including E2-W387, E1-W407, E1-W409 and E1-Y434 (Figure 5A–C), although some of them appear less prominent than the unaveraged densities. Using these side-chain densities as the anchor points, we can determine the registration of Cα positions for E1 and E2 TM helices and further trace the backbones of the entire E1 and E2 endodomains, including their cytoplasmic tails (Figure 5A). Consistent with the secondary structure prediction (Figure 5F), the E1 TM helix (residues 403–442) was modelled as two consecutive helices separated by a kink, while the E2 TM helix (residues 367–401) was modelled as a long, straight helix. Below the kink and towards the nucleocapsid core, the angle between E1 and E2 TM helices is very similar to the characteristic angle associated with a leucine zipper (O’Shea et al, 1991).
Figure 5.
E1 and E2 endodomains and their interactions with the CPs. (A) Our models for E1 (magenta) and E2 (cyan) endodomains and CP (dark blue). The homology model parts and de novo model parts are shown as ribbon and stick, respectively. The asymmetric unit averaged map is shown in transparent grey. Various features are highlighted: E2 Y-R-L motif (red), E1 G415/G416 at the kink region (green), E2 C396/C416/C417 near the inner membrane (yellow) and the helix (residues 115–124) of CP (orange). The disordered densities for the lipid bilayer and genomic RNA are simplified with transparent orange and green lines, respectively. Scale bar: 1 nm. (B, C) The prominent side-chain densities for E1-W407, E1-W409 and E1-Y434 in the averaged density map. (D) Same as (A) with less density transparency showing the E2 C-terminal tail and its interaction with the CP pocket. The blue dashed arrow points to the small C-terminal helix of E2 (residues 409–416). (E) Different viewing angle (rotation of 70° along the z axis) of (D) showing the density for the previously unidentified helix of CP (pointed by blue dashed arrow). (F) Secondary structure prediction for E1 and E2 C-termini from PSIPRED.
In our model, two highly conserved glycines (G415 and G416) (Supplementary Figure S5) are located at the E1 kink region (Figure 5A), which is necessary to bring the two TM helices into close proximity despite the large size of E1 and E2 ectodomains. This conserved GG motif in alphaviruses is different from the canonical GXXXG (with X being any residue) motif that is commonly found at the interface of interacting TM helices, where two glycines are brought to the same side of the helix, allowing close contact between the two helices (Kleiger et al, 2002). Given the register of E1-W407 and E1-W409, and assuming the helix keeps its winding path at the kink region, which is typically the case, these two glycines have to face away from the E2 TM helix. Therefore, it is unlikely that this GG motif is directly involved in the interaction between E1, E2 TM helices. Instead, it may function to alleviate the steric forces at the inner-bending side of the kink. Previous studies have showed that mutations of these glycines to leucines in SFV destabilize the E1-E2 heterodimer and promote the formation of E1 homotrimer during fusion (Sjoberg and Garoff, 2003). Interestingly, this kink leaves space for a small globular density situated between the upper parts of the E1 and E2 TM helices (Figure 2C, blue dashed arrow). This yet-to-be-assigned density is located below the two completely conserved residues E2-Y359 and E2-Y360 (Supplementary Figure S4), but is significantly larger than that expected for a tyrosine side chain.
