Abstract
The transcription factor nuclear factor κB (NF-κB) regulates various cellular processes such as inflammation and apoptosis. The NF-κB essential modulator (NEMO/IKKγ) is indispensable for NF-κB activation by diverse stimuli including genotoxic stress. Here, we show that NEMO linear ubiquitination on K285/309 is critical for genotoxic NF-κB activation. The E3 ligase linear ubiquitin chain assembly complex (LUBAC) facilitates NEMO linear ubiquitination upon genotoxic stress. Inhibiting LUBAC function interrupts the genotoxic NF-κB signalling cascade by attenuating the activation of IKK and TAK1 in response to DNA damage. We further show that the linear ubiquitination of NEMO is a cytoplasmic event, potentially downstream of NEMO nuclear exportation. Moreover, ELKS ubiquitination appears to facilitate linear ubiquitination of NEMO through stabilizing NEMO:LUBAC association upon DNA damage. Deubiquitinating enzyme CYLD inhibits NEMO linear ubiquitination, possibly by disassembling both K63-linked and linear polyubiquitin. We also found that abrogating linear ubiquitination of NEMO significantly increased genotoxin-induced apoptosis, resulting in enhanced sensitivity to chemodrug in cancer cells. Therefore, LUBAC-dependent NEMO linear ubiquitination is critical for genotoxic NF-κB activation and protects cells from DNA damage-induced apoptosis.
Keywords: genotoxic stress, LUBAC, NEMO, NF-κB, ubiquitination
Introduction
The nuclear factor κB (NF-κB) is a family of transcription factors that controls expression of a large number of genes, therefore participates in regulating various physiological and pathological processes (Perkins, 2007; Hayden and Ghosh, 2008). Deregulation of NF-κB signalling has been linked to many diseases such as asthma, arthritis, diabetes, stroke, and cancer (Kumar et al, 2004). NF-κB is kept inactive by association with members of the IκB family of inhibitor proteins that sequester the NF-κB/IκB complex in the cytoplasm of resting cells. A wide variety of extracellular stimuli induces NF-κB activation through the IκB kinase (IKK) complex, which comprises two catalytic subunits, IKKα and IKKβ, and a regulatory subunit IKKγ/NEMO (NF-κB essential modulator) (Hayden and Ghosh, 2008). IKK activation leads to phosphorylation of IκB and its subsequent ubiquitin-dependent proteolysis, which liberates NF-κB for nuclear translocation and transcriptional regulation (Perkins, 2007; Hayden and Ghosh, 2008).
Multiple genotoxic anticancer drugs and ionizing radiation have been shown to activate NF-κB (Baldwin, 2001; Janssens and Tschopp, 2006; Wu and Miyamoto, 2007). Previous studies indicated that NF-κB activation by genotoxic agents is mediated mainly by an IKK-dependent pathway (Baldwin, 2001; Wu and Miyamoto, 2007). However, in contrast to membrane-bound receptor-transduced NF-κB signalling pathways, the genotoxic NF-κB signalling is initiated from the nucleus. How nuclear DNA damage signal induces activation of the cytoplasmic IKK complex has yet to be fully characterized. It was reported that IKK-unbound NEMO forms a nuclear complex, so-called PIDDosome, with p53 induced protein with death domain (PIDD) and receptor-interacting protein kinase 1 (RIP1) in response to DNA damage; and the PIDDosome formation promotes NEMO sumoylation (Janssens et al, 2005). Recently, poly(ADP-ribose) polymerase 1 was shown to assemble another nuclear complex with NEMO, PIASy (the protein inhibitor of activated STAT), and ATM (ataxia telangiectasia mutated), through poly(ADP-ribose) synthesis. This complex facilitates NEMO sumoylation upon genotoxic stimulation in certain cell types (Stilmann et al, 2009). We found sumoylation of NEMO promotes its nuclear accumulation upon genotoxic stress. In the nucleus, DNA damage induces the activation of ATM, which then directly phosphorylates NEMO and promotes NEMO mono-ubiquitination (Huang et al, 2003; Wu et al, 2006). Mono-ubiquitinated NEMO is then exported from the nucleus in association with ATM and finally forms a complex with IKKβ in the cytoplasm, leading to activation of the IKK complex. A recent report also showed that NEMO could be mono-ubiquitinated in cytoplasm in an ATM-dependent manner, which in turn mediated downstream IKK activation upon genotoxic stress (Hinz et al, 2010).
The studies on genotoxic NF-κB activation highlighted the complex and important roles of NEMO post-translational modifications, such as sumoylation, phosphorylation, and ubiquitination, in mediating NF-κB signalling cascade. Ubiquitin has a critical role in regulating multiple NF-κB signalling pathways, by promoting proteasome-dependent proteolysis (e.g., IκBα and p100) and scaffolding kinase activation (e.g., TAK1 and IKK) (Chen, 2005). NEMO has been shown to participate in ubiquitin-mediated NF-κB signalling by either being ubiquitinated or interacting with the polyubiquitin chains of the other signalling components (Skaug et al, 2009). It was shown that ubiquitin forms diverse polychains through isopeptide bond between the Gly76 of a distal ubiquitin molecule and one of seven lysines (K6, K11, K27, K29, K33, K48, or K63) located within a proximal ubiquitin, and distinct functions have been attributed to the different lysine linkages (Hochstrasser, 2006; Chen and Sun, 2009). Recently, a novel form of polyubiquitin linkage was introduced into the NF-κB signalling paradigm, where ubiquitin chains were connected with a peptide bond between Gly76 of one ubiquitin and Met1 of another ubiquitin (Tokunaga et al, 2009). It was shown that NEMO linear ubiquitination is required for efficient NF-κB activation induced by TNFα, which may also depend on the specific recognition of linear polyubiquitin by NEMO (Haas et al, 2009; Lo et al, 2009; Rahighi et al, 2009). However, the molecular mechanisms involved in this process have not been fully elucidated, and the role of NEMO linear ubiquitination in other NF-κB signalling pathways remained to be determined.
The linear polyubiquitin is catalysed by the linear ubiquitin chain assembly complex (LUBAC) E3 ligase, which is composed of HOIL-1L, HOIP, and SHARPIN (Kirisako et al, 2006; Gerlach et al, 2011; Ikeda et al, 2011; Tokunaga et al, 2011). We found that LUBAC was indispensable for NF-κB activation upon genotoxic stimulation, potentially through facilitating NEMO linear ubiquitination on Lys285 and Lys309. DNA damage-induced polyubiquitination of ELKS is crucial for LUBAC to associate with and facilitate NEMO linear ubiquitination. Moreover, abrogating linear ubiquitination of NEMO significantly inhibited the sequential activation of TAK1, IKK, and NF-κB upon genotoxic stress, resulting in increased apoptosis. Altogether, our data indicate that LUBAC-mediated NEMO linear ubiquitination has an important role in transducing nuclear genotoxic signals to the cytoplasmic IKK complex by facilitating sequential activation of TAK1 and IKK, and in turn regulating NF-κB-dependent cell survival.
Results
LUBAC is required for genotoxic NF-κB activation
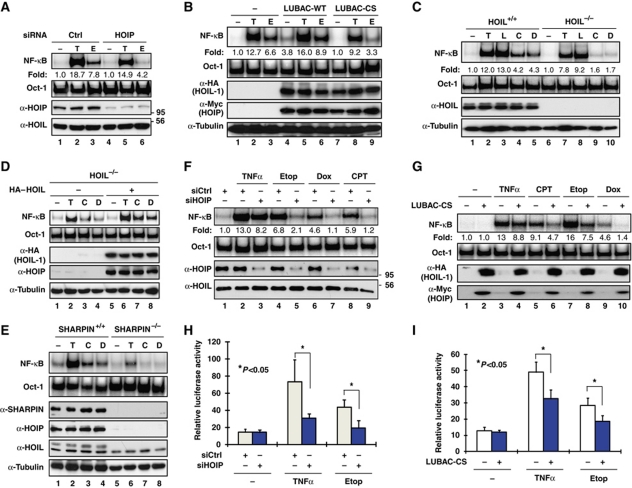
NEMO linear ubiquitination is required for effective NF-κB activation by TNFα and IL-1β, and the ubiquitinated residues within NEMO have been identified as Lys285 and Lys309 (Tokunaga et al, 2009). Coincidently, Lys309 is one of the two NEMO residues that we have shown to be sumoylated and subsequently mono-ubiquitinated in the genotoxic NF-κB signalling pathway (Huang et al, 2003). Also, Lys285 was found to be ubiquitinated upon genotoxic stress (Hinz et al, 2010). To determine whether NEMO linear ubiquitination also has a role in the genotoxic NF-κB signalling pathway, we depleted HOIP, the catalytic subunit of LUBAC ligase complex (Kirisako et al, 2006), to disrupt LUBAC ligase activity in HEK293 cells. We found that knockdown of HOIP substantially reduced NF-κB activation by treatment with genotoxic drug etoposide (Etop) as measured by gel shift assay; while a mild decrease of NF-κB activation by TNFα was also observed (Figure 1A). Consistent with this observation, while transient transfection of wild-type LUBAC slightly enhanced Etop-induced NF-κB activation (Figure 1B, lane 6), overexpression of its dominant-negative mutant LUBACCS (HOIL-1/HOIPCS) significantly inhibited the NF-κB activation in Etop-treated cells (Figure 1B, lane 9). These data indicate that linear ubiquitination ligase LUBAC is required for genotoxic NF-κB activation in HEK293 cells.
