Abstract
Despite great progress in identifying genetic variants that influence human disease, most inherited risk remains unexplained. A more complete understanding requires genome-wide studies that fully examine less common alleles in populations with a wide range of ancestry. To inform the design and interpretation of such studies, we genotyped 1.6 million common single nucleotide polymorphisms (SNPs) in 1,184 reference individuals from 11 global populations, and sequenced ten 100-kilobase regions in 692 of these individuals. This integrated data set of common and rare alleles, called ‘HapMap 3’, includes both SNPs and copy number polymorphisms (CNPs). We characterized population-specific differences among low-frequency variants, measured the improvement in imputation accuracy afforded by the larger reference panel, especially in imputing SNPs with a minor allele frequency of ≤5%, and demonstrated the feasibility of imputing newly discovered CNPs and SNPs. This expanded public resource of genome variants in global populations supports deeper interrogation of genomic variation and its role in human disease, and serves as a step towards a high-resolution map of the landscape of human genetic variation.
The Human Genome Project1, the SNP Consortium2 and the International HapMap Project3 collectively identified ~10 million common DNA variants, primarily SNPs, in a limited set of DNA samples. Knowledge of these SNPs and their linkage-disequilibrium patterns enabled genome-wide association studies, which have successfully identified hundreds of novel genomic loci that influence human diseases4.
Nonetheless, our knowledge of human genetic variation remains limited with respect to variant type, frequency and population diversity. Only common DNA variants (minor allele frequency (MAF) ≥5%) have yet been well studied, even though low MAF variants no doubt contribute to a substantial fraction of hereditary risk for common diseases5. Only recently have systematic studies of other types of variants, in particular copy number variation, begun to guide our knowledge of their frequency spectra, population distributions and patterns of linkage disequilibrium6–10.
To inform efforts aimed at rectifying this, we expanded the public HapMap Phase I and II resource by performing genome-wide SNP genotyping and CNP detection, as well as polymerase chain reaction (PCR) resequencing in selected genomic regions. We collected and studied an extended set of 1,184 samples from 11 populations (Supplementary Information). These included all HapMap Phase I and II samples, along with further samples from the same four populations: individuals from the Centre d’Etude du Polymorphisme Humain collected in Utah, USA, with ancestry from northern and western Europe (CEU); Han Chinese in Beijing, China (CHB); Japanese in Tokyo, Japan (JPT); and Yoruba in Ibadan, Nigeria (YRI). Samples from seven additional populations were also included: African ancestry in the southwestern USA (ASW); Chinese in metropolitan Denver, Colorado, USA (CHD); Gujarati Indians in Houston, Texas, USA (GIH); Luhya in Webuye, Kenya (LWK); Maasai in Kinyawa, Kenya (MKK); Mexican ancestry in Los Angeles, California, USA (MXL); and samples collected in Tuscany, Italy (TSI). These populations were included to provide further variation data from each of the three continental regions represented in HapMap Phase I and II, as well as data from some more admixed populations residing in the US. the specific populations and localities were chosen based on contacts with researchers who worked in those regions and had established trusting relationships with local communities. (See Supplementary Table 1 and the Supplementary Information for more details.)
SNP genotyping
Genotype data were obtained with the Affymetrix Human SNP array 6.0 (interrogating 1,852,600 genomic sites) and the Illumina Human1M-single beadchip (1,199,187 genomic sites), initially applied to 1,486 and 1,284 samples, respectively. Following genotype calling6,11 and initial filtering of low-quality and incomplete data, 909,622 variant SNPs from 1,326 samples (Affymetrix) and 1,055,111 sites from 1,211 samples (Illumina) remained. Data from the two platforms were merged; genotype concordance was 99.5% (across 335,014 overlapping SNPs) at a call rate of 99.8%. Further filters were applied to this merged data set on the basis of population-specific call rates, deviation from Hardy–Weinberg equilibrium and the expected Mendelian inheritance patterns (Supplementary Methods). The consensus genotype set contains 1,440,616 SNPs that are polymorphic in 1,184 individuals from 11 populations. Analysis shows a small but statistically significant bias against rare (MAF = 0.05–0.5%) allele calls (observed in both platforms), consistent with previous reports (Supplementary Information). The data were then phased (Supplementary Information).
Regional sequencing
We selected ten 100-kb regions for direct PCR-Sanger capillary sequencing analysis. These regions included the central 100 kb from five previously sequenced HapMap-ENCODE 500-kb regions12 and five ENCODE regions not previously subject to sequencing in the HapMap Project (Supplementary Table 4). A total of 692 unrelated samples chosen from the ten then available genotyped population samples (ASW, CEU, CHB, CHD, GIH, JPT, LWK, MXL, TSI and YRI) were interrogated and passed quality control metrics (Supplementary Table 1). SNPs were discovered from the raw sequence data using SNP Detector 3.0 software13. Subsequent genotyping showed an overall genotype concordance rate of 99.2% and an 86.8% genotype concordance rate for genotypes with minor alleles (Supplementary Table 5a). Also, a 93.6% genotype concordance rate was found for singleton genotypes with minor alleles and 88% for two to six copies of the minor allele. The higher genotype concordance rate in singletons reflects the higher stringency applied in making singleton calls. (See Supplementary Information and Supplementary Table 5 for details.)
