Abstract
Background
PPARδ (NR1C2) promotes lipid accumulation in human macrophages in vitro and has been implicated in the response of macrophages to vLDL. We have investigated the role of PPARδ in PMA-stimulated macrophage differentiation.
The THP-1 monocytic cell line which displays macrophage like differentiation in response to phorbol esters was used as a model system. We manipulated the response to PMA using a potent synthetic agonist of PPARδ , compound F. THP-1 sub-lines that either over-expressed PPARδ protein, or expressed PPARδ anti-sense RNA were generated. We then explored the effects of these genetic modulations on the differentiation process.
Results
The PPARδ agonist, compound F, stimulated differentiation in the presence of sub-nanomolar concentrations of phorbol ester. Several markers of differentiation were induced by compound F in a synergistic fashion with phorbol ester, including CD68 and IL8. Over-expression of PPARδ also sensitised THP-1 cells to phorbol ester and correspondingly, inhibition of PPARδ by anti-sense RNA completely abolished this response.
Conclusions
These data collectively demonstrate that PPARδ plays a fundamental role in mediating a subset of cellular effects of phorbol ester and supports observations from mouse knockout models that PPARδ is involved in macrophage-mediated inflammatory responses.
Background
The peroxisome proliferator activated receptors (PPARs) function as molecular sensors of dietary fatty acids and serum lipoproteins, and are central to many cellular and metabolic processes including development, proliferation, differentiation and lipid homeostasis. There are three isoforms of: α, γ and δ, with its own tissue-specific distribution, suggesting that different functions can be ascribed to each receptor [1-6].
PPARα plays an important role in lipid homeostasis – agonists upregulate peroxisomal β-oxidation and thus clear circulating lipids [2]. It is also a negative regulator of inflammation, demonstrated by the fact that the PPARα knockout mouse exhibits a prolonged inflammatory response [7]. Activation of PPARα also leads to an upregulation in IκB which prevents nuclear translocation of NF-κB, and thus leads to the inhibition of NF-κB transactivation of a number of pro-inflammatory gene products [8,9].
The role of PPARγ in adipocyte differentiation has been well documented [8-11] and there is also evidence implicating this receptor in the development of the human macrophage [9,12-15]. Both mouse and human atherosclerotic lesions show a high level of PPARγ expression and these findings provoked intense interest in the regulatory actions of PPARs in monocyte-macrophage biology. However, recent studies with PPARγ-null mice and mouse ES cells indicate that this receptor is neither essential for nor substantially affects the development of the mouse macrophage lineage both in vitro and in vivo [12]. Supporting the notion that PPARγ acts as positive regulator of terminal differentiation (at least in some cell types) is the observation that agonists are anti-proliferative [16-20].
The functions of PPARδ are less well characterised, although studies of the δ-null mouse reveal a developmental role [21], and previous work from this laboratory has demonstrated a role in lipid metabolism [22]. Activation of PPARδ in the macrophage leads to lipid accumulation, with an increase in mRNA levels of the scavenger receptors SR-A and CD36. A downregulation of Cyp27 and ApoE, both genes involved in macrophage lipid export is also observed. Furthermore, PPARδ appears to mediate macrophage lipid loading by vLDL [23] and may be the target of oxidised lipids liberated from oxLDL [24]. In agreement with this the deletion of PPARδ in macrophages profoundly decreases atherogenesis in an atherosclerosis prone mouse model [25]. Other studies have also revealed PPARδ to be involved in the proliferation and differentiation of several cell types: while the expression and activation of PPARγ is crucial for the terminal differentiation of the adipocyte, δ is involved in the very early stages of this cascade [26]. It appears that, at the initiation of adipogenesis, PPARδ is pro-proliferative and its activation leads to mitotic clonal expansion. Its role in proliferation is confirmed by its role in intestinal tumorigenesis – expression is up-regulated in human colorectal cancer cells and by β-catenin, whilst expression is down-regulated by the tumour suppressor APC [17,26-28]. PPARδ is also known to be involved in the epidermal wound healing response [29,30]. Pro-inflammatory cytokines such as TNF-α increase PPARδ expression as well as triggering production of endogenous ligands for this receptor. The PPARδ knockout mouse is both severely delayed with respect to inflammatory cytokine-stimulated epidermal differentiation and more sensitive to TNF-α induced apoptosis. PPARδ has been described as "defining the cellular response to stress" [29] with the up-regulation of its expression directing TNF-α signalling away from the apoptotic pathway and towards cell survival and differentiation.
We have shown that PPARδ is a positive effector of lipid accumulation in the macrophage [22] and thus may be involved in the formation of the macrophage-derived foam cell. The initiating event in plaque formation is often considered to be an inflammatory response to endothelial injury, with dyslipidaemia leading to a subendothelial accumulation of modified LDL. This aggregated lipid is scavenged by monocytes which have left the circulation to enter the arterial intima and differentiated into activated macrophages. The resulting lipid accumulation leads to the formation of the macrophage-derived foam cell, and such cells perpetuate the disease state by producing cytokine signals for further monocyte recruitment.
We now present data that implicate PPARδ, not only in lipid accumulation, but also in macrophage differentiation. We have used, as an in vitro model of macrophage differentiation, THP-1 cells, a human monocyte-derived cell line that differentiates in response to phorbol ester. We have demonstrated previously that PMA-induced differentiation is accompanied by a marked upregulation in expression of PPARδ . It has also been shown that PMA is an activator of PPARδ and in this study we have demonstrated that PPARδ activity modulates the differentiation state of PMA-treated THP-1 cells.
