Abstract
Derivation of induced pluripotent stem (iPS) cells requires the expression of defined transcription factors (among Oct3/4, Sox2, Klf4, c-Myc, Nanog and Lin28) in the targeted cells. Lentiviral or standard retroviral gene transfer remains the most robust and commonly used approach. Low reprogramming frequency overall, and the higher efficiency of derivation utilizing integrating vectors compared to more recent non-viral approaches suggests that gene activation or disruption via proviral integration sites (IS) may play a role in obtaining the pluripotent phenotype. We provide for the first time an extensive analysis of the lentiviral integration profile in human iPS cells. We identified a total of 78 independent integration sites (IS) in 8 recently established iPS cell lines derived from either human fetal fibroblasts or newborn foreskin fibroblasts after lentiviral gene transfer of Oct4, Sox2, Nanog, and Lin28. The number of IS ranged from 5 to15 IS per individual iPS clone and 75 IS could be assigned to a unique chromosomal location. The different iPS clones had no IS in common. Expression analysis as well as extensive bioinformatic analysis did not reveal functional concordance of the lentiviral targeted genes between the different clones. Interestingly, in 6 of the 8 iPS clones some of the IS were found in pairs, integrated into the same chromosomal location within six base pairs of each other or in very close proximity. Our study supports recent reports that efficient reprogramming of human somatic cells is not dependent on insertional activation or deactivation of specific genes or gene classes.
Keywords: Induced pluripotent stem cells, gene delivery systems in vivo or in vitro, cellular reprogramming, lentiviral vector, insertional mutagenesis, pluripotent stem cells, transcription factor, cellular therapy
Introduction
The reprogramming of murine fibroblasts into “induced pluripotent stem (iPS) cells” was first achieved by Takahashi and Yamanaka via ectopic retroviral expression of defined transcription factors (Oct4, Sox2, Klf4, and c-Myc) known to be required for normal embryogenesis and to be highly expressed in embryonic stem cells [1]. In an independent screen Yu et al. identified the transcription factors Oct4, Sox2, Nanog, and Lin28 to be sufficient to reprogram human somatic cells into iPS cells [2]. Both approaches were soon adapted to various other cell types, cells from different species as well as to somatic cells from patients [3–10]. The transgenes were introduced into target cells using replication-deficient murine retroviruses or lentiviruses. Although the reprogramming procedure is quite robust and has been replicated in many laboratories, the overall efficiency of obtaining fully functional iPS cells is very low, for instance only 0.02% of transfected cells become iPS cells after retroviral gene transfer [1], indicating that other factors besides the expression of the ectopic transgenes likely play a role in this process. Several studies indicate that the differentiation status of the starting cell population has an impact on the reprogramming efficiency [11], but even when homogeneous undifferentiated murine neural stem cells were used as targets the efficiency remained below 3.6% [12].
The low efficiency of iPS derivation suggests the worrisome possibility that viral vector integration into the genome activates cooperating endogenous proto-oncogenes, and that particular loci must be turned on in this manner to achieve efficient reprogramming. The ability to use episomal vectors for iPS derivation [13] and a recent report showing that protein delivery can induce reprogramming of mouse embryonic fibroblasts suggest that insertional mutagenesis is not required for iPS generation, and are encouraging milestones towards the generation of iPS cells without genetic modification [14]. Due to the higher efficiency and reproducibility of integrating viral gene transfer protocols for reprogramming, this method remains the most common for the derivation of iPS cells. Yu et al. recently optimized the lentiviral based reprogramming strategy and were able to increase the efficiency almost 100 fold to 1% in human foreskin fibroblasts [13]. Therefore, the issue of whether insertional activation contributes to iPS generation utilizing this system remains, and also impacts on assessment of the tumorigenic capacity of cells derived from iPS clones and the use of these vectors to create iPS cells for research applications.
Integrating retroviruses have long been known to be potentially oncogenic via activation of adjacent proto-oncogenes by strong proviral promoter/enhancers. Wild-type replicating viruses were assumed to result in tumors due to repeated random insertions into the genome, eventually resulting in activation of an adjacent gene. Replication-incompetent vectors were presumed to be much less likely to activate oncogenic gene or genes with a limited number of insertions in target cells. However, more recently it has been demonstrated that insertion patterns are far from random, with enhanced integration within actively-expressed genes and surrounding transcription start sites for the Moloney murine leukemia virus (MLV) whereas insertion of lentiviruses such as the human immunodeficiency virus (HIV) or simian immunodeficiency virus (SIV) are overrepresented in expressed transcriptional units [15–18]. Insertional activation of adjacent genes has been shown to result in biologic effects including immortalization, clonal dominance and malignant transformation both in vitro and in vivo, including in several landmark clinical trials targeting hematopoietic stem cells, a cell type that appears to be particularly susceptible [19–23].
In the present study we investigated the distribution of IS in 8 human iPS cell lines derived from fetal fibroblasts (IMR90) or from newborn foreskin fibroblast (FS). These human iPS clones were obtained via transduction utilizing lentivirus vectors expressing Oct4, Sox2, Nanog, and Lin28 transcription factors. We studied the general distribution of integration sites across clones and analyzed whether integration events perturbed expression of interrupted or nearby genes. We also compared integration sites between clones, asking whether specific loci were more likely to be targeted in iPS clones and thus possibly facilitating derivation of cells with primitive characteristics.
Methods
Identification of integration sites
The human iPS cell lines IPS(IMR-90)1–4, IPS(FS) 1–4, IMR90 cells (Cat# CCL-186™, ATCC, Manassas, VA) and human newborn foreskin fibroblast (FS, Cat# CRL-2097™, ATCC) were cultured as previously described [2]. The iPS-clones were derived from either IMR90 or FS cells after lentiviral gene transfer (Backbone: pSin4-EF2-IRES-Pur) of the human cDNA coding for either OCT4, Sox2, Lin28 and Nanog [2]. Detailed vector maps and sequences can be found on www.addgene.org. Genomic DNA was isolated using the DNeasy DNA Purification Kit according to the manufacturer’s instructions (QIAGEN, Valencia, CA).