Unlike SNPs present on microarray platforms, which are intentionally biased towards high frequency by the discovery and selection process, the SNPs discovered by sequencing provide a direct estimate of the underlying allele frequency spectrum in each population. As in previous surveys, common (MAF ≥5%) and low-frequency (MAF = 0.5–5%) variants account for the vast majority of the heterozygosity in each sample, but we also observed a large number of rare (MAF = 0.05–0.5%) and private (singletons and MAF <0.05%) variants (see Supplementary Table 2 for definitions of variant frequency classes). Each population had 42– 66% of sites with a MAF <5%, compared to 10–13% in the genotyping data; 37% of SNPs with a MAF <0.5% were observed in only one population. In total, 77% of the discovered SNPs were new (that is, not in the SNP database (dbSNP) build 129) and 99% of those had a MAF <5%.
Copy number variation
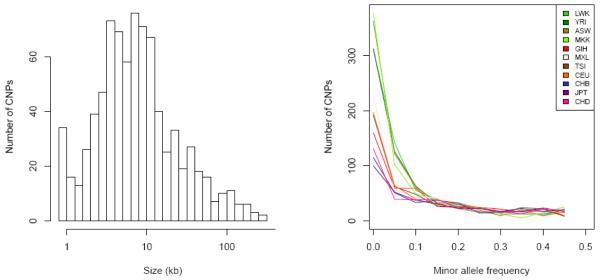
To assess copy number variation we merged and analysed the probe-level intensity data from both the Affymetrix and Illumina arrays, identifying 1,610 genomic segments that probably varied in copy number (CNPs) with an estimated MAF of at least 1% of the cohort (see Methods). Further quality control steps yielded a set of reference genotypes for 856 CNPs with a 99.0% mean call rate and 0.3% Mendelian inconsistency—very high accuracy, but still less than that observed from SNP genotyping (Mendelian inconsistency <0.14% in this data set; Supplementary Information). We estimate that the resolution of this analysis to detect CNVs is at a multi-kilobase scale, but not smaller (Fig. 1a).
Figure 1. Size and frequency spectra of common and rare CNPs.
a, Estimated size distribution of common CNPs calculated from the physical span of the genomic probes supporting each CNP event. b, Allele frequency spectrum for biallelic CNPs calculated from integer CNP genotypes for the samples analysed in this work.
The overall allele frequency spectrum of CNPs resembled that of the SNPs ascertained by resequencing: most variants were at low frequency (Fig. 1b), but most heterozygosity was due to a limited set of common variants. This extends an observation previously made in the original HapMap population samples7 to additional populations. The allele frequency spectrum of common CNPs (MAF >10%) was similar across populations, but differed markedly at lower frequencies. African-ancestry and admixed populations showed by far the greatest number of variants with MAF <5%, and had a higher average number of CNPs differing in copy number between two individuals (160–171) than non-admixed populations without African ancestry (127–142) (Fig. 1b).
At 95% of the CNPs the variation observed was explained by a simple biallelic model obeying Mendelian inheritance and Hardy–Weinberg equilibrium. The remaining 5% of loci showed multi-allelic patterns, somewhat lower than the 15% reported in a recent study7, which may reflect improved resolution of the assays and analyses used in this study. Among the biallelic loci, 92% were deletions (diploid copy numbers ≤2) and 8% were duplications (diploid copy numbers ≥2); the disparity reflects our higher power to detect small deletions than small insertions. The median size of CNPs genotyped in this study was 7.2 kb (Fig. 1a), with biallelic deletions significantly smaller on average than biallelic duplications because of this difference in power.
The 856 genotyped CNPs represent an average of 3.5 megabases of sequence in each individual; this is ≈0.1% of the human genome, and similar to the overall rate of SNP variation. One-third (33.5%) of the genotyped CNPs overlap RefSeq genes, with duplications more likely than deletions to overlap genes (after correcting for the greater average length of duplications (P = 0.006)), which probably reflects greater purifying selection acting on deletions of genes.
Common and low-frequency variation across populations
We used the ENCODE data to assess how well each sample set could serve as a SNP discovery resource for other populations. This is an important practical matter, because it determines the effectiveness of scanning multiple populations for variation discovery as compared to sampling more deeply in a single population. To estimate how informative SNPs discovered in population A were for those present in population B, we counted the fraction of variants found in a sample of 30 A individuals that were also seen in a sample of 30 B individuals. Our measure of informativeness was the ratio of this fraction to that observed for a second, non-overlapping sample of 30 A individuals (Fig. 2a).
Figure 2. SNP discovery informativeness across populations.
a, b, For each of 7 populations for which at least 60 individuals were resequenced, we considered a sample of 30 individuals, another non-overlapping sample of 30 individuals from the same population, and a sample of 30 individuals from each of the 6 other populations (results are averaged over 1,000 random samplings). Out of all SNPs that are either polymorphic (a) or polymorphic with a minor allele with at most two copies in the sample of 30 individuals (b), here we present the fraction that are also polymorphic in a different sample, starting with the other sample from the same population (black bars). The black bars serve as a baseline that accounts for the effect of sampling stochasticity and sequencing errors on SNP discovery. The different y-axis scales used reflect the lower likelihood of a low-frequency variant being seen in a different sample.