Results
Up-regulation of PPARδ message and protein expression accompanies the differentiation of the THP-1 monocyte-derived macrophage
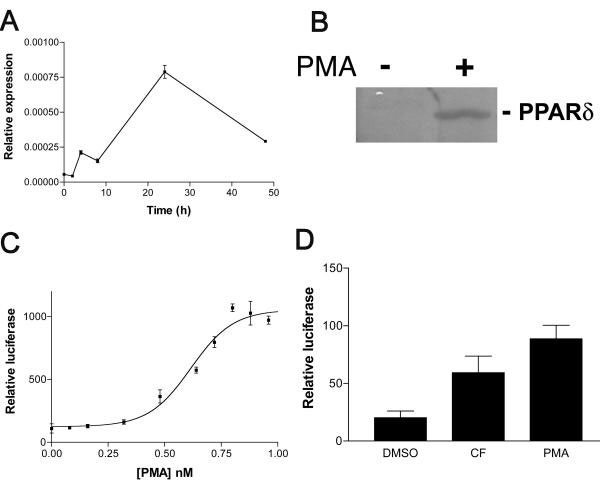
The THP-1 cell line is widely used as a model for the investigation of macrophage biology. These are suspension cells which could be induced to differentiate in the presence of the protein kinase C activator, phorbol ester (PMA), after treatment with which they became adherent to the culture substratum. This process was accompanied by an up-regulation in the PPARδ mRNA (Fig. 1A) beginning within 2 h of the PMA stimulus and reaching a peak at 24 h. This elevation in the mRNA was accompanied by an increase in protein expression (Fig. 1B).
Figure 1.
PPARδ expression and activity is increased by phorbol ester. A: Time course of induction of PPARδ message in response to phorbol ester. THP-1 cells were plated in RPMI medium supplemented with 10% FCS and 1 nM PMA at a density of 1 × 106 per well. At each time point, cells were lyzed and RNA prepared, followed by cDNA synthesis. QPCR analysis was performed using PPARδ-specific primers and probe to determine the amount of message. Results are expressed as total relative to 18S and were performed in triplicate. B: Expression of PPARδ protein by THP-1 cells is up-regulated in response to phorbol ester. THP-1 cells were plated in RPMI medium supplemented with 10% FCS and either 1 nM PMA or vehicle and incubated for 48 h. Cells were then lyzed directly into SDS-PAGE sample buffer for Western blotting. Blots were probed using a rabbit polyclonal anti-serum raised against a GST-PPARδ AB domain fusion protein. C: COS-1 cells were transiently co-transfected with pFABPLuc and pCLDNhPPARδ. Transfection efficiency was controlled for by co-transfection with a plasmid encoding β-galactosidase. Cells thus transfected were treated with increasing concentrations of PMA for 24 h after which luciferase activity was measured. D: PMA-induced δ-activation is mediated through the ligand binding domain. COS-1 cells were transfected with an expression vector encoding a Gal-4/PPARδ ligand-binding domain chimera. Cells were then treated for 24 h with compound F, PMA or vehicle only and luciferase readings obtained.
PMA is an activator of PPARδ
In order to investigate the effect of PMA on PPARδ signalling, we employed a transient transfection approach. COS-1 cells were transfected with a luciferase reporter plasmid containing 4 copies of the liver fatty acid binding protein PPRE in front of a minimal TK promoter and a plasmid directing the expression of full length PPARδ . The ability of increasing concentrations of PMA to activate PPARδ was tested in a dose response (Fig. 1C) and the results indicated that phorbol ester is a very effective activator of the receptor, achieving a 4–5 fold maximal induction compared to the vehicle-only control at approximately 1 nM PMA. This appeared to be due entirely to activation of PPARδ as no increase in PPARδ protein was observed upon treatment of the COS-1 cells with PMA (data not shown). Transient transfection experiments (Fig. 1D) using a chimeric receptor in which the Gal4 DNA-binding domain is fused in frame to the ligand binding domain of PPARδ demonstrated activation by PMA. Thus, activation of the UAS reporter construct therefore reflects PMA signalling mediated by the ligand-binding domain of PPARδ . This is in agreement with the results of Tan et al. [29]
A PPARδ agonist sensitizes THP-1 cells to PMA
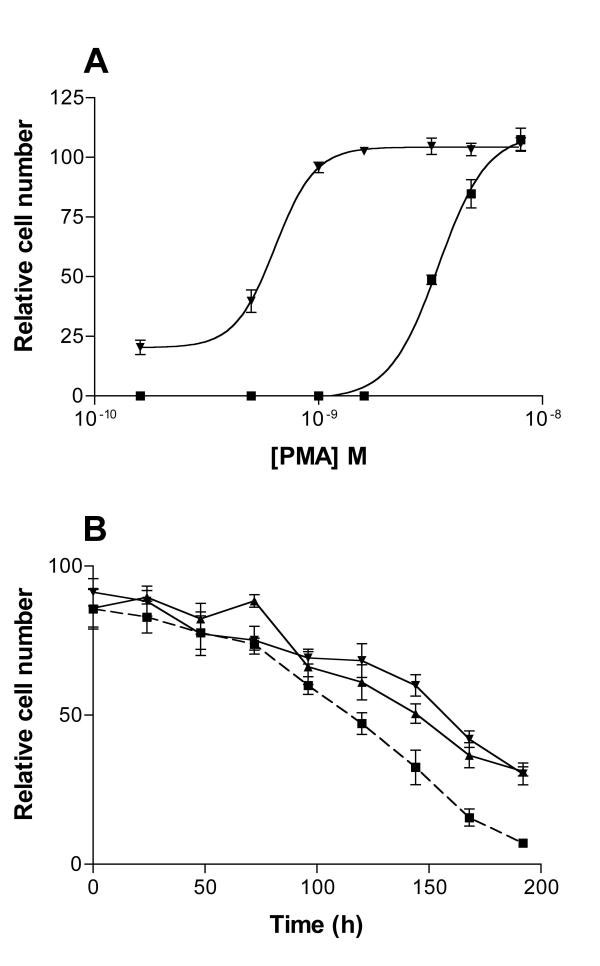
