Abstract
HS (heparan sulfate) has been shown to be an important mediator of Plasmodium sporozoite homing and invasion of the liver, but the role of this glycosaminoglycan in mosquito vector host–sporozoite interactions is unknown. We have biochemically characterized the function of AgOXT1 (Anopheles gambiae peptide-O-xylosyltransferase 1) and confirmed that AgOXT1 can modify peptides representing model HS and chondroitin sulfate proteoglycans in vitro. Moreover, we also demonstrated that the mosquito salivary gland basal lamina proteoglycans are modified by HS. We used RNA interference-mediated knockdown of HS biosynthesis in A. gambiae salivary glands to determine whether Plasmodium falciparum sporozoites that are released from mosquito midgut oocysts use salivary gland HS as a receptor for tissue invasion. Our results suggest that salivary gland basal lamina HS glycosaminoglycans only partially mediate midgut sporozoite invasion of this tissue, and that in the absence of HS, the presence of other surface co-receptors is sufficient to facilitate parasite entry.
Keywords: Anopheles, cell invasion, glycobiology, glycos-aminoglycan (GAG), heparan sulfate (HS), malaria
Abbreviations: AgLDH, Anopheles gambiaeL-lactate dehydrogenase; AgOXT1, Anopheles gambiae peptide-O-xylosyltransferase 1; AMPD, 2-amino-2-methyl-1,3-propanediol; CS, chondroitin sulfate; CSP, circumsporozoite protein; CSPG, CS proteoglycan; DAPI, 4′,6-diamidino-2-phenylindole; dsRNA, double-stranded RNA; dsAgOXT1, AgOXT1 dsRNA; dsGFP, GFP dsRNA; GAG, glycosaminoglycan; GFP, green fluorescent protein; GlcNS, N-sulfated N-acetylglucosamine; HS, heparan sulfate; HSGAG, HS glycosaminoglycan; HSPG, HS proteoglycan; IdoA2S, L-iduronic acid 2-O-sulfate; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; MS/MS, tandem MS; rAgOXT1, recombinant AgOXT1; RNAi, RNA interference; RP-HPLC, reverse-phase HPLC; RT, reverse transcription; scFv, single-chain variable fragment; TRAP, thrombospondin-related adhesion protein; TSR, thrombospondin type 1 repeat domain; VSV, vesicular stomatitis virus; XT-I, xylosyltransferase I; Xyl, xylose
INTRODUCTION
Malaria transmission entails development of the Plasmodium parasite in the mosquito. There are two major tissues that the parasite must traverse during its development in the mos-quito. The first obligatory step requires recognition and attachment of the Plasmodium ookinete stage to lumenal membrane ligands of the mosquito midgut epithelium followed by active cell invasion [1]. We have previously identified the critical interaction between the ookinete and CS (chondroitin sulfate) GAGs (glycosaminoglycans) on the mosquito midgut lumenal proteoglycans. Disruption of AgOXT1 (Anopheles gambiae peptide-O-xylosyltransferase 1) activity in the midgut, which blocks GAG biosynthesis, resulted in 95% inhibition of ookinete invasion of the midgut. These data confirmed the hypothesized role of these anionic polysaccharides in the initial adhesion of the ookinete, directly preceding invasion and establishment of oocysts on the midgut basal wall [2].
The second fundamental step in the sporogonic cycle is the transmission of sporozoites that have developed in the oocyst beneath the midgut basal lamina into a new vertebrate host. To achieve this, sporozoites must first reach, recognize and attach to the salivary glands of the mosquito [3]. Once in the salivary glands, the sporozoites can now be delivered to a new vertebrate host when the mosquito takes another bloodmeal.
There are two major surface molecules that are conserved on both midgut (oocyst-derived) sporozoites and salivary gland sporozoites: CSP (circumsporozoite protein) and TRAP (thrombospondin-related adhesion protein). It has been shown in the vertebrate system that CSP [4] and TRAP are involved in hepatocyte homing and attachment in the vertebrate host [5]. CSP recognizes HSPGs [HS (heparan sulfate) proteoglycans], which extend out through the endothelium from the basolateral surfaces of hepatocytes [6–9]. TRAP also exhibits binding affinity for heparin, HS, dextran and fucoidin, but not for bovine cartilage-derived CS [10–12]. More importantly, both CSP and TRAP have been implicated in salivary gland invasion by midgut sporozoites [13,14]. However, to date, it has not been shown that GAGs are involved in midgut sporozoite interaction with the salivary glands.
It has been hypothesized that although the protein backbone of HSPGs may differ between the basal membrane of hepatocytes and mosquito salivary glands, the GAG chains may be structurally conserved, thus conferring sporozoite tropism for both tissues in two evolutionarily divergent hosts. Previously, it was shown that enzymatic treatment of Anopheles stephensi mosquito salivary glands to release GAG disaccharides identified a HS moiety that is structurally similar to those found on liver HSPGs [15]. Bolstered by this finding, we subsequently hypothesize that the parasite switches ‘preference’ from CSPGs (CS proteoglycans) [2] to HSPGs as binding ligands, probably as a function of the availability of these molecules along the appropriate ‘face’ of polarized epithelia [16] as is found in both the midgut and salivary gland tissues. Ookinetes invade through the apical domain of the midgut, where CSPGs predominate [2], whereas sporozoites can only interact with the basal face of the salivary gland HSPGs (since sporozoites invade from the haemocoel of the mosquito). To test this, we first had to demonstrate that (i) a salivary gland polypeptide-OXT from the malaria vector mosquito Anopheles gambiae can initiate both heparan and chondroitin GAG biosynthesis, and (ii) that HSPGs are localized to the basal lamina of mosquito salivary glands. We report the verification of the enzymatic function of AgOXT1, the successful knockdown of HS biosynthesis in A. gambiae salivary glands by RNAi (RNA interference), which in turn permitted the analysis of the role of HSGAGs (HS glycosaminoglycans) in Plasmodium falciparum sporozoite invasion of the salivary gland.
EXPERIMENTAL
Biological materials
Anopheles gambiae (strain Keele) were reared following standard protocols as described elsewhere [2], with the following modifications. Larvae were raised at a lower density (100–150/pan) to allow for the generation of larger female mosquitoes (with estimated wing length measurements >3 mm and relatively larger body mass). To ensure mosquito survival success for the salivary gland invasion inhibition assay (described below), female mosquitoes of similar metabolic and physiological age were used. Only 5–6-day-old females that met the above size requirements were used for these experiments.
Cloning of xylosyltransferase cDNA fragment and expression in yeast
A cDNA fragment encoding the predicted catalytic domain of AgOXT1 (VectorBase accession code AGAP005811; http://www.vectorbase.org) was isolated by RT (reverse transcription)–PCR using the primers AgOXT1/1/EcoRI (5′-CGGAATTCGACTTTGTGCCGCCGTG-3′) and AgOXT1/2/XbaI (5′-GCTCTAGAACTAGTTCGACGTTACCGATTC-3′) from midgut cDNA and Expand polymerase mix (Roche) in the presence of a ‘GC-rich’ polymerase buffer additive. The ~2.5 kb band was excised from an agarose gel, extracted and ligated into the pGEM-T vector (Promega). The plasmid DNA from one clone was digested with EcoRI and XbaI, and the ~2.5 kb band was excised and extracted prior to ligation into the pPICZαC vector cut with the same enzymes. Plasmid DNA from a selected positive Escherichia coli clone was subjected to DNA sequencing and used to transform Pichia pastoris GS115 cells using Zeocin™ (phleomycin) selection. For selected yeast clones, expression was induced using methanol as described previously [17] and xylosyltransferase assays were performed using the crude supernatant. One clone was used for a larger-scale culture and the PMSF-treated culture supernatant was subjected to ammonium sulfate precipitation. The 40% pellet was dissolved in a 10 mM Hepes, pH 8, buffer (containing 1 mM PMSF as a protease inhibitor and 0.01% sodium azide) and was found to contain the maximal activity; this preparation was desalted using an Ultrafree® centrifugal filtration device and was the source of enzyme for the subsequent enzymatic characterization. The enzyme could be stored at 4 °C for several weeks, retaining at least 50% of its activity after 3 months.