As judged by this measure, informativeness varied greatly for different population pairs. Consistent with the observation that non-African diversity is largely a subset of African diversity14, African samples provided a more complete discovery resource for variant sites in non-African samples than the converse (Fig. 2a). Focusing only on low-frequency variants in the original sample of 30 A individuals (one or two copies, corresponding to allele frequencies of 3.3% or less), even African samples were highly incomplete for diversity outside of Africa, with informativeness ratios dropping to 40–60% in LWK and YRI (Fig. 2b). In general, for low-frequency variants only closely related populations did an adequate job of capturing variation (Fig. 2b), probably reflecting the recent origins of low-frequency variants. Two populations, LWK and GIH, stand out as being poorly captured by any of our other populations, the result of admixture with an ancestral population not closely related to any in our regional sequencing data (Supplementary Methods). (Although the MKK captures similar East African ancestry to that of LWK (Supplementary Fig. 2), it had not been included in the regional sequencing.)
In all cases, FST, a measure of the degree of population differentiation (Supplementary Table 6) correctly predicted the most informative population, despite the FST estimates being based on genotyping array data with SNP ascertainment biases15. However, FST was not a perfect predictor: the correlation coefficient between FST and ascertainment informativeness was highly variable, ranging across populations between −0.67 and −0.99 for all SNPs and between −0.51 and −0.97 for low-frequency SNPs. Furthermore, FST is symmetrical between a pair of populations, whereas informativeness is not. For example, the most informative population for low-frequency GIH SNPs was TSI, with informativeness being only 55% of that of an independent GIH sample (because TSI captures only one of GIH’s ancestral populations; Fig. 2b). Conversely, the informativeness of GIH on low-frequency TSI SNPs was 71% (Fig. 2b).
Within a single population, increasing the sequenced sample size yields diminishing returns of new SNPs. Figure 3 quantifies the number of SNPs discovered by resequencing as a function of sample size; it demonstrates the expected partitioning between populations with genetic proximity to Africa, and therefore higher diversity, and the rest of the populations. The new SNPs are mostly of lower frequency, and account for the majority of the discovered variant sites as the number of interrogated samples is increased (Supplementary Fig. 5).
Figure 3. Effect of sample size on SNP ascertainment.
The number of SNPs discovered as a function of sample size by averaging over 1,000 random samplings. For each population, we randomly sampled without replacement a subset of the individuals of any possible size and considered which SNPs were polymorphic in the resequencing data for that sample. For any given sample size, many more variants are discovered in populations with genetic proximity to Africa (LWK, ASW and YRI), compared to populations of non-African ancestry.
Haplotype sharing
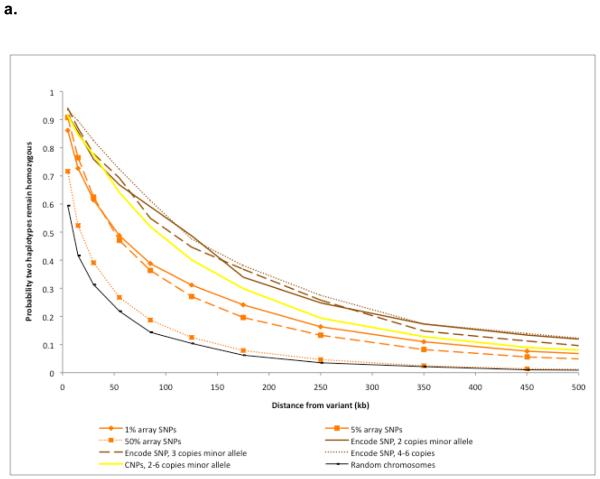
We next characterized the extent to which alleles share haplotype backgrounds as a function of frequency, a question related to the imputation of variants not directly observed in each clinical sample. Population genetic models predict that lower-frequency variants should on average be younger than more common variants, and thus have a longer physical extent of haplotype sharing. We selected from the ENCODE data a set of SNPs observed two to six times in YRI or in CEU; we estimated haplotype phase with high confidence using parent–offspring trio data. After validation using Sequenom genotyping (Supplementary Methods) to ensure highly accurate genotypes, 272 SNPs were examined in YRI and 106 in CEU. For comparison, a set of SNPs from the genotyping arrays with the same frequencies were analysed. Haplotype sharing was measured by calculating the haplotype homozygosity (that is, perfect concordance between haplotypes) using the consensus genotype data around each low-frequency SNP.
In both populations, ENCODE variant alleles had longer shared haplotypes than array-based SNPs of the same frequency, and all low-frequency alleles (whether or not discovered by sequencing) had longer haplotypes than did higher-frequency SNPs (Fig. 4). Shorter haplotypes for array SNPs are expected because of SNP ascertainment, which was biased towards SNPs shared across populations and therefore towards older SNPs with shorter-range linkage disequilibrium. Among the ENCODE SNPs, there was little difference in haplotype sharing between alleles seen twice and those seen four to six times in the sample, indicating that these minor differences in frequency are not good predictors of the age and haplotype sharing of alleles (presumably due to drift and sampling error in the frequency estimate). Haplotype sharing was also greater for derived than for ancestral alleles, although the effect was modest (Supplementary Fig. 6).
Figure 4. Haplotype sharing around SNPs and CNPs.
a, b, Extent of haplotype homozygosity around variant alleles of various frequencies. Shown are SNPs from the ENCODE sequence, CNPs of comparable frequency, SNPs from the arrays and on randomly grouped chromosomes, and (for YRI) the maximum possible sharing for a genotyping error rate of 0.2%. a, CEU. b, YRI.