As PMA appears to modulate both expression and activity of PPARδ, we investigated the dynamics of the THP-1 response to phorbol ester and the PPARδ agonist, compound F. (Fig. 2). PMA induced THP-1 cells to differentiate in a dose-dependent manner, with an EC50 of approximately 3 nM. Co-treatment of the cells with 100 nM compound F resulted in a reduced EC50 of 1 nM PMA (Fig. 2A). If THP-1 cells were treated with low levels of PMA (>1 nM), very few were seen to differentiate, unless simultaneously treated with compound F, when the number of cells undergoing differentiation and adhering to the culture substratum was seen to increase significantly (p < 0.0001) in a compound F dose-dependent manner (Fig. 2B), with maximal activity at approximately 10 nM. PMA-induced differentiation was dependent on continued PMA signalling: when cells were treated with a single dose of PMA (1 nM), and the PMA was subsequently withdrawn 48 h later, the cells began to de-differentiate and detach from the substratum. After 8 days, less than 20% of the initial total remained adherent. When the cells were exposed to 20 nM compound F treatment following the PMA withdrawal, the differentiated phenotype was retained for significantly longer (p < 0.0001), with some 50% of cells still attached (Fig. 2C). These results indicate that PPARδ activity is required for both the initiation and maintenance of adhesion. However, despite the fact that compound F treated THP-1 cells showed acute sensitivity to very low doses of PMA, compound F alone was not sufficient to induce differentiation indicating that other factors in addition to expression and ligand activation of PPARδ are involved in the induction of macrophage differentiation.
Figure 2.
Phorbol ester and compound F co-operate to induce and maintain differentiation of THP-1 cells. A: Compound F sensitizes THP-1 cells to PMA-induced adherence. Dose-responses of THP-1 cells to PMA were prepared in the presence of vehicle alone (squares) or 100 nM compound F(triangles). Cells were plated in RPMI medium (0.1% FCS) and harvested after 48 h. Cell numbers were determined by fluorimetric analysis of SYBR green stained plates. B: Compound F co-operates with phorbol ester in promoting differentiation of THP-1 cells. Cells were plated for 48 h in RPMI medium supplemented with 0.1% FCS and 0.5 nM PMA and a compound F dose-response performed. Results obtained were subjected to non-linear regression analysis. C: Compound F maintains the adherence of THP-1 cells in the absence of a prolonged PMA stimulus. THP-1 cells were plated in RPMI supplemented with 10% FCS and 1 nM PMA. After 48 h, the PMA was removed and replaced with either 100 nM compound F (triangles) or vehicle only (squares). At 48 h intervals up to 6 days, the number of cells remaining adherent to the culture substratum was measured by fluorimetric analysis of SYBR green stained plates and the results compared to a standard curve.
Differentiation markers are regulated by compound F and PMA in a synergistic manner
Cells were treated with increasing doses of PMA for up to 48 h with medium containing 0.5% FCS, in the presence (broken line) or absence (solid line) of compound F. Cells were lyzed and cDNA prepared for QPCR analysis. Several gene targets were analyzed for their known role in macrophage differentiation. CD68 (Fig. 3A) is a classical marker of macrophage differentiation. In the absence of compound F there was a slight increase in CD68 mRNA with increasing concentrations of PMA. In the presence of 20 nM compound F, there was a profound increase in message in response to PMA treatment. Similar effects were observed with IL8 (Fig. 3B), a potent macrophage chemoattractant, known to be important in the development of the inflammatory phenotype. Adipophilin (Fig. 3C) and adipocyte fatty acid binding protein (AFABP) (Fig. 3D) both showed a similar synergistic response following treatment with PMA and compound F. Both of these are crucial to lipid trafficking and storage during the differentiation of macrophages into a foam cells and we and others have shown previously that they are targets of PPARδ [22,23]. Levels of IL1β were not significantly affected (Fig. 3E).
Figure 3.
Compound F modulates phorbol ester activation of gene targets involved in differentiation. Cells were plated in RPMI 1640 supplemented with 0.5% FCS. A PMA dose-response was carried out, in the presence of either vehicle alone (squares, broken lines) or compound F (triangles, solid lines). Cells were lyzed after 24 h (CD68; A, IL8; B, AFABP; C and adipophilin; D) or 48 h (IL1β ; E, Osteopontin; F and MMP-9; G). RNA was prepared from cell lysates and cDNA was prepared for QPCR analysis.