Enzymatic characterization
Xylosyltransferase assays were performed basically as described previously [17–19], routinely using a final concentration of 1 mM syndecan peptide (Syn; DDDSIEGSGGR), 2 mM UDP-Xyl (xylose), 80 mM Hepes, pH 8, and 10 mM MnCl2; one-third of the volume was accounted for by the enzyme preparation. The incubations were normally performed at 30 °C for 2 h in PCR tubes (either 2.5 μl or 10 μl total volume) prior to heat inactivation at 95 °C for 10 min; overnight incubations with three other peptides [Bik (bikunin), Syn2 and Perl (perlecan)] were also performed. For determination of the pH optimum, AMPD (2-amino-2-methyl-1,3-propanediol) buffers were used; for examination of the cation dependency, MnCl2 was replaced with other metal chlorides or by EDTA. The conversion into product was examined by RP-HPLC (reverse-phase HPLC) or by MALDI–TOF-MS (matrix-assisted laser-desorption ionization–time-of-flight MS). For RP-HPLC, a Nucleosil C18 column was used with a gradient of acetonitrile (9–12% acetonitrile from 5–15 min); peptides were detected by measuring A214 and product/substrate ratios were calculated based on integrated peak areas. MALDI–TOF-MS and MS/MS (tandem MS) were performed with a Bruker Ultraflex TOF/TOF instrument with α-cyanohydroxycinnaminic acid as matrix.
Semi-quantitative RT–PCR
The AgOXT1 primers 1557F (5′-CAGCACAAGAAGCTGTTCTTTGGC-3′) and 2337R (5′-AGCTTCCGTGACGAGAAATCGTGT-3′) were used to amplify a 780 bp product corresponding to the C-terminus of AgOXT1 from our A. gambiae midgut cDNA library. The PCR product was cloned into the pJET1.2/blunt vector (Fermentas). Cloned PCR products were confirmed by DNA sequencing. For RT–PCR, 50 ng of total salivary gland RNA (n=8–12 female salivary gland pairs) collected from each time point after injection was reverse-transcribed using RevertAid™ M-MuLV Reverse Transcriptase (Fermentas) following standard protocols. AgOXT1 transcript abundance was determined by using the primers described above. The amplified A. gambiae ribosomal protein gene (AgS7) product was used as a loading control (AgS7F, 5′-GGCGATCATCATCTACTTGC-3′; and AgS7R, 5′-GTAGCTGCTGCAAACTTCGC-3′). Amplification using KlenTaq LA DNA polymerase (Sigma) was performed as follows: 95 °C for 1 min, 94 °C for 30 s and 68 °C for 3 min, with a final extension at 68 °C for 3 min (30 cycles).
RNAi
AgOXT1 dsRNA (double-stranded RNA) was produced by using the primers described above with the T7 polymerase promoter sequence, 5′-TAATACGACTCACTATAGGG-3′, appended to the 5′ termini of each primer and transcribed in vitro using the MEGAscript® RNAi kit (Ambion) as per the manufacturer's protocols. The dsRNA solution was diluted to 6 μg/μl in sterile injection buffer (10 mM Tris/HCl, pH 7.4). A. gambiae females were anaesthetized by cold treatment and inoculated intrathoracically using a mineral oil back-filled borosilicate capillary needle and a Nanoject II instrument (Drummond Scientific) with 400 ng of dsRNA or control dsGFP [GFP (green fluorescent protein) dsRNA]; the injection of dsGFP has been consistently shown to be the best control for injury imparted by dsRNA injection into the mosquito haemocoel via a needle [2,20]. Initially, uninfected mosquitoes were kept for 2–5 days to collect RNA and protein from salivary glands at the prescribed time points. Replicate reproducible knockdown of transcript would provide confidence that during the course of the salivary gland invasion inhibition assay (see below), AgOXT1 message would be likewise repressed. Note that there is no evidence which demonstrates that infection of the mosquito with Plasmodium oocysts results in the loss of efficacy of RNAi-mediated silencing.
SDS/PAGE and semi-quantitative Western blot analysis
Mosquito salivary gland lysates (n=20 female salivary gland pairs) were fractionated on a 10% Tris/glycine gel and transferred to nitrocellulose membranes. Transblots were probed with RB4Ea12 anti-HS scFv (single-chain variable fragment) antibodies (1:25) overnight at 4 °C [21]. For RB4Ea12 detection, a mouse anti-VSV (vesicular stomatitis virus) secondary antibody (1:100; Sigma) was used followed by either Alexa Fluor® 488 (Invitrogen)-conjugated anti-mouse (1: 1000) or an IRDye 800CW (LI-COR) anti-mouse tertiary antibody (1:40000) staining.
We also cloned a 456 bp fragment of the gene encoding AgLDH (A. gambiae L-lactate dehydrogenase; VectorBase accession code AGAP004880) from midgut cDNA using the following primers (AgLDH forward, 5′-CACCATGTCTGAAGTGAAAGCTAAGCTG-3′; and AgLDH reverse, 5′-GACGACCAACAGAATGCAGTCCGGGCT-3′) and expressed the protein using the vector pBAD202 (Invitrogen) in E. coli. Recombinant soluble protein was purified using Ni–agarose beads (Qiagen). Rabbit polyclonal antiserum was generated against the purified recombinant protein (Washington Biotechnology). AgLDH (VectorBase accession code AGAP004880) is expressed in sugar-fed midguts and is predicted to give rise to three splice variants (VectorBase accession codes AGAP004880-RA, -RB and -RC) of approximately 1452 bp. We only cloned and expressed the protein product of AGAP004880-RA. Anti-AgLDH antibodies (1:200) as loading control probes were used to stain the blot overnight at 4 °C and then detected by IRDye 680 goat anti-rabbit antibody (1:40000). Anti-AgLDH antibodies detected a single protein band with the predicted molecular mass of ~35 kDa for all three gene products (results not shown). Transblots were imaged using an Odyssey® Infrared Imaging System (LI-COR).
Immunofluorescence analysis
Whole salivary gland pairs from female A. gambiae were dissected in PBS with protease inhibitor cocktail (Sigma) before being fixed with 4% paraformaldehyde for 30 min on ice. Fixed glands were blocked with 3% BSA/PBS overnight at 4 °C. As was performed for the Western blots, RB4Ea12 scFv antibodies (1:25) were incubated with tissues at 4 °C overnight. After extensive washing, scFv staining was detected by successive incubations with mouse anti-VSV antibody (1:100; Sigma), a second wash step and then the addition of goat anti-mouse Alexa Fluor® 488- or Alexa Fluor® 594-conjugated tertiary antibodies. The slides were mounted with SlowFade® reagent containing DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen), visualized by microscopy (×400) and image-captured by SPOT RT software (v. 3.2).