We performed the same analysis for CNPs, studying variants in the same frequency range (two to six copies) and in the same two populations. To reduce ambiguity, we restricted ourselves to CNPs that had exactly two genotypic states and treated these as biallelic variant sites. We masked out any SNPs within the boundaries of the CNP, and thereafter analysed them in the same way as the SNPs. We found that CNPs and SNPs in the same samples had a similar extent of haplotype sharing (Fig. 4); in CEU, sharing does drop off faster for CNPs, but the difference was not statistically significant with our sample size. This observation, which is consistent with a previous observation that low-frequency CNPs segregate on long shared haplotypes7, suggests that imputation methods should have comparable effectiveness for CNPs as for SNPs, at least for biallelic CNPs that are measured well by our array-based approach.
We examined the subset of SNPs (862) from the ENCODE sequence data that were present at low frequency (two to six occurrences of the alleles) and were also observed in more than one population. These are of special interest as they would be most likely to include examples of independent mutations that occurred since the populations diverged, as opposed to each observed allele being descended from a single ancestral event. In the majority of cases (93%) the rare variants at each site occurred on the same haplotype background, consistent with a single origin, and their current distribution reflects drift. The remaining 51 sites (7%) were observed to have alleles that occurred in more than one haplotype. Furthermore, the different haplotypes occurred in different populations for all except one site. These 51 sites are therefore candidates for independent occurrence of mutation at the same site (Supplementary Information and Supplementary Table 7).
Imputation of untyped variants
Whole-genome sequencing will enable characterization of almost all variants in an individual. However, until this becomes affordable in large collections of samples, genotyping arrays, in concert with statistical imputation of untyped alleles, offer a complementary approach to increase power for previously observed alleles. We therefore evaluated the effect on imputation afforded by the larger HapMap 3 resource and also studied how well imputation performs when applied to lower frequency variants and to CNPs.
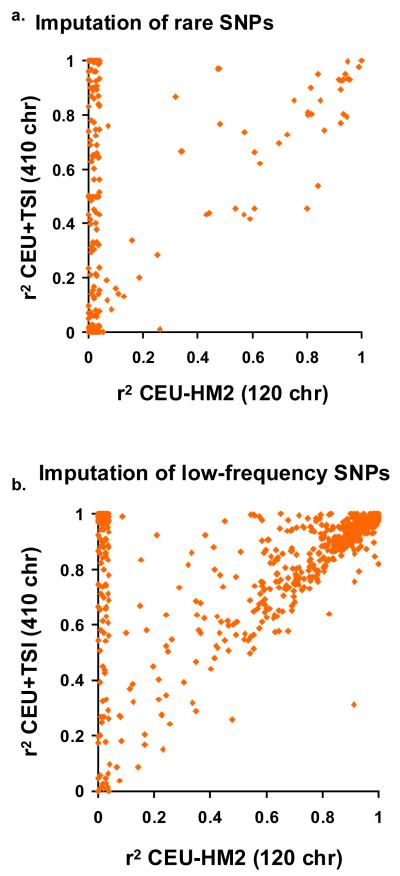
One use of imputation is to combine data for genome-wide association studies performed using different array platforms. Therefore, we first measured the change in performance of imputation for common (array-based) SNPs using a HapMap 3 panel of 410 phased European-ancestry chromosomes (CEU+TSI) in comparison with a HapMap Phase II panel of 120 CEU chromosomes (HMII-CEU). Each panel was used to impute array SNPs in 1,393 Europeans of the 1958 British birth cohort (58BBC), which had previously been genotyped using earlier versions of the Affymetrix and Illumina chips16,17. Using the Illumina array genotypes, we imputed HapMap 3 SNPs on chromosome 20 and calculated the mean r2 between true (called) genotype and imputed genotype dosage for each Affymetrix SNP not on the Illumina chip (Supplementary Table 8).
For common SNPs (MAF ≥5%), the larger HapMap 3 reference panel made only a slight difference to the already excellent performance (mean r2 increased from 0.946 to 0.961). However, as expected there was greater improvement for rare (MAF <0.5%) and low-frequency SNPs (MAF = 0.5–5%). Their combined mean r2 increased from 0.60 to 0.76, driven by a large subset of rare SNPs (41%) and low-frequency SNPs (25%) where r2 increased by at least 0.1, yielding mean r2 improvement for these subsets of 0.62 and 0.49 respectively (Fig. 5a, b and Supplementary Table 8). This improvement occurred mainly at SNPs with unobserved minor alleles in the HMII-CEU reference panel that became informative in the larger CEU+TSI panel (see Supplementary Tables 9 and 10 for the effect of reference panel size on imputation accuracy in other populations).
Figure 5. Imputation accuracy and reference panel size.
a, b, Mean r2 between true and imputed genotype dosage for SNPs imputed from a HapMap-II-sized panel of 120 CEU chromosomes (HMII-CEU) or a HapMap 3 panel of 410 European-ancestry chromosomes (CEU+TSI). Scatter plots show Affymetrix 500K SNPs on chromosome 20 imputed for 1,393 subjects of the 1958 British birth cohort. a, Rare SNPs (MAF <0.5%). b, Low-frequency SNPs (MAF = 0.5–5%).