During alphavirus maturation, the glycoproteins embedded in the plasma membrane use their cytoplasmic tails to interact with the nucleocapsid core in order to form an intact virus particle and bud out of the cell. The molecular mechanism of this process is not well understood, partially due to the lack of high-resolution structures. Here, our cryo-EM structure for the first time clearly resolves the entire cytoplasmic tail (C-terminus) of E2 (residues 402–423), which extends through the inner membrane, then interacts with the hydrophobic pocket of CP (presumably via a previously reported Tyr-X-Leu tripeptide structural motif (Zhao et al, 1994; Owen and Kuhn, 1997)), and finally loops back to the inner membrane (Figure 5A and D). A short, rod-like density is revealed in this ‘hairpin’ region (Figure 5D, blue dashed arrow), consistent with the predicted α-helix for residues 409–416 (Figure 5F). In addition, our model of the E2 C-terminus shows that three completely conserved (Supplementary Figure S4), and presumably palmitoylated (Gaedigk-Nitschko and Schlesinger, 1991; Ivanova and Schlesinger, 1993) cysteines (C396, C416 and C417) are located near the lipid head groups of the inner membrane (Figure 5A). The interactions between their palmitoylated chains and the lipid tails may help anchor the E2 C-terminus to the cytoplasmic side of the viral membrane, thus promoting their interaction with the CPs. Considering that the E2 C-terminus originally adopts a membrane-spanning conformation in ER during biosynthesis (Liljestrom and Garoff, 1991), our observation directly supports the hypothesis (Ivanova and Schlesinger, 1993) that post-translational palmitoylation of cysteines triggers the reorientation of the entire E2 C-terminus.
CP and nucleocapsid core
For the CP, a crystal structure of the protease domain of VEEV TC-83 (residues 120–275) has been reported previously (PDB ID: 1EP5). Fitting of this domain into our cryo-EM density map (Figure 6A) reveals significant connecting density at the intra-capsomere CP/CP interface (Figure 6B), but little connecting density at the inter-capsomere CP/CP interface (Figure 6C). It is likely that during the initial nucleocapsid core assembly process, the pentameric and hexameric capsomeres may be formed first and then brought together by some external forces rather than the lateral CP/CP interactions in the protease domains.
Figure 6.
CP/CP interactions in the nucleocapsid core. (A) Our model of the entire nucleocapsid core fitted into the cryo-EM density. The four copies of CPs in one asymmetric unit are coloured in red, blue, green and yellow. Scale bar: 5 nm. (B) Zoom in view of the intra-capsomere interactions. (C) Zoom in view of the densities around a quasi three-fold axis.
The cryo-EM density of CP also, for the first time, shows the predicted α-helix at residues 115–124 (Figure 6B), which is missing in the crystal structures (Choi et al, 1991, 1997). This helix (Figure 5E, blue dashed arrow) bridges the structured C-terminal protease domain and the unstructured, highly basic N-terminus, which interacts with genomic RNA in the CP/RNA mixture shell (Figure 1C). The sequence of this short helix overlaps with the ‘linker region’ (Wengler, 2009), a stretch of highly conserved residues (109–125 for VEEV, Supplementary Figure S6) that binds to cellular 60S ribosomal subunits, which function to disassemble the nucleocapsid cores after they are released into the cytosol (Wengler et al, 1992). Considering the current location of the linker regions in the mature virus (at the inner surface of the core), the CPs may undergo some conformational changes after its release to the cytosol and expose their linker regions to the cellular factors.
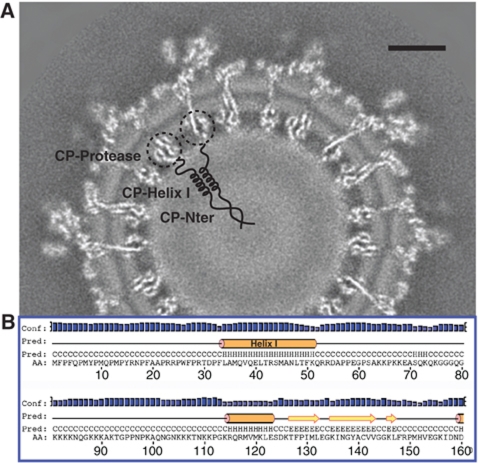
The spatial arrangement of the first ∼120 N-terminal residues of CP in the mature virus has been a mystery. There is a putative helix in this region (helix I, residues 34–51 for VEEV, Figure 7B) that is believed to form a coiled-coil structure between two neighbouring CPs and to stabilize the core (Perera et al, 2001). However, when analysing our map, we did not observe any clear density corresponding to helix I. It is possible that these coiled-coil structures (around 30 Å in length) are located in the low-density shell region (from 95 to 130 Å in radius) between the CP/RNA mixture shell and the central core (Figure 7A). Their distribution may not follow icosahedral symmetry; and therefore, their densities are averaged out in our reconstruction process. Our hypothesis places the first 33 residues of CP (mostly hydrophobic residues, Figure 7B) at the central dense core (radius <95 Å) (Figure 7A), where they would cluster to form a ‘scaffold’ and initiate core assembly. Additionally, since only 10% of the first 50 residues are basic, as opposed to 39% basic ones within residues 51–117, it is likely that the majority of the genomic RNA is confined to the thin CP/RNA mixture shell (Figure 1D), as supported by a theoretical analysis (Belyi and Muthukumar, 2006).