Salivary gland invasion inhibition assay
P. falciparum gametocyte cultures from isolate NF54 (Sanaria) were maintained at the Johns Hopkins Malaria Research Institute Parasite Core Facility. Cultures were harvested 15–17 days after initiation. The culture was washed and raised in normal human serum (Interstate Blood Bank) plus human red blood cells at 0.3% gametocytaemia in a 45% haematocrit. Infective blood was delivered directly into glass water-jacketed membrane feeders warmed to 38 °C. Robust female mosquitoes (5–6 daysold) were allowed to blood feed for 20 min. Unfed mosquitoes were removed from the cups. At 9 days after being fed a blood meal containing P. falciparum gametocytes, infected A. gambiae females that exhibited egg development, as a proxy measure for feeding to repletion, were anaesthetized by cold treatment and inoculated intrathoracically with 400 ng of dsAgOXT1 (AgOXT1 dsRNA) or control dsGFP (see above). Mosquitoes were maintained for an additional 3 days, at which point (day 12 post-infection) surviving mosquitoes were knocked down by cold treatment to collect infected salivary glands. Salivary gland invasion by P. falciparum sporozoites begins at approximately 10–14 days post-midgut infection. Salivary glands were harvested at day 12 post-infection, which essentially captures all sporozoite invasion events that had occurred during a 48 h time period. A minimum of 19–20 salivary gland pairs were dissected for control (dsGFP) and treatment (dsAgOXT1) groups and placed into tubes containing PBS. Each salivary gland pair was crushed with a pestle in a microcentrifuge tube to release the sporozoites. Tissue material was pelleted at 6000 g for 2 min, the supernatant (20 μl) was collected and the sporozoites were counted using a haemocytometer. The isolation was repeated and the supernatants pooled. Two independent experiments (19–24 salivary gland pairs/assay) were performed. The raw data for infection intensity and prevalence in controls were analysed for equality of variance (P=0.3049), demonstrating that these two biological replicates were comparable. The D'Agostino–Pearson omnibus normality test was also performed to determine the most appropriate statistical analysis. Non-parametric statistical analysis was used to evaluate the difference in median sporozoite infection between experimental and control groups (Mann–Whitney U test, one-tailed, α=0.05) using StatView 5.0 software (SAS Institute).
RESULTS
Enzymatic properties of AgOXT1
We have confirmed previously that there is only a single AgOXT1 gene that is transcribed in the A. gambiae genome [2] and, as such, the underlying assumption is that this single peptide OXT is involved in both CS and HS biosynthesis. This would imply that AgOXT1 can modify serine residues within predictable sequons on CSPGs and HSPGs. It was imperative that we confirmed the function of AgOXT1 before pursuing knockdown approaches. If this gene did not modify serine residues on known HSPG acceptor peptides, then it would clearly be an invalid target gene to knock down for our subsequent experiments.
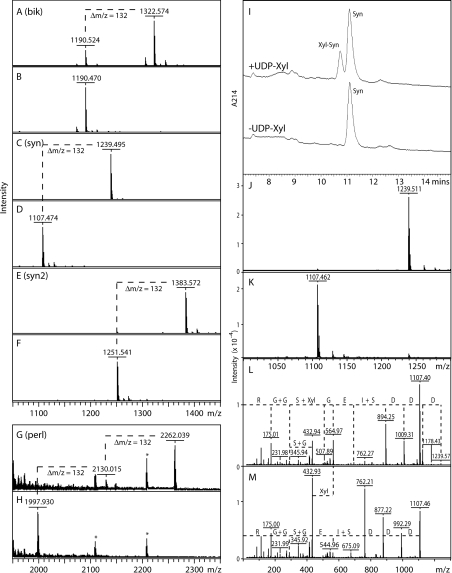
We verified the biochemical function of AgOXT1 in vitro using a truncated form of the enzyme lacking the cytosolic, transmembrane and putative stem regions, but retaining the conserved P-X-C-D/E motif in a Pichia secreted expression system; Pichia cultures have been shown previously to secrete no background xylosyltransferase activity [17,19], which also reflects the restriction of OXT genes to metazoans. Various peptide acceptor substrates were used in the present study: a peptide based on a part of the sequence of bikunin (Bik, QEEEGSGGGGQR), two variants of a sequence based on amino acids 55–65 from Drosophila syndecan (Syn, DDDSIEGSGGR, and the artificial variant Syn2, DDDSIEGSGSGGR, which has two Ser-Gly motifs) and a sequence based on part of human perlecan (Perl, DSISGDDLGSGDLGSGDFQR). Bikunin is a Kunitz-type serine protease inhibitor in vertebrate serum and urine modified by a single CS chain [22], whereas syndecans are modified by HS [23]; perlecans carry both CS and HS chains [24]. All substrates harbour an acidic region as well as a Ser-Gly sequence, which have been described previously for CS and HS modification sites [25,26]. We observed that the rAgOXT1 (recombinant AgOXT1) transferred a single Xyl residue (indicated by a Δm/z of 132) to the Bik, Syn and Syn2 peptides (Figures 1A–1F); the RP-HPLC-based assay and analysis of the relevant collected fractions verifies that Syn accepts one Xyl residue (Figures 1I–1K). The observation that Syn2 is only singly xylosylated is in line with previous assays examining transferase activity of recombinant Caenorhabditis SQV-6, Drosophila OXT and human XT-I (xylosyltransferase I) with Syn2 as an acceptor peptide [19]. Furthermore, in the case of the Perl peptide, rAgOXT1 was observed to transfer two Xyl residues to the acceptor peptide backbone (Figure 1G). Moreover, we were able to confirm that rAgOXT1 modified the second serine residue in the DDDSIEGSGGR peptide only (Figure 1L). These results indicate that AgOXT1 can indeed modify both CS and HS proteoglycan core proteins in the mosquito.
Figure 1. MALDI–TOF-MS and HPLC-based assays for xylosyltransferase activity.
(A–H) Incubation of selected peptides with the recombinant enzyme preparation was performed overnight in the presence (A,C,E,G) or absence (B,D,F,H) of UDP-Xyl as enzyme donor. In all cases, the xylosyltransferase transferred Xyl (indicated by Δm/z=132; one in the case of the Bik, Syn and Syn2 peptides, and two in the case of the Perl peptide); asterisks in (G and H) indicate contaminating peptides originating from the culture medium. (I) A typical RP-HPLC assay is depicted showing the UDP-Xyl-dependent formation over 2 h by AgOXT1 of a product with a slightly earlier elution time; product (Xyl-Syn) and substrate (Syn) peaks were collected and subject to MALDI–TOF-MS (J, K) to show the respective presence of species of m/z 1239 and 1107. The comparative MALDI–TOF-MS/MS data for the two peaks (L, M) verify the transfer of Xyl to the second serine residue of the DDDSIEGSGGR sequence of the Syn peptide (fragments of m/z 564 in D and 432 in both D and E); loss of Xyl directly from the parent ion in D is also apparent. The Δm/z values between peaks correspond to either single amino acids (D, E, G, R), dipeptides (G+G, I+S, S+G) or xylosylserine (S+Xyl).