Figure 1.
LUBAC is required for NF-κB activation by genotoxic agents. (A) HEK293 cells were transfected with control or siRNAs targeting HOIP. After 48 h, cells were treated with etoposide (Etop, 10 μM) for 2 h, TNFα (10 ng/ml) for 30 min or left untreated (−). NF-κB activation was analysed by EMSA using Igκ probe. Total cell lysates were analysed by western blotting using antibodies as indicated. (B) HEK293 cells were mock transfected or transfected with LUBAC-WT (HA–HOIL-1/HOIP–Myc), LUBAC-CS (HA–HOIL-1/HOIPCS–Myc) mutant constructs as shown. Cells were treated and analysed as in (A). (C) HOIL-1+/+ and HOIL-1−/− MEFs were treated with camptothecin/CPT (C, 10 μM), doxorubicin/Dox (D, 2 μg/ml) for 2 h or left untreated. Treatments with TNFα (T, 10 ng/ml, 30 min) and LPS (L, 10 μg/ml, 30 min) were used as control. NF-κB activation was analysed by EMSA. HOIL-1 expression was shown by western blotting. (D) HOIL-1−/− MEFs were mock transfected or transfected with HOIL-1. Cells were treated and analysed as in (C). (E) Wild-type (SHARPIN+/+) and cpdm (SHARPIN−/−) MEFs were treated as in (C), and analysed with EMSA and immunoblotting as indicated. (F) HEK293 cells were transfected with control or siRNAs targeting HOIP, and treated with CPT (10 μM, 2 h), Dox (2 μg/ml, 2 h), Etop (10 μM, 2 h), TNFα (10 ng/ml, 30 min) or left untreated. Total cell lysates were analysed as in (A). (G) HEK293 cells were mock transfected or transfected with LUBAC-CS mutant. EMSA analyses and immunoblotting were carried out as in (F). (H) HEK293 cells were co-transfected with κB-Fluc/hRluc-TK reporter constructs along with control or siRNAs targeting HOIP. After 48 h, cells were treated with Etop (10 μM) or TNFα (10 ng/ml), and cell extracts were prepared and analysed for luciferase activity. The histogram represents the normalized luciferase reporter activity of three independent experiments; shown as mean±s.d. (I) Similar experiments as in (G) were carried out in HEK293 cells co-transfected with κB-Fluc/hRluc-TK reporter constructs only or along with LUBAC-CS mutant.
To determine whether HOIL-1 subunit is also required for LUBAC function in regulating the genotoxic NF-κB signalling, we treated wild-type and HOIL-1-deficient MEF cells with DNA-damaging drugs, camptothecin (CPT) and doxorubicin (Dox). Genetic ablation of HOIL-1 significantly attenuated the NF-κB activation induced by CPT and Dox in MEFs (Figure 1C). Furthermore, reconstitution of wild-type HOIL-1 was able to rescue the deficiency of genotoxic agent-induced NF-κB activation in HOIL−/− MEFs (Figure 1D). Recently, SHARPIN was identified as a subunit of LUBAC complex, which also has a role in mediating TNFα-induced NF-κB signalling (Gerlach et al, 2011; Ikeda et al, 2011; Tokunaga et al, 2011). We found genotoxic agent-induced NF-κB activation was abolished in SHARPIN-deficient MEFs (Figure 1E), which further reinforced the notion that LUBAC is indispensable for effective genotoxic NF-κB activation. We also observed that inhibiting LUBAC function by overexpressing LUBACCS mutant or depleting HOIP with siRNA diminished genotoxic agent-induced NF-κB activation in HT1080 fibrosarcoma cells and U2OS osteosarcoma cells (Supplementary Figure S1A–C), indicating the critical role of LUBAC in mediating genotoxic NF-κB signalling is conserved in cancer cells. Our data further suggest that diverse genotoxic drugs induce NF-κB activation in a LUBAC-dependent manner (Figure 1F and G), and the inhibition of genotoxic NF-κB activation by LUBAC deficiency is not due to the change of the kinetics of NF-κB signalling (Supplementary Figure S1D–F). In addition, the activity of NF-κB-driven luciferase reporter in response to treatment with TNFα or etoposide was significantly reduced in HOIP-depleted cells or cells expressing the LUBACCS mutant (Figure 1H and I). All these results support that LUBAC has a critical role in regulating NF-κB-dependent gene expression in cells exposed to various genotoxic stimuli.
NEMO is ubiquitinated with linear chains in response to DNA damage in a manner dependent on LUBAC
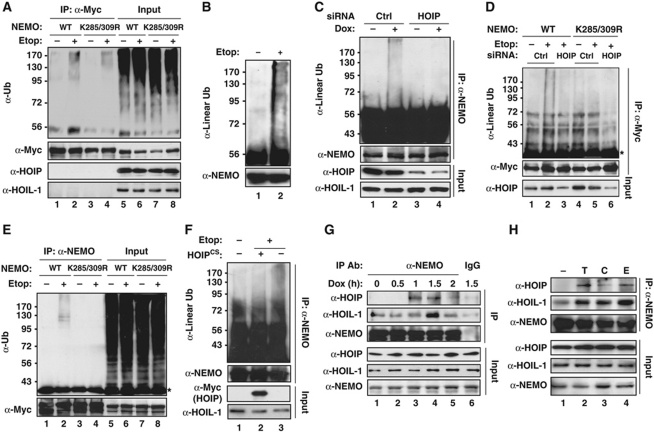
We have shown NEMO can be modified with mono-ubiquitin which is required for its nuclear exportation in genotoxic NF-κB signalling (Wu et al, 2006). To examine whether NEMO can be also polyubiquitinated upon genotoxic stress, HEK293 cells were transiently transfected with NEMO constructs and treated with etoposide. We detected a high molecular weight smear in immunoprecipitated NEMO samples under stringent condition with anti-ubiquitin antibody (Figure 2A), indicating that NEMO was modified with polyubiquitin upon genotoxic stimulation. Interestingly, this NEMO polyubiquitination was remarkably diminished in cells expressing a NEMO mutant harbouring mutation at K285 and K309 (Figure 2A, lane 4). Since K285/309 was identified as residues required for NEMO linear ubiquitination (Tokunaga et al, 2009), we then used an antibody specifically recognizing linear linkage to determine whether the polyubiquitin attached on NEMO upon genotoxic stress was composed of linear chain. We found NEMO was modified with linear polyubiquitin in HEK293 cells treated with etoposide (Figure 2B). Moreover, the NEMO linear ubiquitination was induced by another genotoxic drug doxorubicin in a HOIP-dependent manner (Figure 2C). The requirement of HOIP for NEMO linear ubiquitination was also observed in etoposide-treated cells (Figure 2D), indicating a critical role of LUBAC in mediating NEMO linear ubiquitination upon DNA damage. We also observed the etoposide-induced NEMO polyubiquitination in mouse NEMO-deficient 1.3E2 cells stably reconstituted with wild-type NEMO, but not in those reconstituted with NEMO-K285/309R mutant (Figure 2E), suggesting that the NEMO linear ubiquitination is a conserved stress response to genotoxic treatments.
Figure 2.