MMP9 (Fig. 3F) is a matrix metalloproteinase important in inflammation. MMP9 facilitates the entry of macrophages into peripheral tissues by allowing them to penetrate the basement membrane underlying the endothelium. Osteopontin (Fig. 3G) is another macrophage differentiation marker. Both of these gene targets showed an up-regulation in message in response to PMA, and this effect was still present in the cells treated with the compound F. In this case, the effect of the PPARδ agonist was to blunt the PMA response.
Over-expression of PPARδ is functionally similar to treating cells with compound F
Stable cell lines over-expressing PPARδ were generated as previously described [22]. The effect of this on the PMA-induced differentiation response is shown in Fig. 4A. The PPARδ over-expressing cells differentiated at much lower concentrations of PMA when compared to the parental THP-1 cells. This effect was even greater than that observed with compound F. Over-expression of PPARδ was also able to limit the de-differentiation in the absence of a prolonged PMA stimulus in a manner similar to compound F treated parental cells (p < 0.0001, Fig. 4B).
Figure 4.
Over-expression of PPARδ potentiates the differentiation response. A: Both wild type (squares) and SENSE cells (inverted triangles) were treated with increasing concentrations of PMA. Numbers of adherent cells were quantified by SYBR green staining and compared to a standard curve. B: Over-expression of PPARδ (inverted triangles) enhances the maintenance of adhesion following a single treatment with PMA when compared with the parental cell line (squares). For comparison, the compound F result (triangles; broken line) is also shown.
PPARδ is required for the differentiation of THP-1 monocyte-derived macrophages
Stable cell lines were also generated that expressed PPARδ anti-sense mRNA. These cell lines also appeared to be normal with respect to their routine culture characteristics. Fig. 5A shows western blotting of parental, SENSE and ANTISENSE cell lines, both in the presence and absence of PMA. PPARδ expression is undetectable unless the cells are treated with PMA, while untreated SENSE cells show a level of expression similar to that observed with the PMA-treated parental cells. ANTISENSE cells do not express PPARδ even when stimulated with PMA. Figs. 5B and 5C show the response of the cell lines to treatment with 8 nM PMA compared to the wild type. Cells were plated at a density of 5 × 105/ml and left for 48 h, during which time all three cell lines underwent a doubling in number. At 48 h, the cells were treated with a single dose of PMA. This was left on the cells for a further 48 h, after which the medium was removed and replaced with medium without PMA. The cells were left for a further 96 h. Following PMA treatment, both adherent (Fig. 5B) and non-adherent (Fig. 5C) cell number was determined by SYBR staining at 48 h intervals. Fig. 5C shows that no ANTISENSE cells ever became adherent following PMA treatment, indicating that PPARδ is required for the differentiation of THP-1 cells. When the non-adherent cells are considered (Fig. 5B) it can be seen that, for the duration of the PMA treatment, ANTISENSE cells did not proliferate. Once the PMA was removed, the cells rapidly resumed proliferation, indicating that PMA-induced growth arrest of THP-1 cells is PPARδ-independent.
Figure 5.
PPARδ is necessary for PMA stimulated adherence of THP-1 cells. Wild-type THP-1 cells (squares), SENSE (inverted triangles) and ANTISENSE cells (circles) were plated at a density of 5 × 105 cells/ml. After 48 h the cells were treated with PMA and this was maintained for a further 48 h. After this time the PMA was removed. At all stages, both adherent (A) and non-adherent cells (B) were counted with SYBR green and cell number calculated from a standard curve.
Discussion
Macrophages have a central role as effector cells at sites of chronic inflammation, such as the atherosclerotic plaque, and the differentiation of a monocyte into a macrophage represents a key event in this process; underlying mechanisms may therefore present attractive targets for therapeutic intervention. Work by the group of Wahli [29] has suggested a model for keratinocyte differentiation whereby PMA-induced activation of PKC leads to PLA2-induced liberation of arachidonic acid and subsequent generation of endogenous ligands for PPARδ. Activation of PPARδ leads to modulation of gene targets involved in the differentiated keratinocyte phenotype. This study has examined the hypothesis that PMA signalling in macrophage differentiation involves PPARδ. In this THP-1 model, differentiation is induced by treatment of the cells with the potent inflammatory stimulus, phorbol ester (PMA). Following treatment with PMA, we observed that PPARδ message began to rise almost immediately and this was reflected in the level of protein expression observed at 48 h. PMA represents a potent and pleiotropic inflammatory insult to the cell, and parallels can be drawn with the response to skin wounding (or treatment with PMA) demonstrated by Wahli et al [29,30]. In normal adult skin, PPARδ expression is undetectable, but this is upregulated in response to injury. Treatment with PPARδ agonists induces the expression of keratinocyte markers of differentiation. This up-regulation of PPARδ is associated with necrosis and it appears that it is this receptor, which mediates the necrosis-associated signals that ultimately trigger keratinocyte differentiation. It has also been shown that, in skin, PMA-induced differentiation is PPARδ-dependent: the peak of PPARδ expression coincides with a switch to cell cycle arrest and the onset of differentiation.