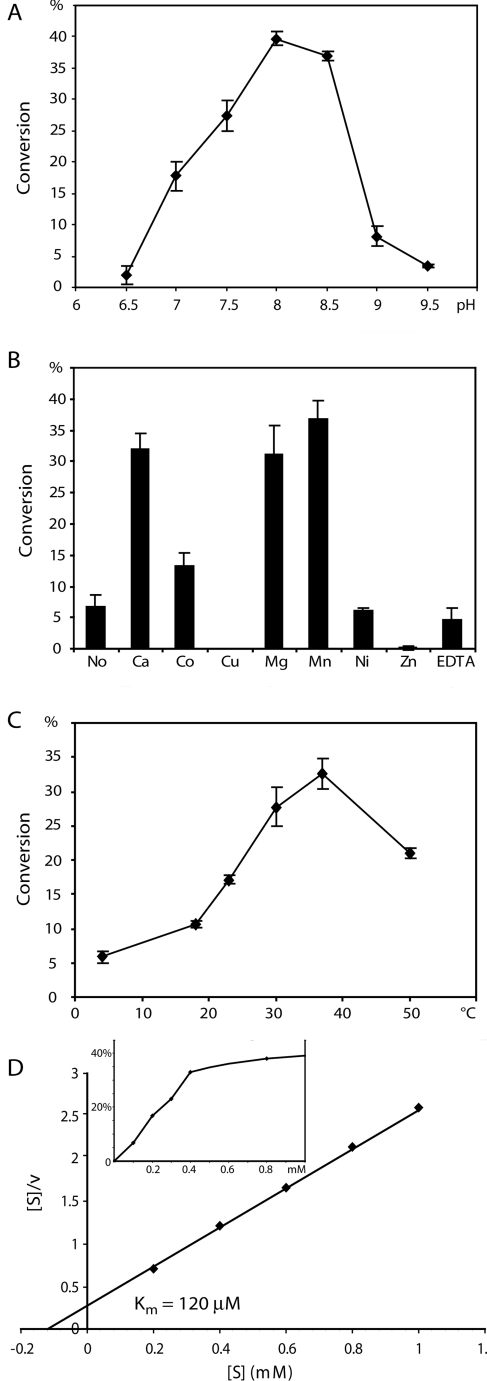
We also determined the activity of rAgOXT1 by RP-HPLC-based assays (Figure 2) and observed a pH optimum of 8, a temperature optimum of 37 °C and a cation dependence of the enzyme that is similar to that for recombinant forms of human XT-I and Drosophila OXT. Using the temperature-dependence data, an Arrhenius plot suggested an activation energy, Ea, of ~39 kJ/mol (~9 kcal/mol). The enzyme is still active in the absence of added cation or in the presence of EDTA, but is significantly activated by Ca(II), Mg(II) and Mn(II) ions, and inhibited by Cu(II) and Zn(II). The Km value for the UDP-Xyl donor is estimated to be 120 μM and is also consistent with values of 100–250 μM for the human and Drosophila homologues [18,19]. We know that mammalian OXT is an intracellular Golgi enzyme [27] and this is likely to be a conserved localization for mosquito OXTs. Although the pH optimum that we observed is more in the range described for the resting state pH for the mosquito midgut [28] and salivary glands of other haematophagic insects [29], the optimum pH described for our in vitro studies may not necessarily be relevant in the in vivo context. Similarly, we observed that rAgOXT1 had greater activity at higher temperatures beyond the recorded ambient temperatures in malaria-endemic regions (28–32 °C). However, the conversion rate data points for temperatures >30 °C overlap, suggesting that this difference is not statistically significant. We speculate that at 30–32 °C AgOXT1 has sufficient glycosylation activity in the salivary glands.
Figure 2. Enzymatic characterization of AgOXT1.
RP-HPLC-based assays were used to determine the percentage conversion over 2 h into product under various conditions to assess (A) optimum pH, (B) cation dependence, (C) optimum temperature and (D) Km value for the donor substrate UDP-Xyl (Hanes–Woolf plot); the inset in (D) shows the kinetic curve plotting UDP-Xyl concentration against percentage conversion.
RNAi-mediated repression of HS biosynthesis
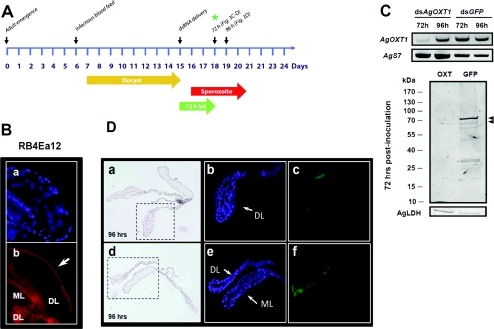
An experimental timeline for RNAi-mediated repression of HS biosynthesis in A. gambiae salivary glands and its effects on P. falciparum sporozoite invasion is outlined in Figure 3(A). The RB4Ea12 scFv antibody stains non-permeabilized salivary glands along the basal surface of the tissue with a more prominent recognition of the distal lateral lobes (Figure 3B, a and b). Note that mouse anti-VSV antibody and goat anti-mouse Alexa Fluor®-labelled antibody did not stain the salivary glands (results not shown). The injection of 400 ng of dsAgOXT1 into adult A. gambiae significantly decreased transcript levels at 72 h, with a strong rebound in transcript abundance at 96 h (Figure 3C, upper panel). RNAi-mediated interference is generally regarded as a leaky method, in that individual mosquitoes that are injected with dsRNA will have varying levels of knockdown, and is known to be particularly challenging when targeting mosquito salivary glands [30]. However, in these small pools of salivary gland pairs from 8–12 mosquitoes, we observed a pronounced reduction in AgOXT1 message at 72 h. We also observed the reduction in HSPG staining in representative experiments using Western blotting (Figure 3C, lower panel) at 72 h post-injection with dsAgOXT1, demonstrating that AgOXT1 activity is repressed, resulting in the loss of GAG modification of proteoglycans that are present on the basal lamina of salivary glands. The loading control anti-AgLDH antibodies recognized a single band of ~35 kDa and of equal intensity across both samples. These results highlight the specificity of the scFv antibody used in the present study, given that in the absence of HS, RB4Ea12 scFv does not bind and cannot be detected by the labelled secondary antibody. Noting the rebound in transcript at 96 h, we further investigated whether repression of HS biosynthesis along the basal lamina of the salivary gland distal lateral lobes was lost at 96 h post-dsRNA delivery (Figure 3D). We observed that the rebound in mRNA observed by semi-quantitative RT–PCR correlates with recovery of HS biosynthesis, as detected by RB4Ea12 scFv staining of the lateral lobes of the salivary glands. Moreover, the success of RNAi-mediated interference for molecules that are already present on the tissue of interest is contingent on the turnover rate of the targeted protein and the physiological state of the mosquito itself. We suspect that the scFv staining observed for the distal lobes of the salivary glands from dsAgOXT1-treated mosquitoes (Figure 3D, a–c) represent basal lamina HSPGs with a lower rate of turnover. This probably corresponds to the barely visible protein bands of ~70 kDa that were observed by Western blotting (Figure 3C, lower panel, arrowheads) and detected using a highly sensitive near-infrared imaging system. Taken together, the results suggest that repression of AgOXT1 activity is lost at 96 h. Therefore, to determine the role of HS on salivary gland invasion by midgut sporozoites, we had to ensure that salivary glands were harvested no later than 72 h post-dsRNA inoculation (as depicted in Figure 3A).
