Abstract
Myc proteins are deeply involved in multiple biological processes including cell proliferation, growth, metabolism, apoptosis, differentiation and tumorigenesis. Paradoxically, Myc proteins have been found to be capable of both inhibiting and facilitating differentiation depending on the biological context. Recently we identified a new mode of Myc regulation in differentiating muscle cells in which c-Myc protein is proteolytically cleaved by calcium-dependent calpains in the cytoplasm. This cleavage serves two purposes. First, it inactivates the transcriptional function of Myc by removing its C-terminus, a region responsible for the interaction of Myc with Max and DNA . Second, it alters cytoskeletal architecture and accelerates muscle differentiation through the activity of the remaining N-terminal cleavage product (termed Myc-nick). Here we discuss the roles and regulation of full-length Myc and Myc-nick in terminal differentiation and propose a model in which calpain-mediated cleavage of Myc operates as a functional switch.
Key words: myc, differentiation, muscle, calpain, tubulin acetylation, acetyltransferase, GCN5
Introduction
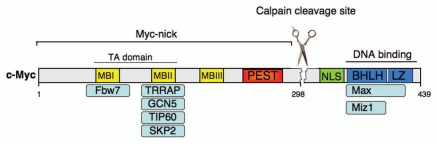
The Myc family of transcription factors is comprised of c-Myc, N-Myc and L-Myc, with c- and N-Myc being essential genes. Myc proteins activate the expression of multiple genes1,2 and microRNAs.3–6 To promote transcriptional activation, Myc proteins form heterodimers with Max and recruit chromatin-modifying complexes to E-box sequences proximal to specific gene promoters. Myc promotes widespread chromatin changes that facilitate transcription initiation and transcript elongation.7–9 In addition, Myc also mediates transcriptional repression by inhibiting the transcriptional activator Miz1.10 Myc proteins contain several conserved domains that are essential for their function as transcription factors (Fig. 1). Myc contains an N-terminally located transactivation domain spanning MBI-MBII (conserved regions known as Myc Boxes). The C-terminal region of Myc harbors a Basic Helix Loop Helix Leucine Zipper (BHLH-LZ) motif responsible for interaction with DNA and Max (the obligatory dimerization partner of Myc).
Figure 1.
Full-length human c-Myc protein and its binding partners. The scissors indicate the major calpain cleavage site within human c-Myc. Myc boxes (MB) and Basic Helix Loop Helix Leucine Zipper (BHLH LZ) domains are highly conserved among Myc family members. NLS, nuclear localization sequence; PEST, region containing amino acids often found in unstable proteins (pro, glu, ser, thr).
While normal Myc levels are essential for embryonic development and cellular homeostasis, aberrant upregulation of Myc can lead to apoptosis or malignant transformation. Indeed, Myc deregulation is closely associated with the genesis of a very broad range of human tumors.11 To avoid outcomes such as apoptosis and oncogenic transformation, cells have developed multiple mechanisms to tightly control the levels of Myc at the transcriptional, translational and posttranslational levels. Normally Myc proteins have a half-life of about 20 minutes,12 being degraded by the ubiquitin proteasome system in the nucleus.13,14 Several E3 ligases including Fbw7, Skp2, HectH9/Huwe1 and TRPC4AP/TRUSS were shown to ubiquitinate Myc and target it for proteasomal degradation.15–19 On the other hand, the ubiquitination of Myc by SCF(β-TrCP) stabilizes Myc by antagonizing its Fbw7-mediated degradation.20 Therefore, Myc abundance is modulated by several ubiquitination complexes that target Myc in response to specific signals or as a function of cell cycle phase.