Linear amplification mediated LAM-PCR was performed as previously described with primers and linker cassettes shown in Table S3 [25]. Three hundred nanograms of genomic DNA were linearly amplified using a HIV-3′-LTR–specific 5′-biotinylated primer. After the second strand synthesis by random priming, the DNA was digested with either ApoI or TasI and ligated to a linker cassette. Nested PCR was performed using HIV-3′-LTR–specific and linker-specific primers. The amplicons were purified from 2.5% low melting point agarose gels (NuSieve GTG, Cambrex, IA) using QIAGEN QIAquick Gel Extraction Kit (Valencia, CA) and cloned into pCR4TOPO vector (Invitrogen, Carlsbad, CA) for sequencing with M13-primers using an ABI Prism Genetic Analyzer (Applied Biosystems, Foster City, CA). Sequences were analyzed by the DNASTAR SeqManII software (Madison, Wisconsin), scanning for the pCR-4 TOPO vector, assembling sequences with lengths greater than 100 base pairs, a match size of minimum 50bp, and a percent match requirement of 95%. The trimmed sequences were aligned to Build 36 of the human genome (hg18) using the BLAT server (http://genome.ucsc.edu/) and a local copy of BLAST.
Integration site specific and allele specific PCR
All PCR analyses were performed on 100ng RNA-free genomic DNA. Each integration site specific PCR was performed using one primer within the 3'LTR and the second within flanking genomic sequences. Primers used are listed in Table S3. After separation on 2.5% Low Melting Temperature agarose gels (NuSieve GTG, Cambrex, IA) the amplicons were purified using QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA) and cloned in the pCR4TOPO vector (Invitrogen, Carlsbad, CA) for further sequencing using M13-primer (Table S3)
Southern Blot
For Southern blot analysis 10ug of genomic DNA was digested with EcoRI and separated on a 0.75% Agarose gel. EcoRI cuts once within the vector genome. Following the transfer to a nylon membrane the DNA fragments were hybridized with a radiolabeled Puromycin cDNA probe generated by PCR from the original pSin EF2 vectors [2] (Primer list Table S3). The labeling reaction was performed according to manufacturer’s recommendations (Amersham Ready –To- Go™ DNA Labelling Beads, GE Healthcare)
Expression Analysis
The previously published Nimblegen expression data on the iPS cell lines was downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL5876). Relative expression levels for each gene interrupted by lentiviral insertions were determined by comparison to all other iPS-clones combined (for instance the data set of gene expression for genes interrupted by vector insertions for IPS(IMR90)-1 was compared to the data set of IPS(IMR90)-2 to 4 and IPS(FS)-1 to 4). A fold ratio and a p-value were calculated for each comparison and p-values less than 0.05 were considered as significant.
Bioinformatics analysis
We compared the IS distribution in the iPS cells to in silico generated control sets of insertions in the human genome. Briefly, 10 000 sets of 75 random IS were designed in silico as follows: A TasI or ApoI site in the genome was selected at random using a random number generator. The in silico IS was placed either upstream or downstream (p=0.5) of the site, at a distance matching the size of one of the sequences obtained experimentally. The in silico IS was validated only when a BLAST alignment of the genomic sequence between the restriction site and the IS returned a unique sequence in the genome. This operation was repeated 75 times to obtain a single matching random dataset, and then again for a total of 10,000 datasets of 75 IS each. These control datasets were subjected to the same analyses as the experimental datasets, and the results were used to generate empiric p-values. p-values of less than 0.05 were considered significant.
Pathway analysis
Two gene lists were generated based on the occurrence of integration sites. The first list comprised of all genes with integration sites within the coding regions. The second list took into account all genes having an integration site within a 30 kb window. The gene lists were analyzed for enrichment of functional pathways using MetaCore™(GeneGo Inc., St Joseph, MI) and Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA). Hypergeometric test was performed to test for equality of observed proportion of genes mapped to a particular pathway between the gene lists and the reference set. Type I error was controlled by using false discovery rate correction for multiple testing (FDR=0.01).
Results
Integration site analysis of iPS clones reveals no common target genes
Genomic DNA samples from previously established iPS cells clones were used for analysis of integration sites. Four iPS clones were derived from IMR90 fetal fibroblasts (IPS(IMR90)-1 to IPS(IMR90)-4) and four iPS clones were derived from foreskin fibroblasts (IPS(FS)-1 to IPS(FS)-4). The Oct4, Sox2, Nanog, and Lin28 transcription factors were transferred to the cells via a standard third generation lentiviral vector. These vectors have much of the viral LTR promoter/enhancer region deleted, but still contain a strong internal promoter to drive transgene expression and residual LTR enhancer elements. All clones were tested in a comparative manner for their ES cell-like phenotype which included telomerase activity, cell surface markers, and genes characterizing human ES cells [2]. They also maintained the developmental potential to differentiate into derivatives of all three primary germ layers. Recently, the clones IPS(IMR90)-4 as well as IPS(FS)-1 were successfully differentiated into in vitro functional cardiomyocytes [24].
To identify the lentiviral integration sites we performed linear amplification–mediated PCR (LAM-PCR, [25]), on genomic DNA from all 8 iPS clones followed by shotgun cloning and sequencing. Valid sequences were mapped to the human genome (Build 36, hg18). In order to identify the complete insertion profile and to minimize restriction enzyme bias [26], we analyzed all samples separately with two different restriction enzymes (Apo1 and Tas1). Furthermore, Southern blot (Figure 1) analysis of each individual clone roughly confirmed the number of IS identified by LAM-PCR, indicating that the vast majority of IS present in each clone had been identified.
Figure 1.