Figure 3. RNAi-mediated knockdown of HS biosynthesis in salivary glands affects P. falciparum sporozoite invasion.
(A) Experimental time line for RNAi experiments shown in Figure 3(C)–3(D) and the salivary gland invasion inhibition assay in Table 1. Each time point represents steps in the experimental process and indicates the age of the mosquito at the time of manipulation, the point of delivery of the P. falciparum infected bloodmeal, and the point of salivary gland harvest, which is 4 days post-dsRNA injection (day 12 post-infectious blood feeding). The gold arrow highlights the oocyst development period on the mosquito midgut. The red arrow highlights the period when sporozoites are escaping the ruptured oocyst and begin invasion of the salivary glands. Note that some oocysts will rupture earlier given the asynchronous nature of oocyst development in the mosquito. The green arrow highlights the 72 h window wherein AgOXT1 mRNA and HS biosynthesis is knocked down (kd). (B) Immunofluorescence staining of the basal lamina of a fixed non-permeabilized A. gambiae salivary gland. DAPI staining identifies nuclei in the tissue and appears blue (panel a). The arrow highlights the surface staining of the medial (ML) and distal lateral (DL) lobes of salivary glands using the RB4Ea12 scFv antibody, which recognizes 6-O-sulfated GlcNS/NAc-containing disaccharides, as detected by mouse anti-VSV secondary antibody and goat-anti mouse Alexa Fluor® 594-conjugated tertiary antibody and appears red (panel b). Close-up images using ×400 magnification. (C) RNAi-mediated knockdown of AgOXT1 transcript in salivary glands measured by semi-quantitative RT–PCR (upper panel) as well as a representative Western blot using RB4Ea12 scFv antibody of HSPGs that remain present on Anopheles gambiae salivary glands at 72 h post-dsRNA delivery. The wild-type salivary gland HS banding profile is represented by the dsGFP control group. Following dsAgOXT1 injection, the HS staining is reduced (lower panel). Antibodies against AgLDH (molecular mass ~35 kDa) were used as a protein loading control. Images were acquired using the Odyssey® near-infrared system. Arrowheads indicate two faint bands that are barely detectable in salivary glands from dsAgOXT1-treated mosquitoes (molecular mass ~70 kDa). (D) Bright-field images and immunofluorescence staining of salivary glands, confirming that repression of HS biosynthesis is lost at 96 h, which correlated with the mRNA rebound observed at 96 h post-dsAgOXT1 inoculation (panels a–c) as compared with dsGFP controls (panels d–f). RB4Ea12 scFv was detected by Alexa Fluor® 488-conjugated secondary antibody, and appears green. The images for bright-field and fluorescence were acquired at ×200 and ×400 magnification respectively, and are representative images of salivary gland matched pairs for mosquitoes from two biological cohorts. Image acquisition and analysis was performed using exactly the same parameters for treatment and control glands. DAPI staining identifies nuclei in the tissue and appears blue. It is clear from the results that the window of opportunity to assess the role of HS in sporozoite invasion of the salivary glands is within the first 72 h following dsRNA delivery (indicated as a green asterisk on the timeline shown in (A). Salivary gland harvest at this time point should capture only the first 2–3 days of sporozoite tissue invasion events.
Salivary gland sporozoite infection intensity is partly reduced by RNAi-mediated silencing of HS biosynthesis
Since we observed that the raw data are skewed (positive kurtosis and non-Gaussian distribution as determined by a normality test), we used the Mann–Whitney U test to compare the median number of sporozoites that invaded the salivary gland in treatment groups compared with control groups for the two biological cohorts (α=0.05). As mentioned previously, sporozoites begin to invade the salivary glands at ~10 days post-infectious blood feeding. Salivary glands were harvested at day 12 post-infection, which captures all sporozoite invasion events that had occurred during a 48 h time period, within the 72 h window for confirmed suppression of AgOXT1 mRNA. The observed trend was towards a reduction (59–79%) in the total sporozoite infection intensity in dsAgOXT1-treated mosquitoes for the two independent biological experiments (Table 1). Of the two biological cohorts, only the second experiment exhibited a statistically significant and patent reduction in sporozoite infection intensity among dsAgOXT1-injected mosquitoes. However, taken together, both experimental replicates do suggest a trend towards a reduction in sporozoite number in the absence of HS biosynthesis during the first 72 h following dsRNA delivery. Moreover, the reduction in infection prevalence between control salivary gland pairs (68–75%) and treatment (50–65%) was also marginal, indicating that although the overall number of sporozoites that invaded the salivary glands was inhibited, a subset of sporozoites was nonetheless successful in invading the tissue. A reasonable expectation is that if HSGAGs play a major role in sporozoite invasion, then not only infection intensity but also infection prevalence in dsAgOXT1-treated mosquitoes should be significantly lower. These results suggest that HS is likely to be only one of several macromolecules that contribute to or mediate sporozoite interaction with the basal lamina of mosquito salivary glands, but its absence alone is not sufficient to block parasite invasion.
Table 1. RNAi-mediated silencing of AgOXT1 activity resulted in a partial reduction in salivary gland sporozoite infection intensity.
The number of sporozoites was determined for each salivary gland pair per mosquito from each treatment and control group from two biological experiments. Independent cohorts of A. gambiae mosquitoes were infected with P. falciparum (NF54) and then inoculated with dsGFP or dsAgOXT1 at day 9 and harvested at day 12 (corresponding to the 72 h time point following dsRNA delivery and the absence of HS biosynthesis; see the time line in Figure 3A and the Western blot in Figure 3C, lower panel). The raw data for the dsGFP control cohorts were analysed for equality of variance (F test, P=0.3049) to demonstrate that these two biological cohorts had similar parasite infection levels in terms of midgut oocysts at day 8 and salivary gland sporozoites at day 12. Statistical significance was determined by Mann–Whitney U test analysis of median sporozoite numbers for control and treatment groups (α=0.05).
| Group | n | Median number of sporozoites per salivary gland pair (range) | Inhibition (%) | Prevalence of infection | P value | |
|---|---|---|---|---|---|---|
| Experiment 1 | dsGFP | 20 | 92000 (0–600000) | − | 75% | − |
| dsAgOXT1 | 20 | 38000 (0–250000) | 59 | 65% | 0.172 | |
| Experiment 2 | dsGFP | 19 | 83421 (0–210000) | − | 68% | − |
| dsAgOXT1 | 24 | 17917 (0–90000) | 79 | 50% | 0.016 |
DISCUSSION
Both Plasmodium CSP and TRAP contain an adhesive TSR (thrombospondin type 1 repeat domain), which is commonly found in extracellular proteins that are involved in immunity, cell adhesion and neuronal development [12,31]. A 20-residue motif called region II of the CSP TSR has been implicated as the probable ligand in the adhesion of sporozoites to liver HSPGs [32–34]. A similar function has been postulated for the TSR domain of TRAP [35,36]. However, TRAP contains a second adhesive motif, the so-called A domain, a member of the vWFA (von Willebrand factor type A) protein family, which is necessary for sporozoite gliding motility, target host cell attachment and invasion [36]. Binding studies with the recombinant A domain and TSR molecules of TRAP [10,12] suggested that both domains interact with GAGs on tissue surface proteoglycans, most probably via clusters of basic and hydrophobic residues.