DNA damage induces NEMO linear ubiquitination on Lys285/309. (A) HEK293 cells were transfected with Myc-tagged NEMO-WT or NEMO-K285/309R and treated with Etop (10 μM, 2 h). Whole cell lysates were subjected to anti-Myc immunoprecipitation under a stringent condition (see details in Materials and methods), and analysed by immunoblotting using antibodies as shown. Input represents 5% of total extracts. (B) HEK293 cells were treated with Etop (10 μM, 2 h) or left untreated. Cells were lysed as in (A) and immunoprecipitated with anti-NEMO antibody, and followed with immunoblotting using anti-linear polyubiquitin antibody. (C) NEMO linear ubiquitination analyses as in (B) were performed in HEK293 cells transfected with control or siRNAs targeting HOIP. (D) HEK293 cells were transfected with Myc-tagged NEMO-WT or NEMO-K285/309R along with control siRNA or siRNA targeting HOIP as shown. Cells were treated and analysed as in (A). *IgG light chain. (E) Myc-NEMO-WT or Myc-NEMO-K285/309R stably reconstituted 1.3E2 cells were treated and analysed as in (A), except for immunoprecipitation using anti-NEMO. (F) HEK293 cells were mock transfected or transfected with HOIP-CS mutant, and treated with Etop (10 μM, 2 h). Whole cell lysates were used for analysing NEMO linear ubiquitination as in (B). (G) HEK293 cells were treated with Doxorubicin (2 μg/ml) for time as indicated. Whole cell lysates were immunoprecipitated with anti-NEMO antibody or IgG control, and immunoblotted with anti-HOIP, anti-HOIL-1, and anti-NEMO antibodies. (H) HEK293 cells were left untreated or treated with TNFα (10 ng/ml, 30 min), CPT (10 μM, 90 min), or Etop (10 μM, 90 min), similar analyses were carried out as in (G).
To further explore the role of LUBAC in regulating NEMO linear ubiquitination, we examined etoposide-induced NEMO linear ubiquitination in HEK293 cells transfected with HOIPCS mutant, which inhibited endogenous LUBAC ligase activity (Tokunaga et al, 2009). The dominant-negative HOIPCS mutant effectively inhibited the linear ubiquitination of NEMO in etoposide-treated HEK293 cells (Figure 2F), suggesting the ligase activity of LUBAC is indispensable for NEMO linear ubiquitination upon genotoxic stress. Furthermore, co-immunoprecipitation analyses indicated that NEMO and LUBAC could form a complex with a delayed kinetics in HEK293 cells treated with genotoxic agents (Figure 2G and H). All these data strongly suggest that LUBAC E3 ligase is essential for DNA damage-induced NEMO linear polyubiquitination, potentially on K285 and K309.
Linear ubiquitination of NEMO is downstream of its nuclear exportation
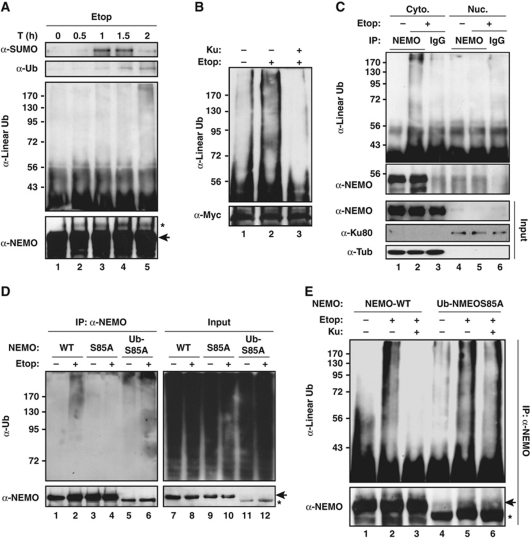
Our previous studies have shown NEMO shuttles between cytoplasm and nucleus upon genotoxic stimulation, and this process is controlled by NEMO sequential modification with SUMO-1, phosphate, and ubiquitin (Miyamoto, 2011). We found HOIP knockdown did not significantly affect sumoylation, phosphorylation, and nuclear translocation of NEMO or ATM activation in Etop-treated HEK293 cells (Supplementary Figure S2A–C). Moreover, sumoylation of NEMO could be detected as early as 60 min after etoposide treatment, which is followed by mono-ubiquitination of NEMO detected at around 90 min after treatment (Figure 3A). In contrast, NEMO linear ubiquitination was more prominent at 2 h after etoposide exposure (Figure 3A), indicating it may take place later than of NEMO mono-ubiquitination. Furthermore, an ATM kinase inhibitor KU55933, which has been shown to abrogate ATM-dependent NEMO phosphorylation and mono-ubiquitination (Wu et al, 2006), effectively inhibited DNA damage-induced NEMO linear ubiquitination (Figure 3B), favouring an upstream role of mono-ubiquitination for subsequent NEMO linear ubiquitination.
Figure 3.
LUBAC catalyses NEMO linear ubiquitination in cytoplasm. (A) Myc–NEMO-WT stable HEK293 cells were treated with Etop (10 μM) for indicated times. Whole cell lysates were analysed by anti-Myc IP followed by immunoblotting using antibodies against SUMO-1, ubiquitin, linear ubiquitin and NEMO. The asterisk indicates a band overlaps with mono-ubiquitinated NEMO. The arrow indicates Myc-tagged NEMO. (B) LUBAC-transfected Myc-NEMO stable HEK293 cells were left untreated or treated with 10 μM Etop for 2 h with or without KU55933 (10 μM). NEMO linear ubiquitination was analysed as in (A). (C) HEK293 cells were treated with Etop (10 μM) for 2 h. Cytoplasmic and nuclear extracts were fractionated and immunoprecipitated with anti-NEMO antibody or IgG control followed by immunoblotting using antibodies as indicated. (D) 1.3E2 cells stably expressing 6 × Myc–NEMO-WT, 6 × Myc–NEMO S85A, and 2 × HA–Ub-NEMO S85A were left untreated or treated with Etop (10 μM, 2 h). Whole cell lysates were analysed by anti-NEMO IP followed by immunoblotting using anti-ubiquitin antibody. The arrow indicates Myc-tagged NEMO; the asterisk indicates HA-tagged NEMO. (E) 1.3E2 cells stably expressing 6 × Myc–NEMO-WT or 2 × HA–Ub-NEMO S85A were left untreated or treated with Etop (10 μM, 2 h) in the presence or absence of KU55933 (10 μM). NEMO linear ubiquitination was examined as in (B).
As the mono-ubiquitination promotes NEMO to export from nucleus, it is plausible the linear ubiquitination of NEMO occurs in cytoplasm. Indeed, we detected a significant increase of NEMO polyubiquitination upon etoposide treatment in cytosolic fraction (Figure 3C), which is consistent with the predominant cytoplasmic distribution of HOIP (Supplementary Figure S2C). Moreover, NEMO polyubiquitination by genotoxic stress was abrogated in cells expressing NEMO-S85A mutant (Figure 3D, lane 4). We have shown Ser85 mutation of NEMO blocked its ATM-dependent phosphorylation, mono-ubiquitination, and subsequent nuclear export in response to DNA damage, while tethering a ubiquitin moiety to NEMO S85A mutant (Ub-S85A) could restore NEMO nuclear export (Wu et al, 2006). In agreement with our previous finding, ubiquitin fusion to NEMO-S85A mutant also recapitulated the NEMO polyubiquitination upon genotoxic stimulation (Figure 3D, lane 6). On the other hand, we found ATM inhibition did not overtly affect linear ubiquitination of NEMO induced by etoposide or doxorubicin in cells expressing NEMO Ub-S85A mutant (Figure 3E; Supplementary Figure S2E), indicating that ATM may have an upstream role in regulating NEMO linear ubiquitination, likely through promoting NEMO nuclear exportation. Altogether, these results suggest that NEMO linear ubiquitination is a cytoplasmic event, potentially downstream of ATM-mediated NEMO nuclear exportation.
A recent report indicated that TNFα played a role in inducing NF-κB activation at a later stage in response to extensive DNA lesions (Biton and Ashkenazi, 2011). Consistent with this report, we found no significant increase of TNFα secretion until 8 h after etoposide treatment (Supplementary Figure S3A). DNA damage-induced NF-κB activation and NEMO linear ubiquitination at 2 h after etoposide exposure was not affected by TNFα neutralizing antibody (Supplementary Figure S3B and C), indicating genotoxic stress-induced NEMO linear ubiquitination, as an early response, is independent of TNFα signalling.