Cytoplasmic Cleavage of Myc by Calcium-Activated Calpains
Recently we found that calcium dependent proteases belonging to the calpain family are also involved in regulating the levels of transcriptionally active c-Myc and N-Myc.21 Unlike the proteasomal degradation of Myc that occurs in the nucleus and leads to total destruction of the protein, the cleavage of Myc by calpains occurs in the cytoplasm and generates a large N-terminal segment of Myc that we named Myc-nick. Upon calpain cleavage the C-terminus of Myc, containing the nuclear localization sequence (NLS) and the BHLH-LZ, is rapidly degraded, while the N-terminal 298 residue segment (Myc-nick) is preserved and can be readily detected in the cytoplasm (Fig. 1). The removal of the C-terminus of Myc, which is essential for binding to DNA and Max, renders Myc-nick transcriptionally inactive. Endogenous Myc-nick is constitutively and ubiquitously expressed in cultured cells. However, the proportion of Myc-nick to full-length Myc increases when cells are cultured at high density and during muscle cell differentiation, conditions that hyperactivate calpains.21 These findings are in agreement with earlier studies in which calpain inactivation was linked to increased Myc levels.22–24
Calpain Activation is Involved in Terminal Differentiation
Calpains are a family of calcium activated cysteine proteases that function at neutral pH (see Sorimachi et al. for a recent review on calpains).25 Numerous proteins have been identified as calpain substrates including cytoskeletal proteins and transcription factors.26 Given the diversity of these substrates, it is not surprising that calpains have been linked to multiple biological processes. Defects in calpain function or expression can cause a variety of dysfunctions including tumorigenesis, muscle dystrophy and lethality. While hyperactivation of calpains is associated with increased tumorigenesis,27 regulated increases in calpain activity are essential for terminal differentiation of multiple cell types. Calpains are upregulated and activated during differentiation of keratinocytes,28 hematopoietic29 and muscle cells,30,31 among others. Importantly, mutations in the muscle-specific calpain 3, which impair its proteolytic activity, are the sole cause of limb girdle muscular dystrophy 2A (LGMD2A).32,33 In addition, loss of function mutation of calpain r (the regulatory subunit of calpains) is lethal due to impaired cardiovascular development.34 Moreover, the targeted deletion of calpain r in cells of the chondrocyte lineage impairs chondrocyte proliferation and differentiation.35 Together these studies imply that calpain activity is critical for terminal differentiation. Our finding that Myc is cleaved by calpains led us to examine a possible function for Myc-nick in differentiation.
Myc Regulates Terminal Differentiation at Several Levels
As a major transcriptional driver of cell growth and proliferation myc gene expression is strongly downregulated during terminal differentiation, consistent with the idea that Myc negatively regulates differentiation. Indeed, many early experiments using cell lines demonstrated that ectopic Myc blocks terminal differentiation. However, more recent work has provided a more complex picture in which, dependent on the biological context, Myc can either block or facilitate differentiation. Below we discuss in vivo and in vitro studies that together suggest a dual role for Myc in terminal differentiation.
Myc attenuates terminal differentiation.
Myc promotes proliferation and maintains pluripotency of stem and progenitor cells.36,37 In addition, numerous studies have established that myc is transcriptionally downregulated during terminal differentiation.38 Consistent with these findings, ectopic expression of Myc inhibits terminal differentiation of multiple cell types in culture and promotes tumorigenesis in vivo when expressed in differentiated cells.39,40 Moreover, the targeted deletion of N-Myc in mice induces the premature differentiation of cortical neuronal progenitor cells.41 Myc is thought to block differentiation at the transcriptional level by activating genes involved in cell proliferation and metabolism and repressing genes involved in differentiation.4,6,42 In addition, in murine ES cells c-Myc induces microRNAs that in turn silence expression of differentiation markers thereby modulating the rate of differentiation.4 Another mechanism of Myc inactivation is likely to relate to the generation of Myc-nick. We found that during myoblast differentiation, calpains are activated and cleave Myc to produce the transcriptionally inactive form of Myc (Myc-nick) as described above. Since calpains do not require ATP, calpain cleavage of Myc is a rapid and energy-efficient way of downregulating the population of transcriptionally functional Myc proteins. We speculate that downregulation of myc gene transcription, ubiquitin-dependent proteolysis, and calpain cleavage may all cooperate to achieve the downregulation of full-length Myc that accompanies terminal differentiation.