Following this line of evidence, midgut sporozoites, which express both CSP and TRAP, have been hypothesized to bind to anionic polysaccharide ligands present along the basal lamina of salivary glands [15]. Previously, it was shown biochemically that A. stephensi salivary gland HS chains actually contained the critical trisulfated disaccharide sequence GlcNS6S (N-sulfated N-acetylglucosamine)-IdoA2S (L-iduronic acid 2-O-sulfate), which is also found in human liver HS [15]. This trisulfated HS disaccharide is the liver receptor for apolipoprotein E and is also hypothesized to be the binding ligand of CSP found on the surface of the Plasmodium sporozoite [14,15]. Our present study further confirms the presence of HSGAGs on the basal lamina of the medial and lateral lobes of A. gambiae salivary glands. The RB4Ea12 scFv epitope was originally described as GlcNS6S-GlcUA-GlcNS3S6S-IdoUA-GlcNS[+/−3S]6S (where GlcUA is glucuronic acid) with an absolute minimal requirement for 6-O sulfation, which would suggest that the protein bands that were observed by immunoblotting probably represent only a subset of salivary gland HSPGs [21,37–39]. Moreover, in these initial studies the presence of IdoA2S can partly inhibit binding of RB4Ea12 to HS [21,37–39]. However, recent data conflict with this initial assessment since it appears that a certain degree of 2-O sulfation is apparently well tolerated in the presence of high 6-O sulfation [40] (T.H. van Kuppevelt, unpublished work). In addition, the degree of polymorphism (or monosaccharide sequence diversity) or density of IdoA2S in a given HS octasaccharide sequence [21] probably have differential inhibitory effects on RB4Ea12 reactivity. In light of these caveats, the HSPGs that are modified with GlcNS6S-IdoA2S HS chains, which are hypothesized to be a conserved ligand for sporozoites in both human and mosquito hosts [15], are likely to be represented among the well-defined as well as the fainter bands observed in the immunoblot (Figure 3C, dsGFP control lane). It should be noted that the proteoglycans themselves remain unaffected by our RNAi strategy. If these proteoglycans can be cycled to the salivary gland surface lacking the HSGAG modifications, then the polypeptide core itself may contribute unidentified binding epitopes for the sporozoite, thus partly explaining our observations. We are presently pursuing HS epitope analysis for RB4Ea12 as well as proteomic analysis of the A. gambiae salivary gland basal lamina to identify the complete repertoire of HS modified proteoglycans in this tissue.
It is likely that HSPGs may not be the only molecules that are important for salivary gland invasion by midgut sporozoites [41]. In a multi-step process, sporozoites invade both the medial lobes and the distal portion of the lateral lobes of the salivary glands [42,43]. However, anecdotal evidence from several laboratories suggests that, by and large, sporozoites are more commonly found only in the distal lateral lobes. Although HSPGs are seemingly ubiquitous along the basal membrane of salivary glands, several basal molecules have also been shown to be involved in sporozoite invasion. A monoclonal antibody recognizing the 100 kDa A. gambiae salivary gland protein saglin was capable of effectively inhibiting (~70–100%) sporozoite invasion [44–46]. The role of saglin in salivary gland invasion is further supported by RNAi experiments, which result in up to 98% inhibition of salivary gland invasion by sporozoites as compared with controls [44]. Saglin does not contain any predictable GAG modification sites and does not appear to be heavily glycosylated with either N- or O-glycans. Moreover, it has been determined that TRAP is the cognate receptor on the sporozoite surface that binds to saglin, since recombinant TRAP protein fragments have been shown to bind both mosquito salivary glands and recombinant saglin in vitro [44]. These results suggest that the interaction of TRAP and saglin significantly influences salivary gland invasion success.
Although we were able to reduce sporozoite infection of the salivary glands by inhibiting HS biosynthesis, it is likely that the unaffected presence of saglin (or another protein) on the dsAgOXT1-treated salivary glands still supported the successful invasion of a subset of sporozoites. Conversely, it cannot be ruled out that salivary gland HSPG core proteins, which remain unaffected in the saglin RNAi experiments, still contribute to the initial adhesion of the sporozoite to the glands, since it has not been shown whether sporozoites can still adhere to the glands, but not necessarily invade them, given the technical hurdles in performing such an assay.
The involvement of several molecules in a multi-step process was described previously for ookinete invasion of the mosquito midgut apical microvilli [47]. Although our results are consistent with a multi-factorial model for tissue invasion, the inherent biological limitations of the study (e.g. the absence of a salivary gland cell line) necessitate caution to avoid overextending the interpretation. Since we cannot measure directly any modulation on invasion kinetics as a result of the loss of HS in our assay, we cannot definitively conclude that HS is not involved, since it could very well facilitate sporozoite cell invasion efficiency. Moreover, we know that the rate of turnover for each HSPG influences the efficacy of RNAi-mediated suppression. Therefore we recognize that although our 72 h time point suggests that salivary gland HSPG turnover is amenable to RNAi, the differential rates of turnover of specific HSPGs during our assay's 48–72 h salivary gland invasion window limit our interpretation of the data. The hypothesized conservation in the stratagem for cell invasion by Plasmodium sporozoites is reasonable, especially in light of the evidence in the literature regarding the role of HSPGs in sporozoite-homing to the liver [32,34,36] and the presence of critical trisulfated ‘conserved’ HS disaccharide moieties on salivary glands [15]. However, if we assume that the loss of RB4Ea12 reactivity in the absence of HS biosynthesis results in the loss of GlcNS6S-IdoA2S HS chains as observed in our present study, then our results suggest indirectly that although HSGAGs may contribute to sporozoite attachment and invasion of salivary glands, their contribution to the process appears to be moderate. We anticipate that the acquisition of the salivary gland proteome will be critical in providing additional insight into the interactions of sporozoites with this tissue.
AUTHOR CONTRIBUTION
Jennifer Armistead, Iain Wilson and Rhoel Dinglasan designed and performed experiments; Iain Wilson, Toin van Kuppevelt and Rhoel Dinglasan conceptualized the study; Iain Wilson and Toin van Kuppevelt provided critical reagents; and Jennifer Armistead, Iain Wilson, Toin van Kuppevelt and Rhoel Dinglasan wrote the paper.
ACKNOWLEDGEMENTS
We thank Natalie Öhl and Anna Gstraunthaler (Department of Biotechnology, University of Natural Resources and Life Sciences, Vienna, Austria) for their assistance with the enzymatic preparation and characterization, as well as Lindsay Parish for generating anti-AgLDH antibodies, Ebrahim Razzazi-Fazeli for access to the Bruker Ultraflex MALDI–TOF mass spectrometer, Anil Ghosh for assistance in salivary gland dissections and Arie Natalie Oosterhof (Department of Biochemistry, Radboud University Medical Centre Nijmegen, Nijmegen, Holland) for the preparation of the scFv antibodies. We are grateful to Sanaria for provision of the P. falciparum NF54 line.
FUNDING
This work was supported in part by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung [grant number L314 (to I.B.H.W)]; the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) [grant number 1K22AI077707]; and start-up funds from the Malaria Research Institute, Johns Hopkins Bloomberg School of Public Health (JHMRI) (to R.R.D.). This publication was also made possible by the NIH National Center for Research Resources [grant number UL1 RR 025005], and by the JHMRI.