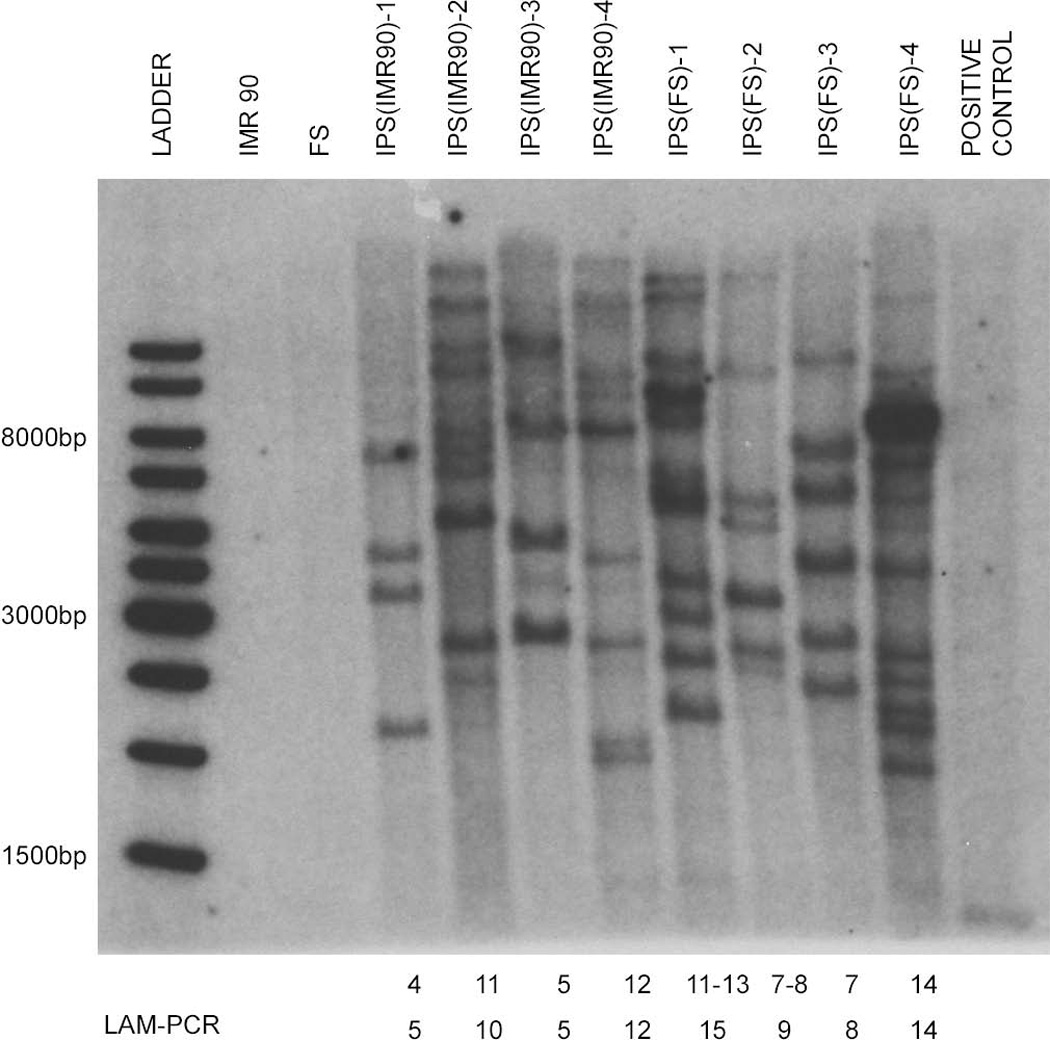
Estimation of the number of proviral insertions into the genome by Southern blot analysis: Genomic DNA of each iPS clone was digested with EcoRI, which cuts once in each reprogramming vector. Puromycin was detected with a p32 labeled probe. At the bottom the number of insertions sites detected by Southern blot or LAM-PCR in each individual clone are indicated.
Using this approach, we were able to identify a total of 78 valid IS ranging from 5 to15 IS per individual iPS clone (Table 1). Valid IS were defined only if the sequence we obtained from the LAM-PCR was correctly juxtaposed to the vector long terminal repeat (LTR) and yielded a best BLAT hit with at least 95% identity over 95% of the length of the sequence (University of California, Santa Cruz [UCSC] Genome Browser, http://genome.ucsc.edu; hg18). Seventy five of the 78 valid IS could be assigned to a unique chromosomal location. Fifty-three out of 75 (70.7%) could be mapped within transcription units (TU) of RefSeq mRNAs, 69.3% in exons and 1.3% in introns, (RefSeq Genes track in the UCSC Genome Browser, Table 1 and Table 2). This IS distribution was significantly different compared to the 10,000 in silico-generated random IS datasets. The profile with an insertional preference for TU was very similar to previous IS analysis for HIV-based vector systems used to transduce other target cell populations [26–28]. There was marginal underrepresentation of IS within genomic long terminal repeat elements (LTR) for the iPS IS (60.0% vs. 75.24%, p ≤ 0.021), and overrepresentation of IS contained within a 5kb window surrounding CpG islands (10.67% vs. 5.33%, p ≤ 0.0444) (Table 1 and Table S1). We also observed a preference for iPS IS within a 30kb window of proto-oncogenes (Table 3) compared to the random controls (10.17% vs. 3.44%, p ≤ 0.0275).
Table 1.