We next investigated imputation across populations. We compared imputation of CEU or TSI using the CEU reference panel, CHD or CHB+JPT using the CHB+JPT reference panel, and YRI or LWK using the YRI reference panel. Imputation into closely related populations worked well for common but not for low-frequency alleles (Supplementary Table 11).
Imputation in admixed populations was examined by comparing reference panels based on either one population, or on mixtures of other populations; one mixture (COSMO1) combined chromosomes from the original three HapMap population panels, whereas the other (COSMO2) included seven populations (CEU, CHB, GIH, JPT, MKK, MXL and YRI, see Methods for details). For ASW, the best reference panel was YRI+CEU, which yielded mean r2 = 0.87 and mean r2 = 0.72 for common and low-frequency SNPs, respectively. For the other admixed populations, the best reference panels were the same-population panel (when available) followed by the diverse reference panel of seven populations (COSMO2) (Supplementary Table 10).
Cross-population imputation can be less effective for low-frequency alleles both because the sets of alleles in the two samples do not overlap perfectly (see earlier), and because haplotype patterns differ between populations. To isolate the effect of differing haplotype patterns, imputation within a population (CEU or YRI) was compared with imputation into a closely related population (TSI or LWK), but restricting the analysis to SNPs that were polymorphic in both target and reference panels (Fig. 6a). Notably, the imputation worked well for low-frequency alleles when using the correct reference panel, with a mean r2 > 0.7 with only two copies of the minor allele in the reference panel, and a mean r2 > 0.6 when imputing from a single copy. Imputation accuracy into a closely related European population (CEU/TSI FST = 0.004) was almost indistinguishable from the accuracy within a single population. For the two African populations, where low-frequency diversity is greater and the populations more diverged (FST = 0.008), the difference between reference and target populations was more substantial, with mean r2 only rising above 0.7 when five copies of the minor allele were in the reference panel. In both cases, however, the cross-population accuracy was much better than that seen in Supplementary Table 10, indicating that cross-population loss of accuracy largely results from the incomplete sharing of low-frequency alleles between reference and target samples, rather than from differences in haplotype backgrounds.
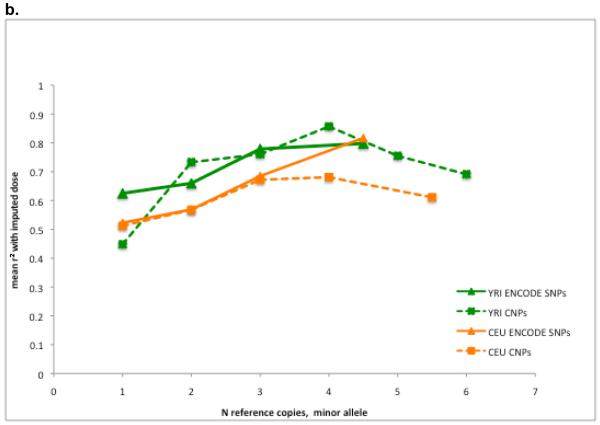
Figure 6. Imputation: new populations, new variants.
a, b, Mean r2 between true and imputed genotype dosage as a function of copies of minor allele in the reference panel. a, The loss in imputation accuracy when the reference population differs slightly from the target population (CEU imputed into CEU compared to CEU into TSI; and YRI into YRI compared to YRI into LWK). b, Imputation accuracy for newly discovered variants (CNPs and ENCODE SNPs).
Using the same approach, we also checked the dependence of imputation accuracy on pedigree information, as trios improve the accuracy of haplotype phasing and therefore imputation. We compared the within-CEU results described earlier to imputation done purely within the TSI sample, with the sample size held fixed. The two populations are closely related, but the CEU samples were genotyped in trios and the TSI samples as individuals. The results were virtually identical (data not shown), indicating that poor phasing was not a problem for our unrelated samples, at least for array SNPs. (Note that pedigree information was used indirectly in our TSI phasing, with phased CEU chromosomes used as a reference panel for phasing TSI.)
In a second set of analyses, we assessed imputation of newly discovered variants using as our test sets SNPs found in CEU and YRI by the complete ENCODE sequencing and CNPs. We created a reference panel of phased haplotypes that incorporated the new variant and the surrounding consensus genotype data, and used it to impute genotypes in additional samples. This models (for example) the imputation into an existing genome-wide association study of new SNPs and CNPs discovered by the 1000 Genomes Project or an exome sequencing project. We assessed imputation accuracy by masking each individual in the sample in turn and imputing its genotype from the rest of the sample, thus preserving the largest reference panel possible. For comparison, we also repeated the analysis by masking randomly selected, frequency-matched array SNPs, rather than newly discovered variants.
The imputation accuracy was quite similar for the SNPs and the CNPs (Fig. 6b), given their similar haplotype properties. Accuracy depended on high SNP density; reducing the set of tag SNPs from the full HapMap 3 set to the subset found on an earlier generation of array (approximately a threefold reduction in density) reduced r2 by roughly a factor of two for low-frequency SNPs (Supplementary Fig. 8). Somewhat unexpectedly, the accuracy was consistently higher for YRI than for CEU for both classes of variant, despite the former’s greater haplotype diversity and the identical panel sizes and SNP frequencies. One possible explanation is that for less common variants, the relationship between frequency and age has been partly obscured by population bottlenecks in the history of European populations, so that minor allele frequency is less effective as a predictor of allele age than in samples from Africa.