LUBAC mediates DNA damage-induced activation of TAK1 and IKK
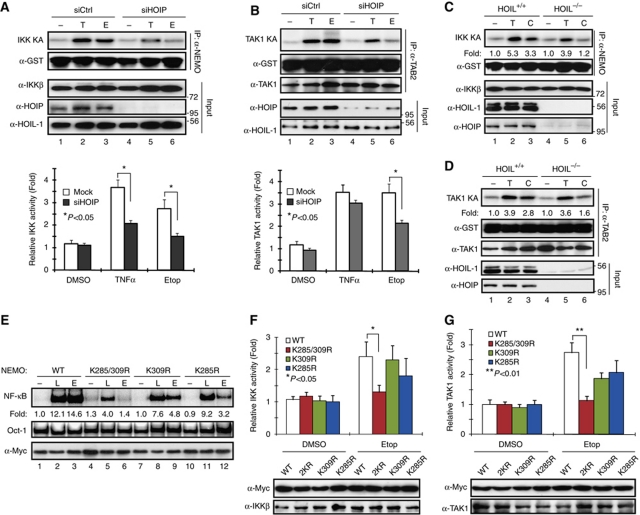
It has been established that genotoxic NF-κB signalling depends on IKK activation (Baldwin, 2001; Wu and Miyamoto, 2007). Recent studies further showed that TAK1 is required for IKK activation upon genotoxic stress (Hinz et al, 2010; Wu et al, 2010; Yang et al, 2011). LUBAC was found in TNFR1-signalling complex upon stimulation and is required for optimal IKK activation by TAK1 in response to TNFα treatment (Haas et al, 2009). To delineate the mechanism of how LUBAC mediates the genotoxic NF-κB signalling cascade, we examined whether LUBAC is required for the activation of IKK and TAK1 by genotoxic agents with in vitro kinase assays. As shown in Figure 4A, HOIP knockdown significantly inhibited Etop-induced IKK activity; a similar effect was also observed in cells treated with TNFα. Moreover, Etop-induced TAK1 activation was greatly decreased in HOIP-depleted cells (Figure 4B). In parallel, we observed the attenuated activation of IKK and TAK1 upon genotoxic stress in HOIL-1−/− MEFs compared with that in wild-type MEFs (Figure 4C and D), which is consistent with the decreased JNK activation in TNFα- or Dox-treated HOIL-1−/− MEFs (Supplementary Figure S4A). These data suggest that LUBAC is critical for activation of IKK and TAK1 upon DNA damage. We found that polyubiquitination had essential roles in mediating TAK1 and IKK activation in response to genotoxic stimulation (Wu et al, 2010). It is plausible that LUBAC promotes linear ubiquitination of NEMO, which participates in the activation of TAK1 and IKK in the genotoxic NF-κB signalling pathway.
Figure 4.
LUBAC is indispensable for activation of TAK1 and IKK upon genotoxic stress. (A, B) HEK293 cells were transfected with control or siRNA targeting HOIP. Cells were treated with Etop (10 μM, 90 min), TNFα (10 ng/ml, 30 min) or left untreated. Total cell lysates were analysed with in vitro kinase assay to assess activity of IKK (A) or TAK1 (B), using GST–IκBα or GST–IKKβ as the substrate, respectively. Immunoprecipitated samples and total cell extracts (input) were analysed by western blotting with antibodies as indicated. Intensity of phosphorylated proteins was quantified with phosphoimager. The histogram represents the quantitative analysis of three independent experiments. The values are presented as mean±s.d. (C) Wild-type (+/+) and HOIL-1-deficient (−/−) MEFs were treated with CPT (10 μM, 90 min), TNFα (10 ng/ml, 30 min), or left untreated. Total cell lysates were analysed with IKK assay and western blotting using antibodies as indicated. Kinase activity was quantified with phosphoimager and shown as fold induction. (D) Similar experiment was carried out as in (C); cell lysates were analysed with TAK1 kinase assay and western blotting using antibodies as indicated. (E) 1.3E2 cells stably expressing NEMO-WT, NEMO-K285/309R, NEMO-K309R, or NEMO-K285R were left untreated or treated with LPS (10 μg/ml, 30 min) or Etop (10 μM, 2 h). Total cell lysates were analysed by EMSA and western blotting as shown. Normalized NF-κB activation was quantified with phosphoimager and shown as fold induction. (F, G) Similar experiments were carried out as in (E). Total cell lysates were analysed with in vitro kinase assay to examine activity of IKK (F) or TAK1 (G), using GST–IκBα or GST–IKKβ as substrate, respectively. Immunoblotting of input samples was shown as indicated. The histograms represent the phosphoimager-quantitated kinase activity induction from three independent experiments. The values are presented as mean±s.d.
It was shown K285 and K309 are required for TNFα treatment-induced NEMO linear ubiquitination (Tokunaga et al, 2009). Our data further suggested that genotoxic stress-induced NEMO linear ubiquitination also depended on these residues (Figure 2). We then generated stable cell lines expressing NEMO-K285R, NEMO-K309R, or NEMO-K285/309R mutant in 1.3E2 cells and examined whether these NEMO lysine residues are required for genotoxic NF-κB signalling. We found, in contrast to mutation on NEMO-K277/309, NEMO sumoylation was still induced by genotoxic stimulation in cells expressing NEMO-K285/309R mutant (Supplementary Figure S4B). Also, DNA damage-induced NF-κB activation was only partially inhibited in single lysine mutants, suggesting that the other intact lysine compensated for the mutated lysine residue (Figure 4E). However, NF-κB activation by etoposide was abrogated in cells expressing NEMO-K285/309R double lysine mutant (Figure 4E), indicating a critical role of linear ubiquitination on NEMO-K285 and K309 in mediating genotoxic NF-κB signalling.
Recent studies have underscored the pivotal roles of NEMO in mediating IKK activation by either interacting with polyubiquitin chain or being ubiquitinated (Skaug et al, 2009). Both mechanisms contribute to stabilizing the protein complex harbouring IKK and its upstream kinases, such as TAK1, leading to efficient IKK activation. To determine whether NEMO linear ubiquitination has a role in mediating IKK activation by DNA damage, we treated NEMO single lysine mutants or double lysine mutant with etoposide and examined IKK activity. In accordance with genotoxic NF-κB activation, double lysine mutation at K285/309 significantly inhibited etoposide-induced IKK activation (Figure 4F), as well as TAK1 activation (Figure 4G), suggesting that NEMO linear ubiquitination may have a pivotal role in regulating TAK1-dependent IKK activation by genotoxic stress.
The zinc finger domain is required for TAB2 association with linear polyubiquitin
The critical role of ubiquitin, especially the K63-linked polyubiquitin, in mediating TAK1 activation has been well established (Adhikari et al, 2007; Skaug et al, 2009). It was shown that TAK1 can be activated by intermolecular phosphorylation between TAK1 complexes docking on K63-linked polyubiquitin chains in a TAB2/3-dependent manner (Kanayama et al, 2004; Xia et al, 2009). Structural analysis revealed that K63-linked and linear polyubiquitin chains share similar open conformation (Komander et al, 2009), and NEMO can bind to either type of polyubiquitin but with distinct affinity (Laplantine et al, 2009; Lo et al, 2009; Rahighi et al, 2009). Although a carboxyl-terminal fragment of TAB2 was not able to pull down linear tetra-ubiquitin in vitro (Komander et al, 2009), it was shown the zinc finger (NZF) domain of TAB2 and TAB3 can directly interact with longer linear polyubiquitin (Tokunaga et al, 2009). As we found NEMO linear ubiquitination was also required for optimal TAK1 activation (Figure 4), it is possible that TAK1/TAB2/3 complex could associate with NEMO linear ubiquitin chain, which may be required for efficient TAK1 activation, especially in cells exposed to genotoxic agents.