Unique Insertion sites of IPS(IMR90)-1,-2,-3,-4 and IPS(FS)-1,-2,-3,-4 Abbreviation: n.l.= not localizable, m.h. = multiple hits, Chr = chromosomal location, Det. = detected in LAM-PCR using A = Apo1, T = Tas1. C = confirmed by conventional PCR
| Clone | Chr | Strand | Coordinate | Gene | RefSeq ID | Det. |
|---|---|---|---|---|---|---|
| IPS(IMR90)-1.1 | 1q31.3 | + | 196439332 | NEK7 | NM_133494 | A,T |
| IPS(IMR90)-1.2 | 1p22.1 | − | 93424957 | CCDC18 | NM_206886 | A,T |
| IPS(IMR90)-1.3 | 20p11.23 | + | 20536159 | C20orf74 | NM_020343 | A |
| IPS(IMR90)-1.4 | 4q28.3 | − | 139094629 | no gene | A | |
| IPS(IMR90)-1.5 | 16p12.3 | + | 18446842 | NOMO2 | NM_001004060 | A |
| IPS(IMR90)-2.1 | 12q24.31 | + | 120845755 | WDR66 | NM_144668 | A,T,C |
| IPS(IMR90)-2.2a | X | − | n.l.; m.h. | A,T | ||
| IPS(IMR90)-2.2b | X | + | n.l.; m.h. | A,T | ||
| IPS(IMR90)-2.3 | 13q33.2 | + | 104669782 | no gene | A,T | |
| IPS(IMR90)-2.4 | 3p24.3 | − | 19970622 | RAB5A | NM_004162 | A,T |
| IPS(IMR90)-2.5 | 8q21.3 | + | 90451996 | no gene | A,T | |
| IPS(IMR90)-2.6 | 11q23.2 | − | 114742187 | CADM1 | NM_014333 | A |
| IPS(IMR90)-2.7 | 3q11.2 | + | 95238084 | ARL13B | NM_144996 | T |
| IPS(IMR90)-2.8 | 11q22.3 | + | 105178329 | GRIA4 | NM_001077243 | T |
| IPS(IMR90)-2.9 | 17q21.33 | + | 45237719 | MYST2 | NM_007067 | T |
| IPS(IMR90)-3.1 | 4q28.3 | + | 137874633 | no gene | A,T | |
| IPS(IMR90)-3.2 | 7p21.1 | − | 17850812 | SNX13 | NM_015132 | A,T |
| IPS(IMR90)-3.3 | 9p21.3 | − | 20349449 | MLLT3 | NM_004529 | A,T |
| IPS(IMR90)-3.4a | 9q33.1 | + | 117124717 | DEC | NM_017418 | A,T,C |
| IPS(IMR90)-3.4b | 9q33.1 | − | 117125516 | DEC | NM_017418 | A |
| IPS(IMR90)-4.1 | 3q26.32 | + | 180217743 | no gene | A,T | |
| IPS(IMR90)-4.2 | 8p23.3 | + | 410727 | no gene | A,T | |
| IPS(IMR90)-4.3 | 15q14 | + | 35015389 | MEIS2 | NM_002399 | A,T |
| IPS(IMR90)-4.4 | 8q13.2 | − | 70277943 | no Gene | A | |
| IPS(IMR90)-4.5 | 3p22.2 | + | 37160889 | LRRFIP2 | NM_006309 | A,T |
| IPS(IMR90)-4.6 | 3q22.2 | + | 137178708 | PPP2R3A | NM_002718 | A,T |
| IPS(IMR90)-4.7 | 3p14.2 | + | 59413559 | no gene | A,T | |
| IPS(IMR90)-4.8 | 1q42.12 | − | 222776208 | no gene | A,T | |
| IPS(IMR90)-4.9a | 12q24.12 | − | 110427921 | ATXN2 | NM_002973 | A,T,C |
| IPS(IMR90)-4.9b | 12q24.12 | + | 110427917 | ATXN2 | NM_002973 | A,T,C |
| IPS(IMR90)-4.10 | 3p24.3 | + | 17418685 | TBC1D5 | NM_001134380 | T |
| IPS(IMR90)-4.11 | 3q13.31 | − | 118700002 | no gene | A,T | |
| IPS(FS)-1.1 | 1q31.2 | − | 190393887 | no gene | A,T | |
| IPS(FS)-1.2 | 14q31.2 | − | 82698434 | no gene | A,T | |
| IPS(FS)-1.3 | 4q23 | − | 99518312 | RAP1 | NM_001100427 | A |
| IPS(FS)-1.4 | 17q22 | − | 52271039 | DGKE, | NM_003647 | A,T |
| IPS(FS)-1.5 | 7p15.3 | + | 24963617 | OSBPL3 | NM_145322 | A,T |
| IPS(FS)-1.6 | 2q22.3 | + | 148328431 | ACVR2A | NM_001616 | A,T |
| IPS(FS)-1.7a | Yq11.21 | − | 13911809 | UTY | NM_007125 | A,T,C |
| IPS(FS)-1.7b | Yq11.21 | + | 13911805 | UTY | NM_007125 | A,T,C |
| IPS(FS)-1.8 | 9q21.32 | + | 85313757 | FRMD3 | NM_174938 | A,T |
| IPS(FS)-1.9 | 3p25.1 | − | 12644639 | RAF1 | NM_002880 | A,T |
| IPS(FS)-1.10 | 20p11.21 | − | 24891470 | no gene | A | |
| IPS(FS)-1.11 | 3p21.31 | + | 48906212 | SLC25A20 | NM_000387 | A |
| IPS(FS)-1.12 | 19p13.3 | − | 1205649 | MIDN | NM_177401 | A |
| IPS(FS)-1.13 | 1p21.2 | − | 101154654 | SLC30A7 | NM_133496 | A |
| IPS(FS)-1.14 | 6_qbl_hap2 | + | 3109385 | EHMT2 | NM_006709 | A,T |
| IPS(FS)-2.1 | 18q21.31 | − | 52596315 | WDR7 | NM_015285 | A,T |
| IPS(FS)-2.2 | 12p12.1 | − | 22574653 | KIAA0528 | NM_014802 | A,T |
| IPS(FS)-2.3 | 13q22.1 | − | 72189883 | C13orf37 | NM_001071775 | A |
| IPS(FS)-2.4a | 11q23.3 | + | 119867974 | no gene | A,T | |
| IPS(FS)-2.4b | 11q23.3 | − | 119869577 | no gene | A,T | |
| IPS(FS)-2.5 | 2q37.3 | + | 237824167 | no gene | A | |
| IPS(FS)-2.6 | 14q23.3 | − | 64977640 | FUT8 | NM_178155 | A,T |
| IPS(FS)-2.7 | 4q21.23 | − | 84577179 | HEL308 | NM_133636 | A |
| IPS(FS)-2.8 | pSinEF2 | Sox2 | T | |||
| IPS(FS)-3.1 | 10q26.13 | − | 127439586 | C10orf137 | NM_015608 | A,T |
| IPS(FS)-3.2 | 3q27.1 | − | 184396400 | MCF2L2 | NM_015078 | A,T |
| IPS(FS)-3.4 | 4q28.3 | − | 134995205 | no gene | A | |
| IPS(FS)-3.5 | 16q21 | − | 65158367 | CMTM1 | NM_181269 | A,T |
| IPS(FS)-3.6 | 17q23.3 | + | 58537848 | TANC2 | NM_025185 | A,T |
| IPS(FS)-3.7 | 11q22.3 | − | 104621207 | no gene | A,T | |
| IPS(FS)-3.8 | 8q24.12 | + | 120699913 | ENPP2 | NM_006209 | A, |
| IPS(FS)-3.9 | 10p14 | − | 77134886 | ITIH5 | NM_001001851 | A |
| IPS(FS)-4.1a | 11q13.4 | − | 72307364 | FCHSD2 | NM_014824 | A,T,C |
| IPS(FS)-4.1b | 11q13.4 | + | 72307360 | FCHSD2 | NM_014824 | A,T,C |
| IPS(FS)-4.2 | 13q21.33 | + | 71102265 | DACH1 | NM_080760 | A,T |
| IPS(FS)-4.3 | 7q31.32 | − | 123131665 | WASL | NM_003941 | A,T |
| IPS(FS)-4.4 | 14q32.11 | − | 9017333 | TTC7B | NM_001010854 | A,T |
| IPS(FS)-4.5 | Xp22.33 | − | 3177763 | no gene | A,T | |
| IPS(FS)-4.6 | 8q23.1 | + | 109326141 | EIF3E | NM_001568 | A,T |
| IPS(FS)-4.7 | 2p25.1 | − | 8823390 | KIDINS220 | NM_020738 | A,T |
| IPS(FS)-4.8 | 12q21.31 | − | 82726819 | no gene | A,T | |
| IPS(FS)-4.9 | 13q33.3 | + | 106009719 | ARGLU1 | NM_018011 | A,T |
| IPS(FS)-4.10 | 14q31.1 | − | 80737156 | GTF2A1 | NM_015859 | T |
| IPS(FS)-4.11 | 3q26.1 | + | 163939363 | no gene | T | |
| IPS(FS)-4.13 | 10q25.1 | − | 108203739 | no gene | T | |
| IPS(FS)-4.14 | 8q24.3 | − | 141421638 | TRAPPC9 | NM_001160372 | T |
Table 2.