Differentiation and proliferation can be considered functional inverses of one another: commitment to differentiation invariably involves the cell entering a state of growth arrest. Differentiation of the macrophage has several distinct phases. Initially the cell will become growth arrested, then it will become activated and attach to the substratum (whether it be the culture vessel or the endothelium) and finally it must maintain its differentiated phenotype. We have shown that PMA was able to induce growth arrest of THP-1 cells and promote adhesion in a dose-dependent manner (EC50 = 3 nM). This effect was potentiated by the highly specific PPARδ agonist, compound F, which reduced the EC50 to 1 nM. Here parallels can be drawn with the adipocyte differentiation cascade: adipocyte differentiation can be induced if the pre-adipocytes are treated by PPARγ agonists, but this effect is dependent on the presence of signalling by insulin [31].
It is apparent that PPARδ is involved not only in the initiation of THP-1 differentiation, but also in its maintenance. PMA-induced molecular events appear, generally, to be short-lived: its effects with respect to differentiation are reversible by its removal. The activation of PPARδ appears to stabilise the PMA response, maintaining the differentiated phenotype even in the absence of prolonged PMA stimulation. Therefore, PPARδ activation may represent a transition from acute effects of PMA to longer-term genomic signalling.
In keratinocytes, both PMA and TNF-α have been shown to elicit differentiation and inflammation via PPARδ [29]. Both stimuli induce PPARδ expression via activation of AP-1 sites in the promoter. This work and that of others [29] has shown that PMA can activate a Gal4PPARδ LBD fusion protein, suggesting the generation of endogenous ligands. However, the identity of a PMA inducible-endogenous ligand has yet to be elucidated.
In keeping with the notion that the effects of phorbol ester on differentiation are mediated through PPARδ, we have demonstrated that PPARδ-specific genes are also targets of PMA, and that the two compounds synergise in regulating expression of genes known to be characteristic of the differentiated phenotype. IL8 is of particular interest because of its role in inflammation and atherosclerosis [32]. Other PPARδ-specific targets, such as adipophilin and AFABP are associated with the differentiation of macrophages into foam cells [22], important in the development of atherosclerosis, another chronic inflammatory condition which can be regarded as a pathological differentiation. That PPARδ is involved in chronic inflammation is perhaps not surprising: this isoform is capable of acting as an intrinsic transcriptional repressor of the other isoforms, PPAR-α and -γ, and it has been suggested that this may account for its ubiquitous expression, acting as a tissue-specific regulator of PPAR target genes [33]. It is known that both PPARα and γ are involved in the limitation of the inflammatory response [7], and it may be that up-regulation of expression of PPARδ allows α- and γ-mediated anti-inflammatory signalling to be overcome, permitting the inflammatory response to continue unchecked. Indeed, PPARγ signalling has been specifically implicated in the resolution of inflammation [34].
We have used both a pharmacological and genetic approach, demonstrating gain- and loss-of-function to implicate PPARδ in macrophage differentiation. Our data is supported by the fact that leukocytes from PPARδ-null mice have an 80% reduction in Mac-1 expression [23] and that PPARδ-null derived bone marrow supports very limited atherogenesis [25]. Our study extends these observations to a human system, and confirms that targeting of the PPARδ signalling pathway may have therapeutic benefits in conditions such as rheumatoid arthritis and atherosclerosis.
Methods
Materials
RPMI 1640, DMEM, fetal bovine serum (FBS) and SuperScript II were purchased from Gibco Life Technologies, UK. Phorbol-12-myristate-13-acetate (PMA) and G418 were obtained from Sigma-Aldrich, UK and the PPARδ ligand 3-propyl-4-(3-(3-trifluoromethyl-7-propyl-6-benz- [35]-isoxazoloxy)-propylthio)phenylacetic acid (compound F) were a gift from GlaxoSmithKline, UK.
Culture and differentiation of THP-1 cells
The human THP-1 cell line was obtained from ATCC and grown in RPMI 1640 supplemented with 10% heat-inactivated FBS and 20 μM β-mercaptoethanol. Cells were maintained at 37°C and 5% CO2. Experiments were performed in 6-well plates where differentiation was induced by resuspending the cells in RPMI containing either 0.5% FBS and PMA as described in the figure legends. Experiments were performed in both the presence, or absence of compound F (either 20 or 100 nM final concentration). All drugs were added with DMSO as a vehicle and were replaced at intervals of 48 hours unless otherwise stated.
Cell counting
At the end of each experiment, adherent THP-1 cells were fixed with a solution of 4% paraformaldehyde, 1% Triton-X for 30 minutes at room temperature, following which the DNA was stained with a 1: 10,000 SYBR green solution (Molecular Probes). Plates were incubated for 24 h at 4°C in the dark. Cells were washed in PBS and the relative numbers determined from a standard curve by fluorimetry using a LabSystems FluoroSkan Ascent FL microplate reader.
Isolation of stable cell lines
PPARδ over-expressing cell lines (SENSE) and THP-1 anti-sense cells (ANTISENSE) were prepared as described previously [22]. Briefly, THP-1 cells were transfected with an expression vector containing the entire human PPARδ coding sequence in either sense or anti-sense orientation respectively. Transfection was by a modified DEAE-dextran procedure [36] and pools of stable cells were selected by maintaining cells in medium containing 1 mg/ml G418, 10% FBS and 10% THP-1 conditioned medium, with vigorous washing to remove dead cells. This procedure was repeated until cell killing stopped and robust growth was observed. Six pools from independent transfections were obtained, each pool displayed a similar phenotype.