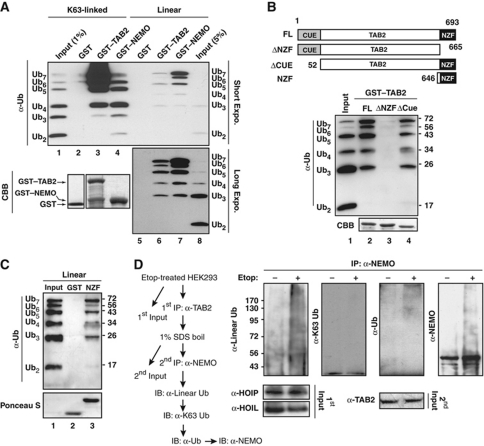
To test this hypothesis, we generated GST-tagged recombinant proteins of full-length NEMO, TAB2 and TAB2 mutants with either NZF or CUE domain deletion (Figure 5B). Consistent with previous reports (Kulathu et al, 2009; Lo et al, 2009; Rahighi et al, 2009), TAB2 showed significantly higher affinity in association with K63-linked polyubiquitin compared with NEMO, which appeared to bind to linear polyubiquitin more efficiently (Figure 5A). However, we were able to detect polyubiquitin with linear linkage associated with TAB2, although to a lesser extent compared with K63-linked polyubiquitin (Figure 5A, lane 6). Further analyses indicated that NZF domain is required and sufficient for the association between TAB2 and linear polyubiquitin (Figure 5B and C).
Figure 5.
TAB2 associates with linear polyubiquitin through its NZF domain. (A) GST, GST–TAB2, and GST–NEMO recombinant proteins were incubated with Ub2−7 mixture with either K63 or linear linkage. In vitro pull-down of ubiquitin chains was visualized by immunoblotting using anti-ubiquitin antibody, and recombinant proteins were stained with Coomassie blue (CBB). Input lanes of K63-linked or linear chain represent 1% or 5% of Ub2–7 mixture, respectively. (B, C) Similar pull-down assay was performed as in (A) using linear Ub2−7 mixture for association with TAB2 full-length/deletion mutants (B) or GST/GST–NZF (C) as illustrated. Input represents 5% of Ub2−7 mixture. (D) HEK293 cells were treated with etoposide (10 μM, 2 h) or left untreated. Whole cell lysates were subjected to immunoprecipitation with anti-TAB2 antibody. Immobilized immunocomplexes were boiled in the presence of 1% SDS, and re-immunoprecipitated with anti-NEMO antibody followed with immunoblotting using antibodies as shown. First input represents 1% of whole cell lysates and second input represents 2% TAB2 immunoprecipitates.
To examine whether TAB2 forms a complex with linear ubiquitinated NEMO in cells upon DNA damage, we treated HEK293 cells with etoposide and analysed the immunocomplex associated with TAB2. We were able to detect a significant increase of NEMO linear polyubiquitination, but not K63-linked polyubiquitination, in TAB2-associated protein complex, suggesting that linear ubiquitinated NEMO may also provide a platform for TAK1 activation in response to genotoxic stimulation (Figure 5D).
DNA damage-induced ELKS ubiquitination enhances linear ubiquitination of NEMO
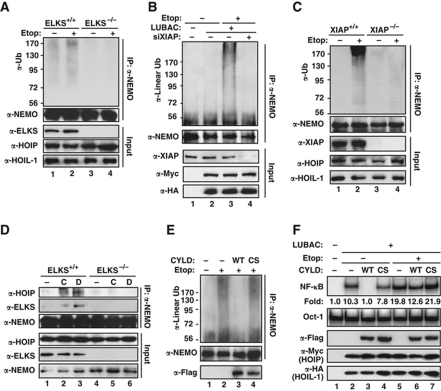
We recently found that ELKS is polyubiquitinated with K63-linked chain upon DNA damage, which is essential for genotoxic NF-κB activation (Wu et al, 2010). It has been shown that K63-linked and linear polyubiquitin collaborate to achieve effective NF-κB activation by TNFα (Haas et al, 2009). We considered the possibility of cross-talking between ELKS K63 ubiquitination and NEMO linear ubiquitination in genotoxic NF-κB signalling. Interestingly, Etop-induced NEMO polyubiquitination was blocked in ELKS-deficient MEFs (Figure 6A), suggesting ELKS has an important role in promoting NEMO linear ubiquitination. Moreover, NEMO linear ubiquitination was diminished in XIAP-depleted HEK293 cells (Figure 6B) and XIAP-deficient MEFs (Figure 6C) upon genotoxic stimulation. As ELKS polyubiquitination upon DNA damage depends on XIAP (Wu et al, 2010), it is plausible that ELKS K63-linked ubiquitination is critical for NEMO linear ubiquitination. LUBAC, especially HOIP, has been found to interact with K63-linked polyubiquitin tethered on cIAPs within TNFR1-signalling complex, which may facilitate NEMO linear ubiquitination in response to TNFα treatment (Haas et al, 2009). Furthermore, NEMO ubiquitin-binding domain appeared to be critical for its linear ubiquitination (Supplementary Figure S4C), and DNA damage-induced association between NEMO and LUBAC was abolished in ELKS-deficient cells (Figure 6D; Supplementary Figure S4D). All this body of evidence suggests that K63-linked polyubiquitin of ELKS may provide a platform for recruiting both LUBAC and NEMO, leading to NEMO linear ubiquitination.
Figure 6.
ELKS polyubiquitination facilitates NEMO linear ubiquitination upon genotoxic stress. (A) Etoposide-induced NEMO polyubiquitination was examined in ELKS wild-type (+/+) and deficient (−/−) MEF cells. (B) HEK293 cells were transfected with LUBAC with or without siRNA targeting XIAP, and treated with etoposide (10 μM, 2 h) as indicated. Whole cell lysates were subjected to NEMO IP and linear ubiquitination was visualized by immunoblotting with indicated antibodies. (C) Similar experiments were carried out as in (A) using wild-type (+/+) and XIAP-deficient (−/−) MEFs. (D) ELKS+/+ and ELKS−/− MEFs were treated with CPT (10 μM, 2 h), Dox (2 μg/ml, 2 h), or left untreated. Whole cell lysates were subjected to IP with anti-NEMO antibody followed by immunoblotting using antibodies as shown. (E) HEK293 cells were transfected with Flag–CYLD wild type (WT) or catalytic-inactive mutant (CS) along with LUBAC as indicated. Etoposide-induced NEMO linear ubiquitination was detected as in (B). (F) HEK293 cells were transfected with CYLD and LUBAC constructs as indicated. Cells were treated with etoposide (10 μM, 2 h) or left untreated, and whole cell lysates were analysed with EMSA and immunoblotting as shown. Fold induction of NF-κB activation was quantified with phosphoimager.
To further investigate the role of ELKS K63 ubiquitination in mediating NEMO linear ubiquitination, we transfected HEK293 cells with either wild-type deubiquitinating enzyme CYLD or its catalytic-inactive mutant (C601S), and examined linear ubiquitination of NEMO by DNA damage. Etoposide-induced NEMO linear ubiquitination was significantly inhibited in cells expressing wild-type CYLD, but not CYLD-C601S mutant (Figure 6E). As CYLD is believed to specifically target K63-linked polyubiquitin (Komander et al, 2008; Sun, 2008), it is possible that deconjugation of K63 polyubiquitin from ELKS had an important role in inhibiting NEMO linear ubiquitination. However, we found expression of CYLD inhibited not only DNA damage-induced NF-κB activation, but also that by overexpression of LUBAC (Figure 6F, lane 3). In addition, CYLD was found to disassemble polyubiquitin with linear linkage in vitro (Komander et al, 2009), and our data further suggest that CYLD may also function as a deubiquitinating enzyme directly targeting linear polyubiquitin in cells exposed to genotoxic stress. Considering the emerging and expanding role of linear ubiquitination in regulating NF-κB signalling, further investigation will be necessary to advance our knowledge into how CYLD regulates NF-κB activation by various stimuli.