References
- 1.Dinglasan R. R., Jacobs-Lorena M. Flipping the paradigm on malaria transmission-blocking vaccines. Trends Parasitol. 2008;8:364–370. doi: 10.1016/j.pt.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinglasan R. R., Alaganan A., Ghosh A. K., Saito A., van Kuppevelt T. H., Jacobs-Lorena M. Plasmodium falciparum ookinetes require mosquito midgut chondroitin sulfate proteoglycans for cell invasion. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15882–15887. doi: 10.1073/pnas.0706340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller A. K., Kohlhepp F., Hammerschmidt C., Michel K. Invasion of mosquito salivary glands by malaria parasites: prerequisites and defense strategies. Int. J. Parasitol. 2010;40:1229–1235. doi: 10.1016/j.ijpara.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinzon-Ortiz C., Friedman J., Esko J., Sinnis P. The binding of the circumsporozoite protein to cell surface heparan sulfate proteoglycans is required for plasmodium sporozoite attachment to target cells. J. Biol. Chem. 2001;276:26784–26791. doi: 10.1074/jbc.M104038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frevert U. Malaria sporozoite–hepatocyte interactions. Exp. Parasitol. 1994;79:206–210. doi: 10.1006/expr.1994.1082. [DOI] [PubMed] [Google Scholar]
- 6.Frevert U., Sinnis P., Cerami C., Shreffler W., Takacs B., Nussenzweig V. Malaria circumsporozoite protein binds to heparan sulfate proteoglycans associated with the surface membrane of hepatocytes. J. Exp. Med. 1993;177:1287–1298. doi: 10.1084/jem.177.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerami C., Frevert U., Sinnis P., Takacs B., Clavijo P., Santos M. J., Nussenzweig V. The basolateral domain of the hepatocyte plasma membrane bears receptors for the circumsporozoite protein of Plasmodium falciparum sporozoites. Cell. 1992;70:1021–1033. doi: 10.1016/0092-8674(92)90251-7. [DOI] [PubMed] [Google Scholar]
- 8.Pancake S. J., Holt G. D., Mellouk S., Hoffman S. L. Malaria sporozoites and circumsporozoite protein bind sulfated glycans: carbohydrate binding properties predicted from sequence homologies with other lectins. Parasitologia. 1993;35(Suppl.):77–80. [PubMed] [Google Scholar]
- 9.Sinnis P. The malaria sporozoite's journey into the liver. Infect. Agents Dis. 1996;5:182–189. [PubMed] [Google Scholar]
- 10.McCormick C. J., Tuckwell D. S., Crisanti A., Humphries M. J., Hollingdale M. R. Identification of heparin as a ligand for the A-domain of Plasmodium falciparum thrombospondin-related adhesion protein. Mol. Biochem. Parasitol. 1999;100:111–124. doi: 10.1016/s0166-6851(99)00052-3. [DOI] [PubMed] [Google Scholar]
- 11.Li F., Templeton T. J., Popov V., Comer J. E., Tsuboi T., Torii M., Vinetz J. M. Plasmodium ookinete-secreted proteins secreted through a common micronemal pathway are targets of blocking malaria transmission. J. Biol. Chem. 2004;279:26635–26644. doi: 10.1074/jbc.M401385200. [DOI] [PubMed] [Google Scholar]
- 12.Akhouri R. R., Bhattacharyya A., Pattnaik P., Malhotra P., Sharma A. Structural and functional dissection of the adhesive domains of Plasmodium falciparum thrombospondin-related anonymous protein (TRAP) Biochem. J. 2004;379:815–822. doi: 10.1042/BJ20031500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myung J. M., Marshall P., Sinnis P. The Plasmodium circumsporozoite protein is involved in mosquito salivary gland invasion by sporozoites. Mol. Biochem. Parasitol. 2004;133:53–59. doi: 10.1016/j.molbiopara.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Wengelnik K., Spaccapelo R., Naitza S., Robson K. J., Janse C. J., Bistoni F., Waters A. P., Crisanti A. The A-domain and the thrombospondin-related motif of Plasmodium falciparum TRAP are implicated in the invasion process of mosquito salivary glands. EMBO J. 1999;18:5195–5204. doi: 10.1093/emboj/18.19.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinnis P., Coppi A., Toida T., Toyoda H., Kinoshita-Toyoda A., Xie J., Kemp M. M., Linhardt R. J. Mosquito heparan sulfate and its potential role in malaria infection and transmission. J. Biol. Chem. 2007;282:25376–84. doi: 10.1074/jbc.M704698200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tveit H., Dick G., Skibeli V., Prydz K. A proteoglycan undergoes different modifications en route to the apical and basolateral surfaces of Madin–Darby canine kidney cells. J. Biol. Chem. 2005;280:29596–29603. doi: 10.1074/jbc.M503691200. [DOI] [PubMed] [Google Scholar]
- 17.Wilson I. B.H. Functional characterization of Drosophila melanogaster peptide O-xylosyltransferase, the key enzyme for proteoglycan chain initiation and member of the core 2/I N-acetylglucosaminyltransferase family. J. Biol. Chem. 2002;277:21207–21212. doi: 10.1074/jbc.M201634200. [DOI] [PubMed] [Google Scholar]
- 18.Voglmeir J., Voglauer R., Wilson I. B.H. XT-II, the second isoform of human peptide-O-xylosyltransferase, displays enzymatic activity. J. Biol. Chem. 2006;282:5984–5990. doi: 10.1074/jbc.M608087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunner A., Kolarich D., Voglmeir J., Paschinger K., Wilson I. B.H. Comparative characterisation of recombinant invertebrate and vertebrate peptide O-xylosyltransferases. Glycoconj. J. 2006;23:543–554. doi: 10.1007/s10719-006-7633-z. [DOI] [PubMed] [Google Scholar]
- 20.Blandin S., Moita L. F., Kocher T., Wilm M., Kafatos F. C., Levashina E. A. Reverse genetics in the mosquito Anopheles gambiae: target disruption of the Defensin gene. EMBO Rep. 2002;3:852–856. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dennissen M. A., Jenniskens G. J., Pieffers M., Versteeg E. M., Petitou M., Veerkamp J. H., van Kuppevelt T. H. Large, tissue-regulated domain diversity of heparan sulfates demonstrated by phage display antibodies. J. Biol. Chem. 2002;277:10982–10986. doi: 10.1074/jbc.M104852200. [DOI] [PubMed] [Google Scholar]
- 22.Sjoberg E. M., Blom A., Larsson B. S., Alston-Smith J., Sjoquist M., Fries E. Plasma clearance of rat bikunin: evidence for receptor-mediated uptake. Biochem. J. 1995;308:881–887. doi: 10.1042/bj3080881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spring J., Paine-Saunders S. E., Hynes R. O., Bernfield M. Drosophila syndecan: conservation of a cell-surface heparan sulfate proteoglycan. Proc. Natl. Acad. Sci. U.S.A. 1994;91:3334–3338. doi: 10.1073/pnas.91.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kokenyesi R., Silbert J. E. Formation of heparan sulfate or chondroitin/dermatan sulfate on recombinant domain I of mouse perlecan expressed in Chinese hamster ovary cells. Biochem. Biophys. Res. Commun. 1995;211:262–267. doi: 10.1006/bbrc.1995.1805. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L., Esko J. D. Amino acid determinants that drive heparan sulfate assembly in a proteoglycan. J. Biol. Chem. 1994;269:19295–19299. [PubMed] [Google Scholar]