Plasmids and transient transfection procedures
COS-1 cells were transfected in 6-well plates using DEAE-dextran as described previously [37]. The reporter construct used was pFABPLuc, which consists of four copies of the human peroxisome proliferator response element from the human liver fatty acid binding protein (FABP) gene in front of the herpes simplex virus thymidine kinase promoter, cloned immediately upstream of the cDNA encoding firefly luciferase in pGLBAS (Promega). The PPARδ expression vector (pCLDNhPPARδ) contained the coding sequence for human PPARδ under the control of the enhancer/promoter of the human cytomegalovirus. The Gal4/LBD plasmid consisted of an in-frame fusion of the Gal4 DNA-binding domain and the ligand-binding domain of PPARδ . The reporter plasmid without the PPRE (TKLuc) and the empty expression vector (pCLDN) were used as controls. pSV β-galactosidase (Promega) was co-transfected in all cases in order to control for transfection efficiency. Following transfection and recovery, the cells were incubated for 18 h with DMEM containing 10% FBS and 1% streptomycin/penicillin, supplemented with vehicle (DMSO), compound F or PMA. Cells were analysed for β-galactosidase and luciferase activities using assay kits as described by the manufacturer (Promega). Data are presented as the ratio of relative light units obtained with the luciferase assay to the absorbance obtained at 415 nm in the β-galactosidase assays (relative luciferase).
Western Blotting
THP-1 wild type, SENSE and ANTISENSE cells were treated with PMA or vehicle for 48 h following which they were lyzed in SDS-PAGE loading buffer and analysed using standard Western blotting procedures. The primary antibody was either PPARδ antiserum (a gift from Dr David Bell), used at a 1:2000 dilution, or an antibody raised against the AB domain (residues 1–100) of PPARδ, used at a dilution of 1:500. The AFAR antiserum (obtained from Prof. John Hayes) was used at a 1:3000 dilution. The secondary detection reagent, a peroxidase conjugated mouse anti-rabbit IgG antiserum (Sigma) was used at a dilution of 1:3000. Bands were visualised using enhanced chemiluminesence (ECL+) as described by the manufacturer (Amersham Pharmacia Biotech).
RNA extraction and analysis
RNA was extracted from differentiated and undifferentiated THP-1 cells using the Qiagen RNeasy kit. cDNA was synthesised from this RNA using SuperScript II. QPCR analysis was performed on triplicate experiments using the standard Applied Biosystems procedures, with primers and probes described in Table 1. 18S RNA was quantified using an assay from Applied Biosystems.
Table 1.
Sequences of primers and probes used for QPCR in Fig. 3.
| Target | Sequence (5'-3') |
| PPARδ | Forward: GGGACCACAGCATGCACTTC Reverse: TGCAGTTGGTCCAGCAGTGA Probe: CCAGCAGCTACACAGACCTCTCCCGG |
| CD68 | Forward: TCCTCGCCCTGGTGCTTA Reverse: GGGCCTGGTAGGCGGAT Probe: TTTCTGCATCATCCGGAGACG |
| IL8 | Forward: AAGGAACCATCTCACTGTGTGTAAAC Reverse: ATCAGGAAGGCTGCCAAGAG Probe: TGACTTCCAGCTGGCCGTGGC |
| AFABP | Forward: AGTAGGAGTGGGCTTTGCCA Reverse: TCACATCCCCATTCACACTGA Probe: AGGAAAGTGGCCGTGGC |
| Adipophilin | Forward: TGGCAGAGAACGGTGTGAAG Reverse: CTGGATGATGGGCAGAGCA Probe: CATCACCTCCGTGGCCATGACCA |
| IL1β | Forward: TGAGCACCTTCTTTCCCTTCA Reverse: GTACAGGTGCATCGTGCACAT Probe: CCTATCTTCTTCGACACATGGGATAACGAGGC |
| Osteopontin | Forward: TGACCCATCTCAGAAGCAGAATC Reverse: CTCTTGTTTAAAGTCATTGGTTTCTTCA Probe: CCTAGCCCCACAGAATGCTGTGTCCTC |
| MMP9 | Forward: GCTCACCTTCACTCGCGTG Reverse: CGCGACACCAAACTGGATG Probe: ACAGCCGGGACGCAGACATCG |
Statistical analysis
All graphs and statistics were prepared using Graphpad Prism for the Macintosh v3.0 (Graphpad Inc., San Diego, CA). p values were calculated using two-way ANOVA, unless otherwise stated.
Authors' contributions
Helen Vosper made the THP-1 cell lines, discovered the requirement of PPARδ in the response to phorbol ester with the anti-sense cell line, performed differentiation and gene expression analysis. Guennadi Khoudoli established the gain-of-function assay using PPARδ ligand and performed differentiation and gene expression analysis. Colin Palmer conceived the study and coordinated its design and implementation. All authors participated in the writing of the manuscript and have read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was entirely supported by the British Heart Foundation project grants 99/020 (GK) and 02/071 (HV). We would also like to thank Mattias Gustafsson for the gift of the expression vector encoding the PPARδ LBD/GAL4 chimera.
Contributor Information
Helen Vosper, Email: vosperh@cancer.org.uk.