Overall, we observed that imputation works well for the newly discovered SNPs, although not as well as for frequency-matched SNPs on the available genotyping arrays, even though newly discovered SNPs show greater haplotype sharing. This difference may be due to an ascertainment bias in the discovery and choice of SNPs on the arrays—most SNPs in HapMap and on arrays were originally detected by sequencing a few individuals, representing a fraction of haplotypes in the population18; these haplotypes are better represented on arrays (which focused on SNPs that served as good proxies) than are newly discovered SNPs. This difference is markedly seen in a comparison of nearby, frequency-matched SNPs from within either the array or ENCODE: looking only at SNPs with two copies of the minor allele, 5% of the time, two frequency-matched ENCODE SNPs are perfect proxies for each other, whereas the fraction is 70–80% for a pair of frequency-matched array SNPs (Supplementary Fig. 9). This highlights the need for caution in extrapolating from low-frequency array SNPs to low-frequency sequencing SNPs.
Natural selection
We searched the larger and more diverse HapMap 3 genotype data for genomic regions showing signals of positive natural selection using a recently published method, the composite of multiple signals (CMS)19. In the three original HapMap populations, CEU, CHB+JPT and YRI, comparing the regions identified in HapMap 3 with published results from HapMap Phase II (Supplementary Methods), we replicated 83% (147 out of 178) of the previous HapMap Phase II candidate regions (Supplementary Fig. 10a–d). Of the 17% of regions that did not replicate, most had lower SNP density in HapMap 3 than in HapMap Phase II; in 20 regions, none of the high-scoring HapMap Phase II SNPs was genotyped in HapMap 3.
Next we sought to identify candidate selection loci in the new HapMap 3 populations TSI, LWK and MKK (that is, all populations except those likely to be recently admixed). First we identified 54 broad candidate regions for selection using long haplotype tests. Applying CMS to these regions, we localized signals to new and intriguing candidates (Supplementary Table 12). In TSI, pigmentation genes were again identified, including KITLG and MLPH23 (Supplementary Fig. 10e, f). We found other signals, like LAMA3, a gene involved in wound healing, and an olfactory receptor cluster. In the Kenyan populations we identified several immune-related genes, such as CD22624, ITGAE12 and DPP7 (Supplementary Fig. 10g–i). A novel signal identified in MKK localized to the gene ANKH; ANKH has a role in bone growth and susceptibility to arthritis, and has previously been identified as being under positive selection in horses25 (Supplementary Fig. 10j). The complete set of new candidates (Supplementary Table 12) may suggest hypotheses regarding natural selection in these populations.
Conclusions and implications
With improvements in sequencing technology, low-frequency variation is becoming increasingly accessible. This greater resolution will no doubt expand our ability to identify genes and variants associated with disease and other human traits. This study integrates CNPs and lower-frequency SNPs with common SNPs in a more diverse set of human populations than was previously available. The results underscore the need to characterize population-genetic parameters in each population, and for each stratum of allele frequency, as it is not possible to extrapolate from past experience with common alleles. As expected, lower-frequency variation is less shared across populations, even closely related ones, highlighting the importance of sampling widely to achieve a comprehensive understanding of human variation.
We find that variants discovered through large-scale sequencing have longer haplotypes than more common variants, and that imputation can perform well for both CNPs and low-frequency SNPs. Success was partial (as compared to common variants), and required a number of conditions: large reference panels, dense and accurate genotyping and good phasing. Moreover, some variants were not well imputed, although it is unclear if this is fundamental or due to a need for improved methods of imputation of lower frequency variants.
Informed by preliminary analyses of these data, the 1000 Genomes Project is studying the collection of samples from five populations within each continental region. Our data suggest that a strategy of identifying polymorphic SNPs and CNPs followed by imputation in densely genotyped samples can provide information even for lower-frequency alleles. Necessary components of such a reference panel include accurate genotyping and characterization of the haplotype background for the alleles (which included here the use of pedigree information to inform phasing), and a broad range of reference populations to capture geographically local variants. The ultimate utility of such a strategy (as compared to a more complete approach using exome or whole genome sequencing) will depend on the as yet poorly characterized distribution of causal alleles across traits, across exons as compared to non-coding regions, and the relative cost and accuracy of sequencing as compared to genotyping followed by imputation. The development of a robust reference panel will be a necessary step in the evaluation of these different strategies across a wide variety of diseases.
METHODS SUMMARY
Genotyping and genotype data quality control
Genotyping was done using Affymetrix 6.0 and Illumina 1.0 Million SNP mass arrays. Data quality control filters were applied as detailed in the text and Supplementary Information.
CNP analysis
For CNP discovery, we combined the genotype data from the Affymetrix and Illumina arrays and applied two algorithms, QuantiSNP27 and Birdseye5. First, approximately 60,000 CNP calls were made by each algorithm (~50 per sample), generally supported by data from both platforms. Shared genomic segments of common CNPs were identified and refined by an algorithm that used cross-sample correlations between nearby probes (Supplementary Information).