Figure 7.
Proposed spatial arrangement of the first ∼120 residues of CP in the nucleocapsid core. (A) In our proposed model, the helix I coiled-coil structure (residues 34–51) between two neighbouring CPs is located at the low-density shell (radius 95–130 Å) between the CP/RNA mixture shell and the central core, while the first 33 hydrophobic residues are located at the central core (radius <95 Å). Scale bar: 10 nm. (B) Secondary structure prediction for CP N-terminus from the PSIPRED results.
In summary, our 4.4 Å resolution cryo-EM density map and derived models reveal many important features not seen in the crystal structures of domains of alphavirus subunits, including the E1 stem loop, E2 subdomain D, E1/E2 TM helices, E2 cytoplasmic C-terminal tail and one additional helix of CP. The presence of E3 in the cleaved form is also unambiguously identified for the first time in the mature VEEV virion. Our structural results suggest a mechanism for the initial stage of nucleocapsid core formation, and shed light on the virulence attenuation, host recognition and neutralizing activities of VEEV as well as other alphavirus pathogens.
Materials and methods
Virus production and purification
Baby hamster kidney cells were prepared to 80–90% confluence and were inoculated with virus at a multiplicity of 0.1 plaque-forming units per cell. Infected cells were incubated at 37°C for 2 days until cytopathic effects appeared; then the supernatant was clarified by centrifuged for 5–10 min at 1000–2000 g to remove cellular debris. The virus was concentrated by precipitation with 7% polyethylene glycol 6000 and 2.3% NaCl at 4°C for >4 h. Then, the virus was centrifuged at ⩾2500 g for 30 min and was gently resuspended in 1–2 ml TEN buffer (0.05 M Tris–HCl, pH 7.4, 0.1 M NaCl and 0.001 M EDTA). The virus suspension was purified by centrifugation through a 20–70% sucrose/TEN continuous gradient for 60 min at 35 000 g. The virus band was harvested using a plastic Pasteur pipette and centrifuged 3 × through Amicon 100 kDa filter (Ultra-4 Cat. No. UFC810024), resuspending each time to maximum load volume with TEN. The purified virus was harvested in the minimal remaining volume after final centrifugation (ca. 50–100 μl).
SDS–PAGE
The SDS–PAGE of VEEV TC-83 was performed using Bio-Rad Mini-PROTEIN TGX 4–20% polyacrylamide gel (Bio-Rad Inc.) and the p7708S molecular size marker (New England Biolabs Inc.).
Electron cryo-microscopy
An aliquot of 2.5 μl purified VEEV TC-83 sample was applied to a 400 mesh Quantifoil R 1.2/1.3 copper grids (hole size 1.2 μm) (Quantifoil Inc.) and were rapidly plunge frozen in liquid ethane by a FEI Vitrobot. In total, eight grids, including two grids with continuous carbon film underneath the samples, were used for imaging in 16 cryo-EM sessions. Cryo-EM images were collected in a JEM3200FSC electron cryo-microscope (JEOL, Tokyo) operated at 300 keV, and at liquid nitrogen specimen temperature. The microscope is equipped with a field emission gun (FEG) and an in-column omega energy filter (a slit width of 10 eV was used for data collection). Approximately 4100 CCD frames were recorded at a detector magnification of × 141 110 (1.07 Å/pixel sampling rate) using a Gatan 4K × 4K CCD camera (model no. 895, Gatan), with a defocus range of 0.5–2.5 μm.