- 26.Brinkmann T., Weilke C., Kleesiek K. Recognition of acceptor proteins by UDP-D-xylose proteoglycan core protein β-D-xylosyltransferase. J. Biol. Chem. 1997;272:11171–11175. doi: 10.1074/jbc.272.17.11171. [DOI] [PubMed] [Google Scholar]
- 27.Schön S., Prante C., Bahr C., Kuhn J., Kleesiek K., Götting C. Cloning and recombinant expression of active full-length xylosyltransferase I (XT-I) and characterization of subcellular localization of XT-I and XT-II. J. Biol. Chem. 2006;281:14224–14231. doi: 10.1074/jbc.M510690200. [DOI] [PubMed] [Google Scholar]
- 28.del Pilar Corena M., VanEkeris L., Salazar M. I., Bowers D., Fiedler M. M., Silverman D., Tu C., Linser P. J. Carbonic anhydrase in the adult mosquito midgut. J. Exp. Biol. 2005;208:3263–3273. doi: 10.1242/jeb.01739. [DOI] [PubMed] [Google Scholar]
- 29.Schewe B., Schmalzlin E., Walz B. Intracellular pH homeostasis and serotonin-induced pH changes in Calliphora salivary glands: the contribution of V-ATPase and carbonic anhydrase. J. Exp. Biol. 2008;211:805–815. doi: 10.1242/jeb.002667. [DOI] [PubMed] [Google Scholar]
- 30.Boisson B., Jacques J. C., Choumet V., Martin E., Xu J., Vernick K., Bourgouin C. Gene silencing in mosquito salivary glands by RNAi. FEBS Lett. 2006;580:1988–1992. doi: 10.1016/j.febslet.2006.02.069. [DOI] [PubMed] [Google Scholar]
- 31.Tucker R. P. The thrombospondin type 1 repeat superfamily. Int. J. Biochem. Cell Biol. 2004;36:969–974. doi: 10.1016/j.biocel.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 32.Sinnis P., Nardin E. Sporozoite antigens: biology and immunology of the circumsporozoite protein and thrombospondin-related anonymous protein. Chem. Immunol. 2002;80:70–96. doi: 10.1159/000058840. [DOI] [PubMed] [Google Scholar]
- 33.Coppi A., Tewari R., Bishop J. R., Bennett B. L., Lawrence R., Esko J. D., Billker O., Sinnis P. Heparan sulfate proteoglycans provide a signal to Plasmodium sporozoites to stop migrating and productively invade host cells. Cell Host Microbe. 2007;2:316–327. doi: 10.1016/j.chom.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tewari R., Spaccapelo R., Bistoni F., Holder A. A., Crisanti A. Function of region I and II adhesive motifs of Plasmodium falciparum circumsporozoite protein in sporozoite motility and infectivity. J. Biol. Chem. 2002;277:47613–47618. doi: 10.1074/jbc.M208453200. [DOI] [PubMed] [Google Scholar]
- 35.Robson K. J., Frevert U., Reckmann I., Cowan G., Beier J., Scragg I. G., Takehara K., Bishop D. H., Pradel G., Sinden R. Thrombospondin-related adhesive protein (TRAP) of Plasmodium falciparum: expression during sporozoite ontogeny and binding to human hepatocytes. EMBO J. 1995;14:3883–3894. doi: 10.1002/j.1460-2075.1995.tb00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matuschewski K., Nunes A. C., Nussenzweig V., Menard R. Plasmodium sporozoite invasion into insect and mammalian cells is directed by the same dual binding system. EMBO J. 2002;21:1597–1606. doi: 10.1093/emboj/21.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lensen J. F., Rops A. L., Wijnhoven T. J., Hafmans T., Feitz W. F., Oosterwijk E., Banas B., Bindels R. J., van den Heuvel L. P., van der Vlag J., et al. Localization and functional characterization of glycosaminoglycan domains in the normal human kidney as revealed by phage display-derived single chain antibodies. J. Am. Soc. Nephrol. 2005;16:1279–1288. doi: 10.1681/ASN.2004050413. [DOI] [PubMed] [Google Scholar]
- 38.ten Dam G. B., Hafmans T., Veerkamp J. H., van Kuppevelt T. H. Differential expression of heparan sulfate domains in rat spleen. J. Histochem. Cytochem. 2003;51:727–739. doi: 10.1177/002215540305100604. [DOI] [PubMed] [Google Scholar]
- 39.Properzi F., Lin R., Kwok J., Naidu M., van Kuppevelt T. H., Ten Dam G. B., Camargo L. M., Raha-Chowdhury R., Furukawa Y., Mikami T., et al. Heparan sulphate proteoglycans in glia and in the normal and injured CNS: expression of sulpho-transferases and changes in sulphation. Eur. J. Neurosci. 2008;27:593–604. doi: 10.1111/j.1460-9568.2008.06042.x. [DOI] [PubMed] [Google Scholar]
- 40.Qu X., Carbe C., Tao C., Powers A., Lawrence R., van Kuppevelt T. H., Cardoso W. V., Grobe K., Esko J. D., Zhang X. Lacrimal gland development and Fgf10–Fgfr2b signaling are controlled by 2-O- and 6-O-sulfated heparan sulfate. J. Biol. Chem. 2011;286:14435–14444. doi: 10.1074/jbc.M111.225003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hegge S., Munter S., Steinbuchel M., Heiss K., Engel U., Matuschewski K., Frischknecht F. Multistep adhesion of Plasmodium sporozoites. FASEB J. 2010;24:2222–2234. doi: 10.1096/fj.09-148700. [DOI] [PubMed] [Google Scholar]
- 42.Sterling C. R., Aikawa M., Vanderberg J. P. The passage of Plasmodium berghei sporozoites through the salivary glands of Anopheles stephensi: an electron microscope study. J. Parasitol. 1973;59:593–605. [PubMed] [Google Scholar]
- 43.Rossignol P. A., Ribeiro J. M., Spielman A. Increased intradermal probing time in sporozoite-infected mosquitoes. Am. J. Trop. Med. Hyg. 1984;33:17–20. doi: 10.4269/ajtmh.1984.33.17. [DOI] [PubMed] [Google Scholar]
- 44.Ghosh A. K., Devenport M., Jethwaney D., Kalume D. E., Pandey A., Anderson V. E., Sultan A. A., Kumar N., Jacobs-Lorena M. Malaria parasite invasion of the mosquito salivary gland requires interaction between the Plasmodium TRAP and the Anopheles saglin proteins. PLoS Pathog. 2009;5:e1000265. doi: 10.1371/journal.ppat.1000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennan J. D., Kent M., Dhar R., Fujioka H., Kumar N. Anopheles gambiae salivary gland proteins as putative targets for blocking transmission of malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13859–13864. doi: 10.1073/pnas.250472597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okulate M. A., Kalume D. E., Reddy R., Kristiansen T., Bhattacharyya M., Chaerkady R., Pandey A., Kumar N. Identification and molecular characterization of a novel protein Saglin as a target of monoclonal antibodies affecting salivary gland infectivity of Plasmodium sporozoites. Insect Mol. Biol. 2007;16:711–722. doi: 10.1111/j.1365-2583.2007.00765.x. [DOI] [PubMed] [Google Scholar]
- 47.Dinglasan R. R., Jacobs-Lorena M. Insight into a conserved lifestyle: protein–carbohydrate adhesion strategies of vector-borne pathogens. Infect. Immun. 2005;73:7797–7807. doi: 10.1128/IAI.73.12.7797-7807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]