For CNP genotyping, we used two algorithms for summarizing the data from the probe sets into a single measurement, followed by clustering the resulting measurements into discrete copy-number classes (Supplementary Information). Although the two approaches agreed on the majority of calls (genotype concordance ≥99% for 96% of common CNPs), wherever they disagreed the approach that yielded the best separated clusters for that particular CNP was preferred. The joint use of the two platforms considerably improved the separation of genotype classes (Supplementary Fig. 1).
Sequence SNPs
Ten ENCODE regions were chosen on the basis of their overlap with previously sequenced ENCODE regions3,10. PCR primers and conventional fluorescent DNA sequencing were used, and the SNPs were identified and filtered as described in Supplementary Information.
Imputation
Imputation was performed using the MACH program26 (http://www.sph.umich.edu/csg/abecasis/MACH/download/). In all analyses, the set of samples whose genotypes were imputed did not overlap the set of samples used to construct reference panels. For the 1958 British birth cohort analysis, we imputed all available SNPs on chromosome 20. The 1958 British birth cohort samples had been previously genotyped on the Affymetrix 500K and Illumina 550K chips, so we used the 1958 British birth cohort Illumina 550K genotypes in tandem with either reference panel (HMII-CEU or CEU+TSI) to impute the known (but masked) Affymetrix 500K SNPs (Supplementary Information).
Supplementary Material
Acknowledgements
We dedicate this work to Leena Peltonen for her vital leadership role in this study, and in memory of a valued friend and colleague. We thank E. Boerwinkle and R. Durbin for critical reading of the manuscript. We thank the USA National Institutes of Health, the National Human Genome Research Institute, the National Institute on Deafness and Other Communication Disorders and the Wellcome Trust for supporting the majority of this work. Funding was also provided by the Louis-Jeantet Foundation and the NCCR ‘Frontiers in Genetics’ (Swiss National Science Foundation). We thank the people from the following communities who were generous in donating their blood samples to be studied in this project: the Yoruba in Ibadan, Nigeria; the Maasai in Kinyawa, Kenya; the Luhya in Webuye, Kenya; the Han Chinese in Beijing, China; the Japanese in Tokyo, Japan; the Chinese in metropolitan Denver, Colorado; the Gujarati Indians in Houston, Texas; the Toscani in Italy; the community of African ancestry in the southwestern USA; and the community of Mexican ancestry in Los Angeles, California. We also thank the people in the Utah Centre d’Etude du Polymorphisme Humain community who allowed the samples they donated earlier to be used for the project. The authors acknowledge use of DNA from the 1958 British birth cohort collection, funded by the UK Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. The Illumina 550K genotype data for the 1958 British birth cohort samples were made available by the Sanger Institute. For the 1958 British birth cohort Affymetrix 500K genotype data, we thank the Wellcome Trust Case Control Consortium (http://www.wtccc.org.uk), which was funded by Wellcome Trust award 076113.
The International HapMap 3 Consortium
Principal investigators David M. Altshuler1, Richard A. Gibbs2, Leena Peltonen‡; Project coordination leaders David M. Altshuler1, Richard A. Gibbs2, Leena Peltonen‡, Emmanouil Dermitzakis3; Manuscript writing group Stephen F. Schaffner1, Fuli Yu2, Leena Peltonen‡, Lisa Brooks5, Emmanouil Dermitzakis3, Penelope Bonnen2, David M. Altshuler1, Richard Gibbs2; HapMap 3 genotyping Paul I. W. de Bakker1, Panos Deloukas5, Stacey B. Gabriel1, Rhian Gwilliam5, Sarah Hunt5, Michael Inouye5, Xiaoming Jia1, Aarno Palotie5, Pamela Whittaker5; ENCODE 3 sequencing and SNP discovery Fuli Yu2, Kyle Chang2, Alicia Hawes2, Lora R. Lewis2, Yanru Ren2, David Wheeler2, Richard Gibbs2, Donna Marie Muzny2; Copy number variation typing and analysis Chris Barnes5, Katayoon Darvishi6, Matthew Hurles5, Joshua M. Korn1, Kati Kristiansson5, Charles Lee6, Steven A. McCarroll1, James Nemesh1; Population analysis Emmanouil Dermitzakis3, Alon Keinan7, Stephen B. Montgomery3, Samuela Pollack1, Alkes L. Price8, Nicole Soranzo5; Low frequency variation analysis Penelope E. Bonnen2, Richard A. Gibbs2, Claudia Gonzaga-Jauregui2, Alon Keinan7, Alkes L. Price6, Fuli Yu2; Linkage disequilibrium and haplotype sharing analysis Verneri Anttila5, Wendy Brodeur1, Mark J. Daly9, Stephen Leslie10, Gil McVean10, Loukas Moutsianas10, Huy Nguyen1, Melissa Parkin1, Stephen F. Schaffner1; Imputation Mohammed J. R. Ghori5, Ralph McGinnis5, Will Mclaren5, Samuela Pollack1, Alkes L. Price8, Stephen F. Schaffner1, Fumihiko Takeuchi5, Qingrun Zhang5; Natural selection Sharon R. Grossman11, Elizabeth B. Hostetter11, Ilya Shlyakhter1, Pardis C. Sabeti11; Community engagement and sample collection groups Clement A. Adebamowo12, Morris W. Foster13, Beborah R. Gordon14, Julio Licinio15, Maria Cristina Manca16, Patricia A. Marshall17, Ichiro Matsuda18, Jean E. McEwen19, Duncan Ngare20, Vivian Ota Wang19, Deepa Reddy21, Charles N. Rotimi22, Charmaine D. Royal23, Richard R. Sharp14 & Changqing Zeng24