Image processing
We carefully screened all the CCD frames from which 3558 images with evident signals up to 1/5 Å−1 in their 1D power spectra were selected for subsequent processing. A total of 37 315 virus particles were automatically boxed out using ethan (Kivioja et al, 2000), among which ∼10 000 particles contained continuous carbon film. The contrast transfer function parameters for each CCD image were manually determined using ctfit in EMAN1 (Ludtke et al, 1999). An initial model at ∼7 Å resolution was quickly obtained by MPSA (Liu et al, 2007). The structure was further refined by EMAN1 using standard projection matching method with progressively decreasing angular step size (with a final value of 0.4°). After each iteration, the non-icosahedral part, including the lipids and the RNA, in the reconstruction was removed by a soft-edged mask, which defines the outline of the icosahedrally organized, low-pass filtered ‘protein-only’ content in the map. This masked map then served as the reference model for the next iteration. The resolution of the final reconstruction was estimated to be 4.8 Å based on the 0.5 criterion of the Fourier shell correlation (FSC) between two independent reconstructions (Van Heel, 1987; Supplementary Figure S1).
Averaging subunits within an asymmetric unit
To improve the resolvability in our density map, we computationally segmented out the densities for the four unique sets of E1-E2-E3-CP molecules in one asymmetric unit using Chimera (Goddard et al, 2007). The four molecules (E1, E2, E3 and CP) in each set were treated as an intact unit during the segmentation. We then used foldhunterP program (Baker et al, 2007) in EMAN1 to align the four pieces of segmented densities and used proc3d in EMAN1 to compute the average. To estimate the resolution of our averaged map, we applied the same averaging technique to the two independent reconstructions that were used to calculate the 4.8-Å resolution for the original density map, and then calculated the FSC between the two resulting averaged maps.
Model building and refinement
To model the VEEV E1, E2 ectodomains and E3 (E1: residues 1–389; E2: residues 1–341 and E3: residues 1–59), the sequence alignment and subsequent homology modelling was performed by MODELLER (Eswar et al, 2006), using the crystal structure of its CHIKV homologue (Voss et al, 2010) (PDB ID: 3N40) as the template. The missing parts in the crystal structure (E1: residues 390–442; E2: residues 342–423) were modelled de novo by first tracing the backbones using GORGON (Baker et al, 2011), with several visible side-chain densities serving as the anchor points.
To model the TM helices, in particular, we generated the Cα models of two consecutive helices (residues 403–412 and 415–442) separated by a kink for E1, and a long straight helix (residues 367–401) for E2. These residue assignments are based on both the secondary structure predictions from PSIPRED online server (McGuffin et al, 2000; Figure 5F) and our cryo-EM density map. The three helical models were placed in the corresponding densities in GORGON, and the registration of their Cα positions was determined by the evident bulky side-chain densities along the helices (e.g., E1-W407, E1-W409, E1-Y434 and E2-W387).
Next, we converted all the de novo traced Cα models to their corresponding all-atom models of VEEV using SABBAC online server (Maupetit et al, 2006), and then stitched the homology and de novo portions together in COOT (Emsley and Cowtan, 2004) to generate the initial full-length E1 and E2 atomic models.
Our model for the CP protease domain is taken directly from the previous crystal structure of VEEV TC-83 (PDB ID: 1EP5: A, residues 120–275), and was fitted into the density map as a rigid body using CHIMERA's Fit to Map function. The α-helix of CP (residues 115–124) was modelled de novo in the same way as the E1 and E2 TM helices.