Distribution of lentiviral integration sites from all investigated iPS clones in comparison to in silico-generated random integration sites. Numbers in bold represent significant differences (p-value ≤ 0.05) between detected insertion sites and in silico-generated random controls.
| Insertion sites (n=75) | In silico | p-value | ||
|---|---|---|---|---|
| Transcription units | 70.7% | 37.5% | p < 0.0001 | |
| Exons | 1.3% | 1.6% | p ≤ 0.6738 | |
| Introns | 69.3% | 36.0% | p < 0.0001 | |
| within 30 kb upstream of 1 or more genes | 1.3% | 9.5% | p ≤ 0.0046 | |
| within 30 kb downstream of 1 or more genes | 6.7% | 5.4% | p ≤ 0.3830 | |
| IS within 30kb window of transcription units | 78.7% | 52.4% | p < 0.0001 | |
| Integration site contained w/in CpG island | 0.0% | 0.1% | p ≤ 0.9146 | |
| Integration site contained w/in CpG island, or within 1 kb window on either side | 2.7% | 1.1% | p ≤ 0.1875 | |
| Integration site contained w/in CpG island, or within 5 kb window on either side | 10.7% | 5.3% | p ≤ 0.0444 | |
| Insertion sites relative to gene density w/in a 1Mb window | 0 – 10 genes | 89.3% | 85.8% | p ≤ 0.2438 |
| 11 – 20 genes | 6.7% | 10.4% | p ≤ 0.1993 | |
| 21 genes and up | 4.0% | 3.9% | p ≤ 0.5594 | |
Table 3.
Proximity of vector insertions to oncogenes. Numbers in bold represent significant differences (p-value ≤ 0.05) between detected insertion sites and in silico-generated random controls.
| Insertion sites (n=75) | In silico | p-value | |
|---|---|---|---|
| Number of oncogenes within 30 kb of one or more integration sites | 5.0 | 1.4 | p ≤ 0.013 |
| Number of genes within 30 kb of one or more integration sites | 82.0 | 51.0 | p ≤ 0.002 |
| % genes that are oncogenes within 30 Kb of integration site | 6.1% | 2.7% | p ≤ 0.0854 |
| Number of integration sites within 30 kb of one or more oncogenes | 6.0 | 1.4 | p ≤ 0.0027 |
| Number of integration sites within 30 kb of one or more genes | 59.0 | 39.3 | p < 0.0001 |
| % of sites within 30 Kb of a gene that is an oncogene | 10.2% | 3.4% | p ≤ 0.0275 |
The most important finding was the lack of any common integration sites shared between the different IPS clones. The closest distance between two integration sites found in different clones was 557kb (IPS(IMR90)-2.8 and IPS(FS)-3.7).
Impact of insertions on gene expression
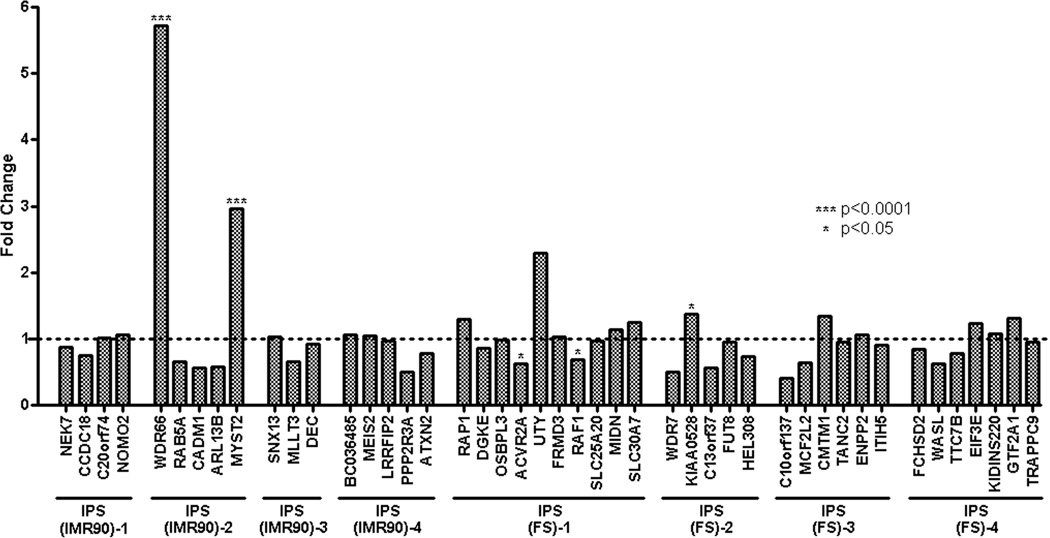
To determine if the lentiviral insertions resulted in a pattern of differential expression of interrupted or nearby genes, we used microarray data from IPS(IMR90)-1 to -4 and IPS(FS)-1 to -4 (GEO: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL5876). The expression arrays (NimbleGen Systems) utilized allowed comparative expression analysis for 45 out of the 53 genes with lentiviral insertions. The expression of genes with IS from each individual clone was compared to either the profile of the remaining 3 clones derived from the same parental cells, or all 7 iPS clones combined. For example, the expression of the genes interrupted by insertions in IPS(IMR90)-1 was compared either to the expression of the same genes in IPS(IMR90)-2, IPS(IMR90)-3 and IPS(IMR90)-4 respectively or IPS(IMR90)-2 to -4 and IPS(FS)-1 to-4. Out of 45 evaluable genes only three (WDR66 and MYST2 in clone IPS(IMR90)-2, p<0.0001 and KIAA0528, p=0.03 in clone IPS(FS)-2) were significantly over-expressed. The expression of two genes in clone FS-1 (ACVR2A, p=0.01 and RAF1, p=0.02) were decreased compared to the expression of the genes in the other clones combined (Figure 2 and Table S2).