Guennadi A Khoudoli, Email: g.a.khoudoli@dundee.ac.uk.
Colin NA Palmer, Email: colin.palmer@cancer.org.uk.
References
- Hihi AK, Michalik L, Wahli W. PPARs: transcriptional effectors of fatty acids and their derivatives. Cell Mol Life Sci. 2002;59:790–798. doi: 10.1007/s00018-002-8467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latruffe N, Malki MC, Nicolas-Frances V, Clemencet MC, Jannin B, Berlot JP. Regulation of the peroxisomal beta-oxidation-dependent pathway by peroxisome proliferator-activated receptor alpha and kinases. Biochem Pharmacol. 2000;60:1027–1032. doi: 10.1016/S0006-2952(00)00416-0. [DOI] [PubMed] [Google Scholar]
- Green S. PPAR: a mediator of peroxisome proliferator action. Mutat Res. 1995;333:101–109. doi: 10.1016/0027-5107(95)00136-0. [DOI] [PubMed] [Google Scholar]
- Green S, Wahli W. Peroxisome proliferator-activated receptors: finding the orphan a home. Mol Cell Endocrinol. 1994;100:149–153. doi: 10.1016/0303-7207(94)90294-1. [DOI] [PubMed] [Google Scholar]
- Devchand PR, Ijpenberg A, Devesvergne B, Wahli W. PPARs: nuclear receptors for fatty acids, eicosanoids, and xenobiotics. Adv Exp Med Biol. 1999;469:231–236. doi: 10.1007/978-1-4615-4793-8_34. [DOI] [PubMed] [Google Scholar]
- Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20:649–688. doi: 10.1210/er.20.5.649. [DOI] [PubMed] [Google Scholar]
- Devchand PR, Keller H, Peters JM, Vazquez M, Gonzalez FJ, Wahli W. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- Neve BP, Corseaux D, Chinetti G, Zawadzki C, Fruchart JC, Duriez P, Staels B, Jude B. PPARalpha Agonists Inhibit Tissue Factor Expression in Human Monocytes and Macrophages. Circulation. 2001;103:207–212. doi: 10.1161/01.cir.103.2.207. [DOI] [PubMed] [Google Scholar]
- Delerive P, Gervois P, Fruchart JC, Staels B. Induction of IkappaBalpha expression as a mechanism contributing to the anti-inflammatory activities of peroxisome proliferator-activated receptor-alpha activators. J Biol Chem. 2000;275:36703–36707. doi: 10.1074/jbc.M004045200. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Hu E, Kim JB, Brun R. PPAR gamma and the control of adipogenesis. Biochimie. 1997;79:111–112. doi: 10.1016/S0300-9084(97)81500-3. [DOI] [PubMed] [Google Scholar]
- Vidal-Puig AJ, Considine RV, Jimenez-Linan M, Werman A, Pories WJ, Caro JF, Flier JS. Peroxisome proliferator-activated receptor gene expression in human tissues. Effects of obesity, weight loss, and regulation by insulin and glucocorticoids. J Clin Invest. 1997;99:2416–2422. doi: 10.1172/JCI119424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Barak Y, Nagy L, Liao D, Tontonoz P, Evans RM. PPAR-gamma dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat Med. 2001;7:48–52. doi: 10.1038/83336. [DOI] [PubMed] [Google Scholar]
- Marx N, Sukhova G, Murphy C, Libby P, Plutzky J. Macrophages in human atheroma contain PPARgamma: differentiation-dependent peroxisomal proliferator-activated receptor gamma(PPARgamma) expression and reduction of MMP-9 activity through PPARgamma activation in mononuclear phagocytes in vitro. Am J Pathol. 1998;153:17–23. doi: 10.1016/s0002-9440(10)65540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, Rosen ED, Fitzgerald ML, Randow F, Andersson LP, Altshuler D, Milstone DS, Mortensen RM, Spiegelman BM, Freeman MW. The role of PPAR-gamma in macrophage differentiation and cholesterol uptake. Nat Med. 2001;7:41–47. doi: 10.1038/83328. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93:241–252. doi: 10.1016/S0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- Haydon RC, Zhou L, Feng T, Breyer B, Cheng H, Jiang W, Ishikawa A, Peabody T, Montag A, Simon MA, He TC. Nuclear receptor agonists as potential differentiation therapy agents for human osteosarcoma. Clin Cancer Res. 2002;8:1288–1294. [PMC free article] [PubMed] [Google Scholar]
- Bastie C. [PPARdelta and PPARgamma: roles in fatty acids signalling, implication in tumorigenesis] Bull Cancer. 2002;89:23–28. [PubMed] [Google Scholar]
- Itami A, Watanabe G, Shimada Y, Hashimoto Y, Kawamura J, Kato M, Hosotani R, Imamura M. Ligands for peroxisome proliferator-activated receptor gamma inhibit growth of pancreatic cancers both in vitro and in vivo. Int J Cancer. 2001;94:370–376. doi: 10.1002/ijc.1488. [DOI] [PubMed] [Google Scholar]
- Ricote M, Huang J, Fajas L, Li A, Welch J, Najib J, Witztum JL, Auwerx J, Palinski W, Glass CK. Expression of the peroxisome proliferator-activated receptor gamma (PPARgamma) in human atherosclerosis and regulation in macrophages by colony stimulating factors and oxidized low density lipoprotein. Proc Natl Acad Sci U S A. 1998;95:7614–7619. doi: 10.1073/pnas.95.13.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami S, Arai G, Hayashi T, Fujii Y, Xia G, Kageyama Y, Kihara K. PPARgamma ligands suppress proliferation of human urothelial basal cells in vitro. J Cell Physiol. 2002;191:310–319. doi: 10.1002/jcp.10099. [DOI] [PubMed] [Google Scholar]