Finally, we used ROSETTA (DiMaio et al, 2009) to refine the full-length E1, E2 and the structured part of CP. The E3 homology model was not further refined due to the less-resolved quality of our E3 density. ROSETTA uses the cryo-EM density as a restraint, along with energy minimization to eliminate the steric clashes and assure proper molecular geometry. In total, two separate refinements were performed. First, one set of E1-E2-CP molecules was refined against the asymmetric unit averaged map. Second, four copies of the models from the first round were placed at the T=4 related positions within the asymmetric unit, and these four sets of E1-E2-CP molecules were refined together against the original cryo-EM density map to produce our final model.
Accession numbers
Atomic coordinates of our refined model of E1-E2-CP and unrefined homology model of E3 within one asymmetric unit have been deposited with the Protein Data Bank under the accession codes 3J0C and 3J0G, respectively. A cubic portion of the original 3D density map (covering one asymmetric unit) and the asymmetric unit averaged map have been deposited in the Macromolecular Structure Database at the European Bioinformatics Institute under the accession codes 5275 and 5276, respectively.
Supplementary Material
Acknowledgments
We thank Dr Félix A Rey for providing the crystal structure of CHIKV E1-pE2 complex before publication. We also thank Drs Michael F Schmid, Steve J Ludtke, Donghua Chen, Qinfen Zhang and Patrick Barth for their valuable advice. This research has been supported by NIH grants (P41RR002250 and R01GM079429) and Robert Welch Foundation grant (Q1242).
Author contributions: SCW prepared and purified the viruses. RZ and JJ collected the cryo-EM images. RZ and XL processed the data and did the 3D reconstruction. RG performed the SDS–PAGE of VEEV TC-83 samples. RZ, CFH, YC and MLB built and refined the models. RZ, CFH, YC, MLB, SCW and WC analysed the density map and models and contributed to the final manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andersson H, Barth BU, Ekstrom M, Garoff H (1997) Oligomerization-dependent folding of the membrane fusion protein of Semliki Forest virus. J Virol 71: 9654–9663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anishchenko M, Bowen RA, Paessler S, Austgen L, Greene IP, Weaver SC (2006) Venezuelan encephalitis emergence mediated by a phylogenetically predicted viral mutation. Proc Natl Acad Sci U S A 103: 4994–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Abeysinghe SS, Schuh S, Coleman RA, Abrams A, Marsh MP, Hryc CF, Ruths T, Chiu W, Ju T (2011) Modeling protein structure at near atomic resolutions with Gorgon. J Struct Biol 174: 360–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Ju T, Chiu W (2007) Identification of secondary structure elements in intermediate-resolution density maps. Structure 15: 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth BU, Wahlberg JM, Garoff H (1995) The oligomerization reaction of the Semliki Forest virus membrane protein subunits. J Cell Biol 128: 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi VA, Muthukumar M (2006) Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci U S A 103: 17174–17178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge TO, Banks IS, Tigertt WD (1961) Attenuation of Venezuelan equine encephalomyelitis virus by in vitro cultivation in guinea pig heart cells. Am J Hyg 73: 209–218 [Google Scholar]
- Brault AC, Powers AM, Ortiz D, Estrada-Franco JG, Navarro-Lopez R, Weaver SC (2004) Venezuelan equine encephalitis emergence: enhanced vector infection from a single amino acid substitution in the envelope glycoprotein. Proc Natl Acad Sci U S A 101: 11344–11349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronze MS, Huycke MM, Machado LJ, Voskuhl GW, Greenfield RA (2002) Viral agents as biological weapons and agents of bioterrorism. Am J Med Sci 323: 316–325 [DOI] [PubMed] [Google Scholar]
- Byrnes AP, Griffin DE (2000) Large-plaque mutants of Sindbis virus show reduced binding to heparan sulfate, heightened viremia, and slower clearance from the circulation. J Virol 74: 644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Lu G, Lee S, Wengler G, Rossmann MG (1997) Structure of Semliki Forest virus core protein. Proteins 27: 345–359 [DOI] [PubMed] [Google Scholar]
- Choi HK, Tong L, Minor W, Dumas P, Boege U, Rossmann MG, Wengler G (1991) Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature 354: 37–43 [DOI] [PubMed] [Google Scholar]
- DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D (2009) Refinement of protein structures into low-resolution density maps using rosetta. J Mol Biol 392: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A (2006) Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics (Suppl 15): 5.6.1–5.6.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaedigk-Nitschko K, Schlesinger MJ (1991) Site-directed mutations in Sindbis virus E2 glycoprotein's cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology 183: 206–214 [DOI] [PubMed] [Google Scholar]
- Garoff H, Wilschut J, Liljestrom P, Wahlberg JM, Bron R, Suomalainen M, Smyth J, Salminen A, Barth BU, Zhao H (1994) Assembly and entry mechanisms of Semliki Forest virus. Arch Virol Suppl 9: 329–338 [DOI] [PubMed] [Google Scholar]
- Gavel Y, von Heijne G (1990) Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng 3: 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DL, Vaney MC, Roussel A, Vigouroux A, Reilly B, Lepault J, Kielian M, Rey FA (2004) Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427: 320–325 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE (2007) Visualizing density maps with UCSF Chimera. J Struct Biol 157: 281–287 [DOI] [PubMed] [Google Scholar]
- Hunt AR, Frederickson S, Maruyama T, Roehrig JT, Blair CD (2010) The first human epitope map of the alphaviral E1 and E2 proteins reveals a new E2 epitope with significant virus neutralizing activity. PLoS Negl Trop Dis 4: e739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova L, Schlesinger MJ (1993) Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J Virol 67: 2546–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain SK, DeCandido S, Kielian M (1991) Processing of the p62 envelope precursor protein of Semliki Forest virus. J Biol Chem 266: 5756–5761 [PubMed] [Google Scholar]
- Johnson BJ, Brubaker JR, Roehrig JT, Trent DW (1990) Variants of Venezuelan equine encephalitis virus that resist neutralization define a domain of the E2 glycoprotein. Virology 177: 676–683 [DOI] [PubMed] [Google Scholar]
- Kinney RM, Chang GJ, Tsuchiya KR, Sneider JM, Roehrig JT, Woodward TM, Trent DW (1993) Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J Virol 67: 1269–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivioja T, Ravantti J, Verkhovsky A, Ukkonen E, Bamford D (2000) Local average intensity-based method for identifying spherical particles in electron micrographs. J Struct Biol 131: 126–134 [DOI] [PubMed] [Google Scholar]
- Kleiger G, Grothe R, Mallick P, Eisenberg D (2002) GXXXG and AXXXA: common alpha-helical interaction motifs in proteins, particularly in extremophiles. Biochemistry 41: 5990–5997 [DOI] [PubMed] [Google Scholar]
- Klimstra WB, Ryman KD, Johnston RE (1998) Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol 72: 7357–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko VA, Jakana J, Liu X, Haddow AD, Aung M, Weaver SC, Chiu W, Lok SM (2011) The 6Å resolution cryo-EM Barmah Forest virus structure shows detailed transmembrane proteins architecture and interactions. J Virol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Wengler G, Rey FA (2001) The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105: 137–148 [DOI] [PubMed] [Google Scholar]
- Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG (2010) Structural changes of envelope proteins during alphavirus fusion. Nature 468: 705–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljestrom P, Garoff H (1991) Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J Virol 65: 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang W, Jakana J, Chiu W (2007) Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-Path Simulated Annealing optimization algorithm. J Struct Biol 160: 11–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- Mancini EJ, Clarke M, Gowen BE, Rutten T, Fuller SD (2000) Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol Cell 5: 255–266 [DOI] [PubMed] [Google Scholar]
- Maupetit J, Gautier R, Tuffery P (2006) SABBAC: online Structural Alphabet-based protein BackBone reconstruction from Alpha-Carbon trace. Nucleic Acids Res 34: W147–W151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Zhang W, Gabler S, Chipman PR, Strauss EG, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG (2006) Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M, Brown DT (1996) Assembly of the Sindbis virus spike protein complex. Virology 219: 125–132 [DOI] [PubMed] [Google Scholar]