NEMO linear ubiquitination-dependent genotoxic NF-κB activation promotes cell survival in response to DNA damage
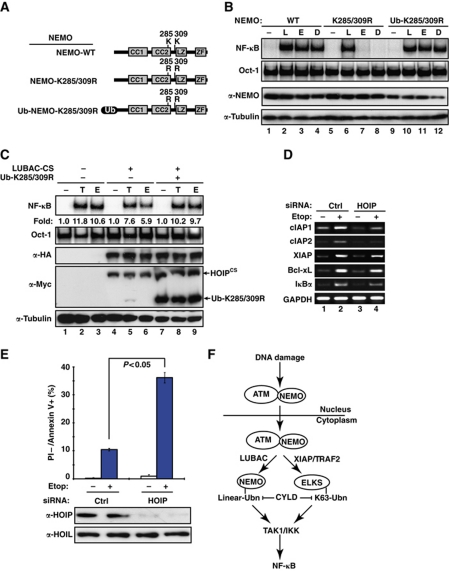
In our previous studies, we fused an SUMO or ubiquitin moiety to NEMO mutants in order to bypass the deficiency of NEMO sumoylation or ubiquitination caused by mutation (Huang et al, 2003; Wu et al, 2006). Following the same strategy, we fused an ubiquitin molecule at the N-terminal of NEMO-K285/309R mutant and examined whether the ubiquitin fusion could restore genotoxic NF-κB activation (Figure 7A). Indeed, both etoposide and Dox induced robust NF-κB activation in cells expressing Ub-NEMO-K285/309R, whereas this activation was absent in NEMO-K285/309R cells (Figure 7B). We also found Ub-NEMO-K285/309R mutant primarily localized in cytoplasmic compartment, while NEMO-WT or NEMO-K285/309R showed both nuclear and cytoplasmic distribution in transiently transfected cells (Supplementary Figure S5C). In addition, the inhibition of genotoxic NF-κB activation by overexpressing the dominant-negative mutant of LUBAC was rescued, at least in part, by co-transfecting the Ub-NEMO-K285/309R mutant in HEK293 cells (Figure 7C). These results indicate that the K285 and K309 lysine residues are required mainly for NEMO ubiquitination in cytoplasm to transduce genotoxic NF-κB signalling. On the contrary, neither SUMO nor ubiquitin fusion alone to NEMO-K277/309R reconstituted DNA damage-induced NF-κB activation, which can only be recapitulated in cells expressing NEMO-K277/309R tethered with both SUMO and ubiquitin moieties, supporting the dual requirement of these two lysine residues for both sumoylation and mono-ubiquitination (Supplementary Figure S5A and B).
Figure 7.
Abrogation of NEMO linear ubiquitination promotes apoptosis in cells exposed to genotoxic drugs. (A) Schematic representation of constructs encoding NEMO-WT, NEMO-K285/309R mutant or ubiquitin fusion to NEMO-K285/309R (ZF, zinc finger; CC, coiled coil; LZ, leucine zipper; Ub, ubiquitin). (B) 6 × Myc–NEMO-WT, 6 × Myc–NEMO-K285/309R, and 3 × Myc–Ub-NEMO-K285/309R reconstituted 1.3E2 cells were left untreated or treated with LPS (L, 10 μg/ml, 30 min), etoposide (E, 10 μM, 2 h), or Dox (D, 2 μg/ml, 2 h). NF-κB activation was analysed by EMSA. Total cell lysates were analysed for NEMO and α-tubulin by western blotting. (C) HEK293 cells were transfected with LUBAC-CS mutant with or without Ub-NEMO-K285/309R construct as shown. Cells were treated with TNFα (T, 10 ng/ml, 30 min), etoposide (E, 10 μM, 2 h) or left untreated, and whole cell lysates were analysed with EMSA and immunoblotting as indicated. Fold induction of NF-κB activation was quantified with phosphoimaging. (D) HEK293 cells were transfected with control or HOIP-specific siRNAs. After 48 h, cells were treated with Etop (10 μM) or left untreated for 4 h. Total RNA was isolated and the mRNA of specific genes was analysed by semi-quantitative RT–PCR. PCR products were visualized by electrophoresis on 1.5% agarose gel. (E) HT1080 cells were transfected with control or HOIP-specific siRNAs and treated with Etop (10 μM, 16 h). Treated cells were stained with propidium iodide (PI) and Annexin V-FITC, and analysed by flow cytometry to detect the percentage of apoptotic cells (PI−/Annexin V+). Histogram bars represent the mean of three independent experiments. Total cell extracts were immunoblotted with indicated antibodies. (F) A model depicting the role of LUBAC-mediated NEMO linear ubiquitination in genotoxic NF-κB signalling.
To further delineate the function of LUBAC in regulating cellular response to genotoxic stimulation, we depleted HOIP with siRNAs and examined the expression of a number of NF-κB target genes in cells treated with etoposide. RT–PCR analysis indicated that a cohort of anti-apoptotic genes whose upregulation upon DNA damage was inhibited in HOIP knockdown cells (Figure 7D). In accordance with these data, significant increase of apoptosis upon genotoxic treatment was observed in HOIP-depleted HT1080 cells compared with the cells maintaining functional LUBAC (Figure 7E). Similar phenotypes in anti-apoptotic gene expression and chemodrug-induced cell death were also observed in cells expressing NEMO-K285/309R mutant, but not in wild-type or Ub-NEMO-K285/309R cells (Supplementary Figure S5D and E).
Discussion
We have shown NEMO is sequentially modified by SUMO-1, phosphate, and ubiquitin in response to genotoxic stimulation, which is essential for transducing the nuclear DNA damage signal to the cytoplasm, leading to activation of the IKK complex. The mono-ubiquitination of NEMO on K277/309 is essential for NEMO nuclear exportation (Wu et al, 2006). In this study, we introduced another form of NEMO post-translational modification, linear ubiquitination, into this complex genotoxic NF-κB signalling cascade, which may in part elucidate the mechanisms involved in IKK activation regulated by nuclear-exported NEMO and ATM. We found LUBAC could facilitate NEMO linear polyubiquitination on K285/309 in response to genotoxic stress, which appeared to be downstream of NEMO nuclear exportation. In cytoplasm, XIAP-mediated ELKS K63 ubiquitination promoted the association between NEMO and LUBAC to facilitate NEMO linear ubiquitination. NEMO was shown to bind to linear polyubiquitin with a higher affinity compared with K63-linked polyubiquitin (Lo et al, 2009; Rahighi et al, 2009), it is plausible that the NEMO-attached linear polyubiquitin may serve as binding platform for further recruitment of cytoplasmic NEMO-associated IKK complexes. Previous studies demonstrated that TAB2 preferentially binds to K63-linked polyubiquitin (Kulathu et al, 2009), and we found TAB2 may also interact with linear ubiquitin chain. Therefore, TAK1/TAB2 complex may primarily associate with the K63 polyubiquitin chains on ELKS, and with NEMO linear polyubiquitin to a lesser extent, upon genotoxic stress. The intertwined association could promote IKKβ/NEMO, TAK1/TAB2, K63 polyubiquitin and linear polyubiquitin to form a stabilized framework for optimal activation of both kinases, leading to efficient NF-κB activation upon genotoxic stimulation (Figure 7F).
The functions of the ubiquitin system involved in regulating NF-κB activation have been studied extensively (Skaug et al, 2009). The degradative role of K48-linked polyubiquitin and the scaffolding role of K63-linked polyubiquitin in multiple NF-κB signalling pathways have been well documented (Skaug et al, 2009; Wertz and Dixit, 2009). However, the role of linear ubiquitin chains in NF-κB signalling is just emerging. The first evidence that linear ubiquitin chain is involved in regulating NF-κB activation came from the study on LUBAC function in canonical NF-κB signalling (Iwai and Tokunaga, 2009). It was found that LUBAC bound to NEMO and conjugated linear polyubiquitin chains onto its Lys285 and Lys309 residues. Abrogation of LUBAC-mediated NEMO linear ubiquitination suppressed NF-κB activation by pro-inflammatory cytokines such as TNFα and IL-1β (Tokunaga et al, 2009). Nevertheless, the mechanism of how NEMO linear ubiquitination regulates canonical NF-κB signalling was not well understood.
Recent in vitro studies on the structure of NEMO ubiquitin-binding domain (NOA/UBAN/NUB) shed some light on potential mechanisms of how linear polyubiquitin promotes NF-κB signalling (Lo et al, 2009; Rahighi et al, 2009). It was found that the NEMO NOA domain bound to linear polyubiquitin in vitro with higher affinity than to K63-linked polyubiquitin, suggesting a potential scaffolding role of the linear polyubiquitin similar to that of K63 polyubiquitin in mediating IKK activation. However, the role of linear polyubiquitination in regulating NF-κB signalling cascade in vivo remains to be fully elucidated. It was shown that the zinc finger domain (ZF) of NEMO also binds to polyubiquitin (Laplantine et al, 2009). While neither NOA nor ZF shows any preference for K63-linked chains, they form a bipartite ubiquitin-binding domain with high affinity to K63-linked polyubiquitin. It is plausible that both K63-linked and linear polyubiquitin chains are intertwined through association with various ubiquitin-binding domains of members of a signalosome, which renders a stabilized environment for optimal IKK activation. This hypothesis is supported by the evidence that LUBAC was found in TNFR-super complex and LUBAC-facilitated NEMO linear ubiquitination enhanced its interaction with the TNFR-super complex, leading to effective IKK and NF-κB activation (Haas et al, 2009). HOIP was also identified as a component of CD40 activating complex which is required for CD40 ligation-induced NF-κB activation, suggesting a similar role LUBAC may play in mediating alternative NF-κB signalling pathway (Hostager et al, 2010). Our recent investigation indicated ELKS is ubiquitinated with K63-linked chain upon genotoxic stress, which is required for IKK activation (Wu et al, 2010). Here, we showed that ELKS K63 ubiquitination promoted NEMO linear polyubiquitin in cytoplasm, which is also indispensable for effective IKK activation in cells exposed to genotoxic agents. It is likely that both K63 polyubiquitination of ELKS and linear polyubiquitination of NEMO are critical for optimized IKK activation upon DNA damage. Further studies on the interaction between polyubiquitin with different linkages and the responsible ubiquitin-binding domain in members of this signalling complex are needed to delineate the underlying mechanisms.