- Peters JM, Lee SS, Li W, Ward JM, Gavrilova O, Everett C, Reitman ML, Hudson LD, Gonzalez FJ. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta) Mol Cell Biol. 2000;20:5119–5128. doi: 10.1128/MCB.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosper H, Patel L, Graham TL, Khoudoli GA, Hill A, Macphee CH, Pinto I, Smith SA, Suckling KE, Wolf CR, Palmer CN. The peroxisome proliferator-activated receptor delta promotes lipid accumulation in human macrophages. J Biol Chem. 2001;276:44258–44265. doi: 10.1074/jbc.M108482200. [DOI] [PubMed] [Google Scholar]
- Chawla A, Lee CH, Barak Y, He W, Rosenfeld J, Liao D, Han J, Kang H, Evans RM. PPARdelta is a very low-density lipoprotein sensor in macrophages. Proc Natl Acad Sci U S A. 2003;100:1268–1273. doi: 10.1073/pnas.0337331100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shureiqi I, Jiang W, Zuo X, Wu Y, Stimmel JB, Leesnitzer LM, Morris JS, Fan HZ, Fischer SM, Lippman SM. The 15-lipoxygenase-1 product 13-S-hydroxyoctadecadienoic acid down-regulates PPAR-delta to induce apoptosis in colorectal cancer cells. Proc Natl Acad Sci U S A. 2003;100:9968–9973. doi: 10.1073/pnas.1631086100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Chawla A, Urbiztondo N, Liao D, Boisvert WA, Evans RM. Transcriptional repression of atherogenic inflammation: modulation by PPARdelta. Science. 2003;302:453–457. doi: 10.1126/science.1087344. [DOI] [PubMed] [Google Scholar]
- Hansen JB, Zhang H, Rasmussen TH, Petersen RK, Flindt EN, Kristiansen K. Peroxisome proliferator-activated receptor delta (PPARdelta )-mediated regulation of preadipocyte proliferation and gene expression is dependent on cAMP signaling. J Biol Chem. 2001;276:3175–3182. doi: 10.1074/jbc.M005567200. [DOI] [PubMed] [Google Scholar]
- Roberts-Thomson SJ. Peroxisome proliferator-activated receptors in tumorigenesis: targets of tumour promotion and treatment. Immunol Cell Biol. 2000;78:436–441. doi: 10.1046/j.1440-1711.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- He TC, Chan TA, Vogelstein B, Kinzler KW. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99:335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan NS, Michalik L, Noy N, Yasmin R, Pacot C, Heim M, Fluhmann B, Desvergne B, Wahli W. Critical roles of PPAR beta/delta in keratinocyte response to inflammation. Genes Dev. 2001;15:3263–3277. doi: 10.1101/gad.207501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik L, Desvergne B, Tan NS, Basu-Modak S, Escher P, Rieusset J, Peters JM, Kaya G, Gonzalez FJ, Zakany J, Metzger D, Chambon P, Duboule D, Wahli W. Impaired skin wound healing in peroxisome proliferator-activated receptor (PPAR)alpha and PPARbeta mutant mice. J Cell Biol. 2001;154:799–814. doi: 10.1083/jcb.200011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougald OA, Lane MD. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- Kraan MC, Patel DD, Haringman JJ, Smith MD, Weedon H, Ahern MJ, Breedveld FC, Tak PP. The development of clinical signs of rheumatoid synovial inflammation is associated with increased synthesis of the chemokine CXCL8 (interleukin-8) Arthritis Res. 2001;3:65–71. doi: 10.1186/ar141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Hon M, Evans RM. The peroxisome proliferator-activated receptor delta, an integrator of transcriptional repression and nuclear receptor signaling. Proc Natl Acad Sci U S A. 2002;99:2613–2618. doi: 10.1073/pnas.052707099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy DW, Colville-Nash PR, Willis D, Chivers J, Paul-Clark MJ, Willoughby DA. Inducible cyclooxygenase may have anti-inflammatory properties. Nat Med. 1999;5:698–701. doi: 10.1038/9550. [DOI] [PubMed] [Google Scholar]
- Baillie G, MacKenzie SJ, Houslay MD. Phorbol 12-myristate 13-acetate triggers the protein kinase A-mediated phosphorylation and activation of the PDE4D5 cAMP phosphodiesterase in human aortic smooth muscle cells through a route involving extracellular signal regulated kinase (ERK) Mol Pharmacol. 2001;60:1100–1111. doi: 10.1124/mol.60.5.1100. [DOI] [PubMed] [Google Scholar]
- Wu H, Moulton K, Horvai A, Parik S, Glass CK. Combinatorial interactions between AP-1 and ets domain proteins contribute to the developmental regulation of the macrophage scavenger receptor gene. Mol Cell Biol. 1994;14:2129–2139. doi: 10.1128/mcb.14.3.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]