- O’Shea EK, Klemm JD, Kim PS, Alber T (1991) X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254: 539–544 [DOI] [PubMed] [Google Scholar]
- Owen KE, Kuhn RJ (1997) Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology 230: 187–196 [DOI] [PubMed] [Google Scholar]
- Paredes AM, Heidner H, Thuman-Commike P, Prasad BV, Johnston RE, Chiu W (1998) Structural localization of the E3 glycoprotein in attenuated Sindbis virus mutants. J Virol 72: 1534–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R, Owen KE, Tellinghuisen TL, Gorbalenya AE, Kuhn RJ (2001) Alphavirus nucleocapsid protein contains a putative coiled coil alpha-helix important for core assembly. J Virol 75: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnev SV, Zhang W, Mukhopadhyay S, Fisher BR, Hernandez R, Brown DT, Baker TS, Rossmann MG, Kuhn RJ (2001) Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice CM, Strauss JH (1981) Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc Natl Acad Sci U S A 78: 2062–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Hesse R, Weaver SC, de Siger J, Medina G, Salas RA (1995) Emergence of a new epidemic/epizootic Venezuelan equine encephalitis virus in South America. Proc Natl Acad Sci U S A 92: 5278–5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel A, Lescar J, Vaney MC, Wengler G, Wengler G, Rey FA (2006) Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 14: 75–86 [DOI] [PubMed] [Google Scholar]
- Salminen A, Wahlberg JM, Lobigs M, Liljestrom P, Garoff H (1992) Membrane fusion process of Semliki Forest virus II: cleavage-dependent reorganization of the spike protein complex controls virus entry. J Cell Biol 116: 349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MB, Weaver SC (2010) Structure of the recombinant alphavirus Western equine encephalitis virus revealed by cryoelectron microscopy. J Virol 84: 9775–9782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoberg M, Garoff H (2003) Interactions between the transmembrane segments of the alphavirus E1 and E2 proteins play a role in virus budding and fusion. J Virol 77: 3441–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG (1994) The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58: 491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heel M (1987) Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy 21: 111–123 [DOI] [PubMed] [Google Scholar]
- Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA (2010) Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468: 709–712 [DOI] [PubMed] [Google Scholar]
- Wahlberg JM, Garoff H (1992) Membrane fusion process of Semliki Forest virus. I: Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol 116: 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Ferro C, Barrera R, Boshell J, Navarro JC (2004) Venezuelan equine encephalitis. Annu Rev Entomol 49: 141–174 [DOI] [PubMed] [Google Scholar]
- Wengler G (2009) The regulation of disassembly of alphavirus cores. Arch Virol 154: 381–390 [DOI] [PubMed] [Google Scholar]
- Wengler G, Wurkner D, Wengler G (1992) Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology 191: 880–888 [DOI] [PubMed] [Google Scholar]
- Wolf M, Garcea RL, Grigorieff N, Harrison SC (2010) Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A 107: 6298–6303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SR, Haag L, Sjoberg M, Garoff H, Hammar L (2008) The dynamic envelope of a fusion class II virus. E3 domain of glycoprotein E2 precursor in Semliki Forest virus provides a unique contact with the fusion protein E1. J Biol Chem 283: 26452–26460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Fisher BR, Olson NH, Strauss JH, Kuhn RJ, Baker TS (2002) Aura virus structure suggests that the T=4 organization is a fundamental property of viral structural proteins. J Virol 76: 7239–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Heil M, Kuhn RJ, Baker TS (2005) Heparin binding sites on Ross River virus revealed by electron cryo-microscopy. Virology 332: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH (2010) 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141: 472–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N (2008) Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A 105: 1867–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Lindqvist B, Garoff H, von Bonsdorff CH, Liljestrom P (1994) A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J 13: 4204–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W (2000) Seeing the herpesvirus capsid at 8.5 A. Science 288: 877–880 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.