The K285 of NEMO was recently found to be modified, presumably, with mono-ubiquitin after NEMO exportation from nucleus in response to genotoxic stimulation (Hinz et al, 2010). It is interesting to note that the mono-ubiquitination of K285 requires the intact ubiquitin-binding domain of NEMO, which is consistent with our finding that NEMO association with polyubiquitin chain on ELKS is critical for linear ubiquitination of NEMO on K285/309. Since the mono-ubiquitination on NEMO-K285 appeared to take place earlier than linear ubiquitination of NEMO in genotoxic signalling, it is possible the single ubiquitin molecule attached to NEMO-K285 may serve as a cornerstone for further extension with linear chain in genotoxic NF-κB signalling. Moreover, the mono-ubiquitin may also promote LUBAC-dependent NEMO linear ubiquitination on additional site, such as K309, by facilitating NEMO:LUBAC association. This model is in part supported by a recent publication showing that mono-ubiquitination of TRIM25 makes it susceptible for LUBAC-dependent polyubiquitination (Inn et al, 2011). Additional investigation will be needed to further delineate the underlying mechanisms.
We found genotoxic stress-induced TAK1 activation was affected by inhibiting NEMO linear ubiquitination (Figure 4). The mechanism of TAK1 activation remained ambiguous until a recent study showed that TAK1 can be activated by intermolecular autophosphorylation depending on TAK1 association with K63-linked polyubiquitin chains through its binding partner TAB2/3 (Xia et al, 2009). Structural analysis has revealed that both K63-linked and linear ubiquitin dimers adopted equivalent open conformation with a high flexibility, and the ubiquitin-binding domains of TRABID and cIAP1 can interact with either linkage formed ubiquitin polymers (Komander et al, 2009). However, in vitro analyses suggested that the ubiquitin-binding domain of TAB2 (NZF, similar to that of TRABID) pertain significantly higher affinity for di- and tetra-ubiquitin with K63 linkage than those with linear linkage (Komander et al, 2009; Sato et al, 2009). This may be partially explained by that TAB2-NZF binds adjacent K63-linked ubiquitin moieties with two distinct binding surfaces which may not be adopted by linear linkage (Kulathu et al, 2009). Nevertheless, while the NZF domain of TAB2 is sufficient to interact with polyubiquitin in vitro, the other region of TAB2 may also contribute to the affinity and stability of interaction between TAB2 and polyubiquitin chains in vivo. Moreover, TAB2 was found to pull down long linear polyubiquitin chain (approximately Ub7) in vitro (Tokunaga et al, 2009). We also found TAB2 can interact with a mixture of ubiquitin polymer (Ub2–Ub7), preferentially the longer polyubiquitin, through its NZF domain (Figure 5). Since only long polyubiquitin chains activates TAK1 and IKK (Xia et al, 2009), it is possible that TAB2 may be able to bind to long linear polyubiquitin chain which participates in TAK1 activation upon DNA damage. In accordance, we found TAB2 associated with linear ubiquitinated NEMO in cells exposed to genotoxic drug (Figure 5D), and DNA damage-induced TAK1 activation was attenuated in cells defective in NEMO linear ubiquitination (Figure 4). However, long linear polyubiquitin chain alone was insufficient for activation of TAK1 and IKK in vitro (Xia et al, 2009), which suggests that linear polyubiquitin chain may collaborate with K63-linked polyubiquitin to achieve optimal activation of TAK1. Nevertheless, the significance of such collaboration in regulating TAK1 activation may vary depending on stimuli used. We did not observe significant inhibition of TAK1 activation by TNFα in LUBAC-deficient cells. It may be explained by that TNFα treatment induces quick and robust ubiquitination of TRAF2 (Li et al, 2009) and RIP1 with multiple forms of linkage including K63, K11, and linear linkages (Dynek et al, 2010; Gerlach et al, 2011), which may be sufficient for TAK1 activation. The functional significance of the interaction between TAB2 and NEMO linear ubiquitin chain may be more prominent in cells treated with genotoxic agents, which induce relatively weak NF-κB activation with a delayed signalling kinetics. Additional genetic, biochemical, and structural studies will be required to elucidate underlying mechanisms.
We found either mutation on NEMO-K285/309 or disrupting LUBAC diminished genotoxic NF-κB activation, which in turn attenuated upregulation of several key anti-apoptotic genes upon genotoxic stimulation, resulting in increased apoptosis in response to genotoxic drug treatment. Although the changes were consistent, we observed much stronger phenotypes in NEMO-K285/309R mutant cells than in HOIP-depleted cells, indicating the mutation on K285/309 may not only abrogated NEMO linear ubiquitination but also interfered additional functions of these critical residues such as sumoylation and mono-ubiquitination upon genotoxic stress. Altogether, these data indicate an important role of NEMO linear ubiquitination in facilitating the pro-survival function of genotoxic NF-κB activation. As many cancer cells, either epithelial or haematopoietic, can achieve resistance to genotoxic anticancer drugs, radiation and death cytokines by activating NF-κB (Baldwin, 2001), our study suggests that LUBAC may serve as a novel target for pharmacological intervention to improve the efficacy of conventional cancer therapeutic regimes.
Materials and methods
Plasmids and reagents
The lysine-substituted NEMO mutants, K309R, K285R, and K285/309R, were generated by quick change PCR. The expression constructs of HA-HOILWT, Myc-HOIPWT, and Myc-HOIPCS, as well as antibodies against HOIL-1, HOIP, and linear polyubiquitin were generously provided by Dr K Iwai (Kirisako et al, 2006; Tokunaga et al, 2009). Antibodies against NEMO (FL419), ubiquitin (P4D1), Myc (9E10), TAK1 (M579), TAB2 (H300), IKKα/β (H470), and HA (Y11) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody against IKKβ was from Cell Signaling Technology (Danvers, MA). Antibodies against SUMO-1 (21C7) and Tubulin (DM1A) were purchased from Invitrogen (Carlsbad, CA) and Calbiochem (Gibbstown, NJ), respectively. Smartpool siRNAs targeting HOIP/RNF31 were obtained from Dharmacom (Lafayette, CO). Ub2−7 protein mixtures with K63 or linear linkage were purchased from Biomol (Plymouth Meeting, PA).
Immunoprecipitation and immunoblotting
Cell lysates were resolved by SDS–PAGE and immunoblotted with the appropriate antibodies. For immunoprecipitation experiments, cells were harvested and lysed in immunoprecipitation lysis buffer. 20mM Tris [pH 7.0], 250mM NaCl, 3mM EDTA, 3mM EGTA, 0.5% NP-40, 2mM DTT, 0.5mM PMSF, 20mM beta-glycerol phosphate, 1mM sodium orthovanadate, 1μg of leupeptin/ml, 1μg of aprotinin/ml, 10mM p-nitrophenyl phosphate [PNPP], 10mM sodium fluoride. To detect ubiquitinated or sumoylated form of NEMO, cells were lysed with 1% SDS in immunoprecipitation lysis buffer mentioned above, incubated at 95 °C for 30 min, and then diluted to 0.1% SDS with immunoprecipitation lysis buffer. NEMO was immunoprecipitated from diluted lysates using antibodies against NEMO or Myc-tag at 4 °C for 1 h, followed by incubation with protein G-conjugated beads for 3 h. Immunoprecipitates were resolved on SDS–PAGE and transferred onto nitrocellulose blot. The transferred blot was further boiled in water for ∼30 min, then probed with antibodies against ubiquitin, linear chain-specific ubiquitin or SUMO-1.