1Broad Institute, 7 Cambridge Center, Cambridge, Massachusetts 02138, USA. 2Baylor College of Medicine, Human Genome Sequencing Center, Department of Molecular and Human Genetics, One Baylor Plaza, Houston, Texas 77030, USA. 3University of Geneva, Medical School, Department of Genetic Medicine and Development, Faculty of Medicine, Geneva 1211, Switzerland. 4Genetic Variation Program, National Human Genome Research Institute, National Institutes of Health, Building 31, Room B2B07, 31 Center Drive, MSC 2032, Bethesda, Maryland 20892-2033, USA. 5Wellcome Trust Sanger Institute, Department of Human Genetics, Wellcome Trust Genome Campus, Cambridge CB10 1HH, UK. 6Harvard Medical School, Brigham and Women’s Hospital, Department of Pathology, Boston, Massachusetts 02115, USA. 7Cornell University, Department of Biological Statistics and Computational Biology, 102A Weill Hall, Ithaca, New York 14853, USA. 8Harvard School of Public Health, Departments of Epidemiology and Biostatistics, 665 Huntington Avenue, Building 2 Room 211, Boston, Massachusetts 02115, USA. 9Massachusetts General Hospital, Center for Human Genetic Research, Simches Research Center, 185 Cambridge Street, Boston, Massachusetts 02114, USA. 10University of Oxford, Department of Statistics, 1 South Parks Road, Oxford, OX1 3TG, UK. 11Harvard University, Department of Organismic and Evolutionary Biology, Center for Systems Biology, 52 Oxford Street, Room 469, Cambridge, Massachusetts 02215, USA. 12University of Maryland School of Medicine, Department of Epidemiology and Preventative Medicine, N406 Institute of Human Virology, 725 West Lombard Street, Baltimore, Maryland 21201, USA. 13University of Oklahoma, Department of Anthropology, 455 West Lindsey Room 505C, Norman, Oklahoma 73019, USA. 14The Cleveland Clinic, Department of Bioethics, 9500 Euclid Avenue JJ60, Cleveland, Ohio 44124, USA. 15The Australian National University, John Curtin School of Medical Research, Garran Road, Building 131, Canberra, ACT2603, Australia. 16Institute for Oncological Study and Prevention, Florence 50139, Italy. 17Case Western Reserve University, Department of Bioethics, School of Medicine TA200, 10900 Euclid Avenue, Cleveland, Ohio 44106-4976, USA. 18Health Sciences University of Hokkaido, 1757 Kanazawa, Tobetsu-cho, Ishikari-gun, Hokkaido 061-0293, Japan. 19National Human Genome Research Institute, Ethical, Legal, and Social Implications Research Program, 5635 Fishers Lane, Sutie 4076, MSC 9305, Bethesda, Maryland 20892-9305, USA. 20Moi University, Department of Population and Family Health, PO Box 4606, Eldoret 30100, Kenya. 21University of Houston at Clear Lake, Department of Anthropology, 2700 Bay Area Boulevard, PO Box 295, Houston, Texas 77058-1098, USA. 22National Human Genome Research Institute, Center for Research on Genomics and Global Health, 12 South Drive, MSC 5635, Building 12A, Room 4047, Bethesda, Maryland 20892-5635, USA. 23Duke University, Institute for Genome Sciences and Policy, 450 Research Drive, PO Box 91009, LSRC B-Wing, Room 320B, Durham, North Carolina 27708, USA. 24Beijing Institute of Genomics, Chinese Academy of Science, Beijing Airport Industrial Zone B-6, Beijing 101300, China.
‡Deceased.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information The HapMap 3/ENCODE 3 data set has been deposited at http://www.hapmap.org. The sequence traces of ENCODE 3 can be accessed at http://www.ncbi.nlm.nih.gov/Traces/trace.cgi by submitting the query : species_code=“HOMO SAPIENS” and CENTER_NAME = “BCM” and CENTER_PROJECT = “RHIAY”.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.The Internation SNP Map Working Group A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 3.The International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 5.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nature Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nature Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 8.Barnes C, et al. A robust statistical method for case-control association testing with copy number variation. Nature Genet. 2008;40:1245–1252. doi: 10.1038/ng.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teo YY, et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics. 2007;23:2741–2746. doi: 10.1093/bioinformatics/btm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Internatinal HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, et al. SNPdetector: a software tool for sensitive and accurate SNP detection. PLOS Comput. Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. doi:10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu. Rev. Genomics Hum. Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nature Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Heel DA, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nature Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pe’er I, et al. Biases and reconciliation in estimates of linkage disequilibrium in the human genome. Am. J. Hum. Genet. 2006;78:588–603. doi: 10.1086/502803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossman SR, et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327:883–886. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- 20.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 21.Lamason RL, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 22.Akey JM. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 2009;19:711–722. doi: 10.1101/gr.086652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson CS, et al. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005;15:1553–1565. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu J, et al. A genome scan for positive selection in thoroughbred horses. PLoS ONE. 2009;4:e5767. doi: 10.1371/journal.pone.0005767. doi:10.1371/journal.pone.0005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Abecasis GR. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- 27.Colella S, et al. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.