Figure 2.
Normalized mRNA Microarray data of the investigated IPS clones were obtained from the NCBI Gene Expression Omnibus (GEO) and used to analyze the impact of proviral integration on gene regulation. The expression data of vector targeted genes of each individual clone were compared to the mRNA expression profile of the remaining 7 IPS clones studied combined. Assuming the same variance between the different data sets expression changes with a p-value ≤ 0.05 were considered as significant. *** p<0.0001, * p<0.05
Functional Analysis of genes targeted by lentiviral integration
The lentiviral-tagged genes were investigated for over-representation of functional pathways in MetaCore™ (GeneGo Inc., St Joseph, MI) and Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA). MetaCore and IPA are web-based software suites incorporating proprietary, manually curated database of biological pathways, networks, interactions and disease biomarkers. Two gene lists were considered for the analysis: genes with integration sites within the coding regions and those having an integration site within a 30kb window. A false discovery rare (FDR) cut-off of p < 0.01 failed to identify significantly enriched biological pathways, indicating that no functional pathways were preferentially targeted by the lentiviral integrations (Figures S1A, S1B, S1C, and S1D).
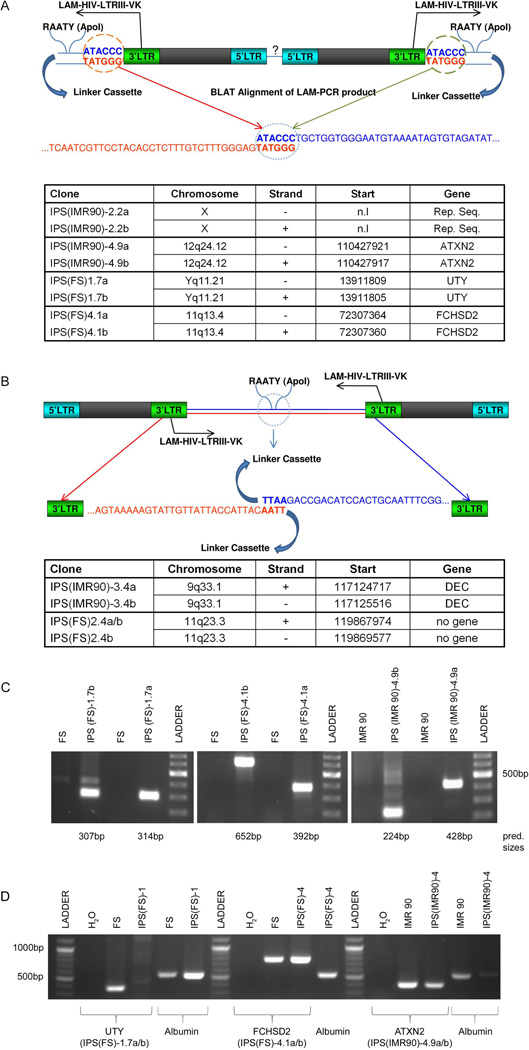
Dual insertion of lentiviral vectors in human iPS cells
Surprisingly, in 6 of 8 iPS clones two independent insertions were found within the same or very close chromosomal location. In what we have termed variant 1 double insertions (IPS(IMR90)-2.2a/b, IPS(IMR90)-4.9a/b, IPS(FS)-1.7a/b, IPS(FS)-4.1a/b) the two proviruses were integrated in the exact same location in reverse orientation head-to-head with the 3’-LTR junctions showing the expected 4–6bp duplication at the integration site (Figure 3 A) [29]. In variant 2 double insertions the integrations were very close but not at identical sites, and were in a tail-to-tail (3’LTR to 3’LTR) configuration, found in IPS(IMR90)-3.4a/b, and IPS(FS)-2.4a/b (Figure 3 B). In order to confirm our LAM-PCR results and the existence of these unusual IS we performed conventional PCR on 3 clones (IPS(IMR90)-4, IPS(FS)-1 and IPS(FS)-4) harboring double insertions using primers within the LTR of the provirus and the gene affected by its integration. In all cases we were able to amplify a specific product of the expected size. Subsequent sequencing of these products confirmed the LAM-PCR results for the location of both integrants in these pairs (Figure 3 C).
Figure 3.
Schematic illustration of the proviral dual insertions. Panel A diagrams the configuration for Variant 1 and Panel B for Variant 2. The table at the bottom of each panel shows the corresponding coordinates of the proviral insertions. Panel C: Confirmation PCR of 3 Variant 1 insertions using one primer within the proviral LTR and one genomic primer within each genomic insertion region identified by LAM-PCR. Panel D: PCR to check the integrity of the genomic loci containing the double insertion sites (same ones as in panel C) in order to verify that each double insertion has both proviruses interrupting the same allele. Primers were chosen to span the proviral insertion sites. Since IPS(FS)-1.7a/b (UTY) is on the Y chromosome, as expected no product was detected. For IPS(FS)-4.1a/b (FCHSD2) and IPS(IMR90)-4.9a/b (ATXN2), both encoded on non-sex chromosomes, the presence of a normal-sized product indicates that only a single allele was disrupted.
Abbreviation: LTR = long terminal repeat, LAM-HIV-LTRIII-VK = primer 2nd exponential PCR (nested PCR), n.l.= not localizable, n.g. = no gene
Although the characteristic duplication at the IS of the variant 1 double insertions indicate that they are truly IS on a single allele, along with the finding of set on a single Y chromosome (IPS(FS)-1.7a/b), we further confirmed the single allele nature of the double integrants by performing PCR using primers spanning the potential site of insertion in IPS(IMR90)-4, IPS(FS)-1and IPS(FS)-4. In all cases we were able to detect an amplicon of the expected size (Figure 3 D) and again confirmed the genomic sequences. This result proves that one allele had no insertions, and thus further suggesting that the double insertions must be on the other allele.