In vitro kinase assay
For TAK1 and IKK kinase assays, treated cells were lysed in immunoprecipitation lysis buffer. Cell lysates were immunoprecipitated using anti-TAB2 antibody or anti-NEMO antibody, respectively, and immobilized on protein G-conjugated beads. Kinase reactions were carried out in kinase buffer containing 32P-ATP for 45 min at 30 °C. Recombinant GST–IKKβ (166–197) and GST–IκBa (1–56) were used as substrates for TAK1 and IKK kinase assays, respectively. The reaction samples were resolved on SDS–PAGE and phosphorylated proteins were quantitated using Cyclone phosphoimager.
In vitro pull-down assay
In vitro pull-down assay of NEMO/TAB2 with various ubiquitin chains was carried out as reported (Komander et al, 2009). Briefly, GST-tagged NEMO/TAB2/TAB2 deletion mutants or GSTs (10 μg) were incubated with 20 μl glutathione sepharose 4B (GE Lifescience) for 30 min in 450 μl pull-down buffer (150 mM NaCl, 50 mM Tris (pH 7.5), 5 mM DTT, and 0.1% NP-40). Then, 1.5 μg of Ub2−7 protein mixture with K63 or linear linkage was subsequently added to each sample and further incubation was carried out for overnight at 4 °C. The beads were washed with pull-down buffer, boiled with 2 × SDS loading buffer and resolved on 12.5% SDS–PAGE. Western blotting was performed with anti-ubiquitin antibody (Santa Cruz).
Flow cytometry
Cells were harvested and then washed with PBS once. To quantitate the percentage of cells that are actively undergoing apoptosis, cells were stained with FITC conjugated Annexin V and propidium iodide (PI) following the manufacturer’s instruction, then measured with Flow Cytometer (C6, Accuri Cytometers). For cell-cycle analysis, cells were fixed in cold 75% ethanol at 4 °C overnight, then pelleted by centrifugation and stained with PI (50 μg/ml, Sigma). The percentage of cells in different cell-cycle phases was analysed by flow cytometer (LSR II, BD BioSciences).
Supplementary Material
Acknowledgments
We thank Drs L Pfeffer, R Laribee (UTHSC), and S Miyamoto (UW-Madison) for stimulating discussion and critical reading of the manuscript. This work was supported by R01CA149251 from NIH and research grant from Leukemia Research Foundation (to ZW)
Author contributions: ZW conceived and designed experiments; JN, YS, and ZW performed experiments; JN and ZW analysed data and wrote the manuscript; KI provided reagents and comments on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adhikari A, Xu M, Chen ZJ (2007) Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26: 3214–3226 [DOI] [PubMed] [Google Scholar]
- Baldwin AS (2001) Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest 107: 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton S, Ashkenazi A (2011) NEMO and RIP1 control cell fate in response to extensive DNA damage via TNF-[alpha] feedforward signaling. Cell 145: 92–103 [DOI] [PubMed] [Google Scholar]
- Chen ZJ (2005) Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol 7: 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell 33: 275–286 [DOI] [PubMed] [Google Scholar]
- Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS, Vucic D (2010) c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J 29: 4198–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WWL, Nachbur U, Gangoda L, Warnken U, Purcell AW, Silke J, Walczak H (2011) Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471: 591–596 [DOI] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, Koschny R, Komander D, Silke J, Walczak H (2009) Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 36: 831–844 [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S (2008) Shared principles in NF-kappaB signaling. Cell 132: 344–362 [DOI] [PubMed] [Google Scholar]
- Hinz M, Stilmann M, Arslan SC, Khanna KK, Dittmar G, Scheidereit C (2010) A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. Mol Cell 40: 63–74 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (2006) Lingering mysteries of ubiquitin-chain assembly. Cell 124: 27–34 [DOI] [PubMed] [Google Scholar]
- Hostager BS, Fox DK, Whitten D, Wilkerson CG, Eipper BA, Francone VP, Rothman PB, Colgan JD (2010) HOIL-1L interacting protein (HOIP) as an NF-kB regulating component of the CD40 signaling complex. PLoS ONE 5: e11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S (2003) Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell 115: 565–576 [DOI] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJL, Goswami P, Nagy V, Terzic J, Tokunaga F, Androulidaki A, Nakagawa T, Pasparakis M, Iwai K, Sundberg JP, Schaefer L, Rittinger K, Macek B, Dikic I (2011) SHARPIN forms a linear ubiquitin ligase complex regulating NF-αB activity and apoptosis. Nature 471: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inn K-S, Gack MU, Tokunaga F, Shi M, Wong L-Y, Iwai K, Jung JU (2011) Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell 41: 354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K, Tokunaga F (2009) Linear polyubiquitination: a new regulator of NF-[kappa]B activation. EMBO Rep 10: 706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J (2005) PIDD mediates NF-kappaB activation in response to DNA damage. Cell 123: 1079–1092 [DOI] [PubMed] [Google Scholar]
- Janssens S, Tschopp J (2006) Signals from within: the DNA-damage-induced NF-[kappa]B response. Cell Death Differ 13: 773–784 [DOI] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ (2004) TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell 15: 535–548 [DOI] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25: 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D (2008) The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell 29: 451–464 [DOI] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JDF, Odenwaelder P, Wilkinson KD, Barford D (2009) Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep 10: 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D (2009) Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol 16: 1328–1330 [DOI] [PubMed] [Google Scholar]
- Kumar A, Takada Y, Boriek AM, Aggarwal BB (2004) Nuclear factor-kappaB: its role in health and disease. J Mol Med 82: 434–448 [DOI] [PubMed] [Google Scholar]
- Laplantine E, Fontan E, Chiaravalli J, Lopez T, Lakisic G, Veron M, Agou F, Israel A (2009) NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J 28: 2885–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang L, Dorf ME (2009) PKC phosphorylation of TRAF2 mediates IKK[alpha]/[beta] recruitment and K63-linked polyubiquitination. Mol Cell 33: 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YC, Lin SC, Rospigliosi CC, Conze DB, Wu CJ, Ashwell JD, Eliezer D, Wu H (2009) Structural basis for recognition of diubiquitins by NEMO. Mol Cell 33: 602–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S (2011) Nuclear initiated NF-[kappa]B signaling: NEMO and ATM take center stage. Cell Res 21: 116–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND (2007) Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol 8: 49–62 [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136: 1098–1109 [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamashita M, Yamagata A, Fukai S (2009) Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J 28: 3903–3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ (2009) The role of ubiquitin in NF-kB regulatory pathways. Annu Rev Biochem 78: 769–796 [DOI] [PubMed] [Google Scholar]
- Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C (2009) A nuclear poly(ADP-Ribose)-dependent signalosome confers DNA damage-induced IkB kinase activation. Mol Cell 36: 365–378 [DOI] [PubMed] [Google Scholar]
- Sun S-C (2008) Deubiquitylation and regulation of the immune response. Nat Rev Immunol 8: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S-i, Tanaka K, Nakano H, Iwai K (2011) SHARPIN is a component of the NF-kB-activating linear ubiquitin chain assembly complex. Nature 471: 633–636 [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K (2009) Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol 11: 123–132 [DOI] [PubMed] [Google Scholar]
- Wertz IE, Dixit VM (2009) Regulation of death receptor signaling by the ubiquitin system. Cell Death Differ 17: 14–24 [DOI] [PubMed] [Google Scholar]
- Wu ZH, Miyamoto S (2007) Many faces of NF-kappaB signaling induced by genotoxic stress. J Mol Med 85: 1187–1202 [DOI] [PubMed] [Google Scholar]
- Wu ZH, Shi Y, Tibbetts RS, Miyamoto S (2006) Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science 311: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Wu ZH, Wong ET, Shi Y, Niu J, Chen Z, Miyamoto S, Tergaonkar V (2010) ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol Cell 40: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ (2009) Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461: 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xia F, Hermance N, Mabb A, Simonson S, Morrissey S, Gandhi P, Munson M, Miyamoto S, Kelliher MA. (2011) A cytosolic ATM/NEMO/RIP1 complex recruits TAK1 to mediate the NF-{kapp}B and p38 MAP kinase/MAPKAP-2 responses to DNA damage. Mol Cell Biol 31: 2774–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.