Discussion
Since the pioneering work from Takahashi and Yamanaka [1] first demonstrated the possibility of converting differentiated murine somatic cells into iPS cells with similar characteristics and functional properties as embryonic stem cells via the introduction of a set of transcription factors via viral gene transfer, several groups have confirmed the general principle of reprogramming by applying modified methods to cells of different origins and species, including human and non-human primate cells [2–5,8–10,30]. Even though the reprogramming strategies are rapidly evolving and there are recent reports documenting generation of iPS cells without the use of integrating viral vectors [13,14,31,32], the transfer and expression of the crucial transcription factors via retro- or lentiviral gene transfer remains the most commonly-utilized reprogramming strategy in the preclinical and laboratory setting, due to its robustness and reproducibility. Moreover, the reprogramming efficiency in the episomal and protein transduction studies was substantially lower in comparison to the viral gene delivery methods, raising the possibility that a selection of de novo mutations in the parental cell population contributed to the induction of the iPS phenotype [13,14].
Whether activation or dysregulation of cellular genes caused by genomic vector insertions contributes to or modifies the process of iPS cell generation or properties are important to fully investigate, even if non-integrating delivery systems are eventually utilized for future clinical applications. Are specific integration sites found repeatedly in iPS cell lines, indicating that dysregulation of one or more genes greatly enhance the chance of iPS generation? Or specific gene classes? Are genes near integration sites reproducibly up-or down-regulated?
Our study is the first to investigate the vector insertion profile in human iPS- cell lines. Prior studies reported on integration sites found in murine iPS cells generated utilizing standard MLV retroviral vectors, and did not reveal any recurrent integration sites shared by different iPS clones [33, 34]. Given differences in the ease of transformation of murine versus human cells of all types, and the lower efficiency of iPS generation from human versus murine cells, it is important to carry out insertion studies specifically in human iPS cell lines. Human iPS cells were initially created utilizing either standard MLV or lentiviral HIV vectors [1, 2, 4]. Because MLV vectors have been associated more directly with genotoxicity and insertional proto-oncogene activation in various clinical and experimental systems than lentiviral vectors, we felt iPS cells created utilizing MLV vectors were unlikely to move forward into clinical applications or be utilized by most laboratory investigators [35–37].
We therefore focused on analysis of IS in 8 human iPS cell lines created via transfer of four transcription factors utilizing safety-modified HIV-based lentiviral vectors [2,38]. These vectors drive gene expression with an internal promoter and have the LTR enhancer regions deleted. However, this type of vector has still been shown to perturb the expression of adjacent genes [39, 40]. Even if iPS cells shut down expression of the vector transgenes during the derivation process, it is possible that dysregulation of adjacent genes might persist and impact on iPS properties.
In order to circumvent possible restriction enzyme bias and to increase the probability of identifying all IS present in these clones, we performed independent LAM-PCR reactions using two restriction enzymes. Using this approach, we independently validated 66% of the IS; for the remaining IS the alternative restriction site was either too far or too close from the LTR-genome junction to allow amplification or an unambiguous mapping. The number of IS detected by LAM-PCR was also in very close approximation to the number of IS predicted by Southern blotting (Figure 1 and Table1), providing reassurance that virtually all IS were identified. It is critical with LAM-PCR to use stringent criteria for identification of IS, including presence of the appropriate LTR junctions and at least 95% matching to the human genome for the entire length of the cloned fragment [41]. We found no IS shared between the 8 examined cell lines. Furthermore, mRNA expression analysis did not reveal any overlapping dysregulation of genes near insertion sites. Pathway and network studies showed a wide functional diversity of the lentivirally tagged genes, all indicating that insertional mutagenesis is unlikely to systematically contribute to the reprogramming process.
Proto-oncogenes were over-represented in the list of insertion sites in iPS cells, similar to analysis of the murine dataset obtained by Aoi et al. [33, 39]. The target cells were highly proliferative embryonic or foreskin fibroblasts, and proto-oncogenes may be highly expressed in these target cells and therefore more likely targets. HIV vectors are known to preferentially integrate in genes that are highly expressed [42].
Although we did not detect any IS shared between independent iPS clones, 6 of the 8 clones were found to harbour unusual “double” insertions, with two proviral insertions located very close to each other on the same chromosomal allele (Figure3 A and B). To our knowledge, this phenomenon has not been previously reported in any insertion site survey, neither in our own published analysis of 702 MLV IS and 501 SIV IS in rhesus macaques [43,44], nor in insertion site analyses of cells transduced with HIV vectors similar to those used to create iPS clones [45]. The loci with double insertions did not fall into any particular class of genes, nor were they shared between clones or differentially expressed between clones. This suggests no direct functional link with the success of reprogramming and achieving pluripotency, but instead an epiphenomenon of the lentiviral reprogramming procedures. In order to obtain iPS clones, there is strong positive selection for cells that were able to be simultaneously transduced with four different vectors, unlike previous MLV and HIV experiments, where integration profiles were carried out on cell populations harbouring fewer copies, and without selection pressure for a high copy number per cell. The target cells were exposed to high titer stocks of all vectors, and only those that successfully integrated and expressed proviruses carrying all four transgenes were able to grow out as iPS cells. Most of the cells had more than one copy of each vector. Possibly in cells successfully transduced simultaneously with so many vector copies events occur during integration process that somehow favor nearby dual integrants, for instance more than one provirus utilizing the same molecular site as part of the integrase complex, resulting in adjacent integration after tethering to chromatin. Successful reprogramming to fully functional iPS cells not only requires the correct stoichiometrical expression of the factors, but also the subsequent silencing of the viral vectors [46]. It is possible that one or both requirements are better achieved with proviruses integrated close together [47].
Conclusion
In conclusion, although the sample size of our study as well as of prior studies in the murine system [33, 34] does not unequivocally rule out a contribution of insertional mutagenesis via proviral insertion on the reprogramming of somatic cells to iPS cells, we found no evidence of shared integration site selection between independent iPS clones, and no pattern of gene expression perturbation linked to vector insertions. Thus reprogramming of human somatic cells likely does not depend on activation/deactivation of specific genes induced by lentiviral integration. Our study supports the continued utilization of lentiviral vectors for generation of iPS cells, at least for non-clinical applications, given their improved efficiency compared to most non-integrating vector or protein transfer approaches. Further research is needed to explore the nature and the influence of the double insertions.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Programs of the National Heart, Lung, and Blood Institute and the National Human Genome Research Institute, National Institutes of Health.
Footnotes
Author contributions: T.W.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; A.C.: collection and assembly of data, data analysis; J.Y.M: collection and assembly of data; X.X: collection and assembly of data; A.D.N. and B.B.: assembly of data, data analysis and interpretation; J.E.AB.: provision of study materials. T.G.W. assembly of data, data analysis and interpretation, J.A.T: provision of study materials, conception and design and final approval of manuscript. C.E.D: conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript
Conflict of interest: J.E.AB.: Cellular Dynamics International: Consultancy, Equity Ownership. J.A.T.: Cellular Dynamics International: Equity Ownership, Membership on entity’s Board of Directors. Tactics II Stem Cell Ventures: Consultancy. The other authors have no conflicts of interest to declare.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Liu H, Zhu F, Yong J, et al. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 6.Dimos JT, Rodolfa KT, Niakan KK, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 7.Park IH, Arora N, Huo H, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye Z, Zhan H, Mali P, et al. Human induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009 doi: 10.1182/blood-2009-04-217406. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haase A, Olmer R, Schwanke K, et al. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5:434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 10.Ebert AD, Yu J, Rose FF, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim JB, Zaehres H, Wu G, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 13.Yu J, Hu K, Smuga-Otto K, et al. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou H, Wu S, Joo JY, et al. Generation of Induced Pluripotent Stem Cells Using Recombinant Proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsukahara T, Agawa H, Matsumoto S, et al. Murine leukemia virus vector integration favors promoter regions and regional hot spots in a human T-cell line. Biochem Biophys Res Commun. 2006;345:1099–1107. doi: 10.1016/j.bbrc.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Hematti P, Hong BK, Ferguson C, et al. Distinct Genomic Integration of MLV and SIV Vectors in Primate Hematopoietic Stem and Progenitor Cells. PLoS Biology. 2004;2:2181–2190. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bushman F, Lewinski M, Ciuffi A, et al. Genome-wide analysis of retroviral DNA integration. Nat Rev Micro. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell RS, Beitzel BF, Schroder ARW, et al. Retroviral DNA Integration: ASLV, HIV, and MLV Show Distinct Target Site Preferences. PLoS Biology. 2004;2:1127–1137. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Dullmann J, Schiedlmeier B, et al. Murine Leukemia Induced by Retroviral Gene Marking. Science. 2002;296:497–497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- 21.Seggewiss R, Pittaluga S, Adler RL, et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood. 2006;107:3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kustikova O, Fehse B, Modlich U, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- 23.Hacein-Bey-Abina S, von KC, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Wilson GF, Soerens AG, et al. Functional Cardiomyocytes Derived From Human Induced Pluripotent Stem Cells. Circ Res. 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt M, Schwarzwaelder K, Bartholomae C, et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat Meth. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 26.Wang GP, Garrigue A, Ciuffi A, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49–e61. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends in Genetics. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Wang GP, Levine BL, Binder GK, et al. Analysis of Lentiviral Vector Integration in HIV+ Study Subjects Receiving Autologous Infusions of Gene Modified CD4+ T Cells. Mol Ther. 2009;17:844–850. doi: 10.1038/mt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coffin JM. Retroviridae: The Viruses and Their Replication. In: Fields BN, Knipe DM, Howley PM, editors. Virology. 3rd ed. Philadelphia: Lippincott-Raven; 1996. pp. 1767–1847. [Google Scholar]
- 30.Carey BW, Markoulaki S, Hanna J, et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proceedings of the National Academy of Sciences. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okita K, Nakagawa M, Hyenjong H, et al. Generation of Mouse Induced Pluripotent Stem Cells Without Viral Vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 32.Stadtfeld M, Nagaya M, Utikal J, et al. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aoi T, Yae K, Nakagawa M, et al. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 34.Varas F, Stadtfeld M, De Andres-Aguayo L, et al. Fibroblast derived induced pluripotent stem cells show no common retroviral vector insertions. Stem Cells. 2008;27:300–306. doi: 10.1634/stemcells.2008-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montini E, Cesana D, Schmidt M, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotech. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 36.Montini E, Cesana D, Schmidt M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Ma Y, Ramezani A, Lewis R, et al. High-Level Sustained Transgene Expression in Human Embryonic Stem Cells Using Lentiviral Vectors. Stem Cells. 2003;21:111–117. doi: 10.1634/stemcells.21-1-111. [DOI] [PubMed] [Google Scholar]
- 39.Hawley RG. Does Retroviral Insertional Mutagenesis Play a Role in the Generation of Induced Pluripotent Stem Cells? Mol Ther. 2008;16:1354–1355. doi: 10.1038/mt.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hargrove PW, Kepes S, Hanawa H, et al. Globin Lentiviral Vector Insertions Can Perturb the Expression of Endogenous Genes in [beta]-thalassemic Hematopoietic Cells. Mol Ther. 2008;16:525–533. doi: 10.1038/sj.mt.6300394. [DOI] [PubMed] [Google Scholar]
- 41.Gabriel R, Eckenberg R, Paruzynski A, et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med. 2009;12:1431–1436. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- 42.Schroder AR, Shinn P, Chen H, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 43.Calmels B, Ferguson C, Laukkanen MO, et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim YJ, Kim YS, Larochelle A, et al. Sustained high-level polyclonal hematopoietic marking and transgene expression 4 years after autologous transplantation of rhesus macaques with SIV lentiviral vector-transduced CD34+ cells. Blood. 2009;113:5434–5443. doi: 10.1182/blood-2008-10-185199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cattoglio C, Facchini G, Sartori D, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- 46.Papapetrou EP, Tomishima MJ, Chambers SM, et al. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc.Natl.Acad.Sci.U.S.A. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hotta A, Ellis J. Retroviral vector silencing during iPS cell induction: an epigenetic beacon that signals distinct pluripotent states. J Cell Biochem. 2008;105:940–948. doi: 10.1002/jcb.21912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.