For almost 20 years, different labs have reported a change in Myc localization from nuclear to cytoplasmic in diverse types of differentiating cells. For neuronal cells, cytoplasmic Myc signal was detected in neural crest, retinal ganglion cells, neurons of spinal ganglia43,44 and Purkinje cells.44,45 Similar observations were made in chondrocytes where c-Myc was nuclear in proliferating cells, but was primarily cytoplasmic in mature chondrocytes.46 In endometrial carcinomas, c-Myc was nuclear in poorly differentiated forms and cytoplasmic in well-differentiated carcinomatous endometrium.47 c-Myc was also detected in the cytoplasm of differentiating ML-1 human myeloid leukemia cells.48 Most of these observations were largely based on immunohistochemistry studies and did not describe the apparent molecular size or other characteristics of the Myc protein expressed in each tissue. Based on our experiments, we posit that Myc-nick is the major form of Myc previously reported to accumulate in the cytoplasm of differentiated cells.
Myc promotes differentiation.
While downregulation of Myc has been found to be important for terminal differentiation, there is considerable evidence that Myc is required for cells to progress along a differentiation lineage. This was shown in systems where commitment to a specific lineage is linked to increased proliferation.49–52 For example, by employing targeted deletions of murine c- and N-myc, Habib et al. demonstrated that Myc is essential for pre-B-cell proliferation and differentiation. This was found to involve a Myc-dependent increase in the levels of intracellular calcium. In addition, while the conditional knockout of c-Myc in hematopoietic stem cells (HSC) leads to diminished differentiation and an increase in HSCs, the overexpression of Myc induces HSC differentiation.51 In this system, Myc is believed to downregulate the adhesion of progenitor cells to the stem cell niche, allowing them to detach and differentiate. One of the best-documented examples of Myc-mediated differentiation occurs in keratinocytes where the overexpression of Myc was shown to stimulate keratinocyte differentiation in vivo.49,50,53,54 Interestingly, keratinocytes are not sensitive to Myc induced apoptosis49 and the increased differentiation was suggested to function as a protective mechanism against uncontrolled proliferation induced by Myc.55 Further evidence that high levels of Myc are conducive to differentiation comes from the observation that neuroblastoma cells with amplified N-Myc can still undergo differentiation.56 Moreover, the knock down of c-Myc in Xenopus results in the failure to form neural crest-derived structures. Here the block to differentiation caused by the downregulation of Myc was found to be independent of cell proliferation or death.57 Together these studies provide strong evidence for a positive role of Myc in driving differentiation. Since we found that Myc-nick is the form of Myc that is increased during differentiation of muscle cells, we explored the possibility that Myc-nick plays a positive role in terminal differentiation.
Myc-nick Regulates the Microtubule Cytoskeleton and Promotes Muscle Differentiation
As shown in Figure 2, extracts from a number of differentiated tissues are capable of converting full-length c-Myc into Myc-nick. Among these tissues, skeletal muscle extracts display the highest cleavage activity towards Myc (Fig. 2), which was blocked by calpain inhibitors (data not shown). We also found elevated levels of Myc-nick in skeletal muscles.21 Importantly, we observed that overexpression of Myc-nick in both primary and cultured myoblasts accelerates their differentiation and increases expression of muscle specific markers such as tropomyosin, troponin, myosin heavy chain and desmin. Moreover, while the expression of MyoD is sufficient to transdifferentiate wild type fibroblasts into muscle cells, MyoD is unable to transdifferentiate myc-null fibroblasts.21 This indicates that the presence of the myc gene is necessary for MyoD induced transdifferentiation. However, MyoD was capable of inducing transdifferentiation in myc-null fibroblasts expressing Myc-nick, indicating that Mycnick is sufficient to compensate for the lack of the myc gene in this biological setting.
Figure 2.
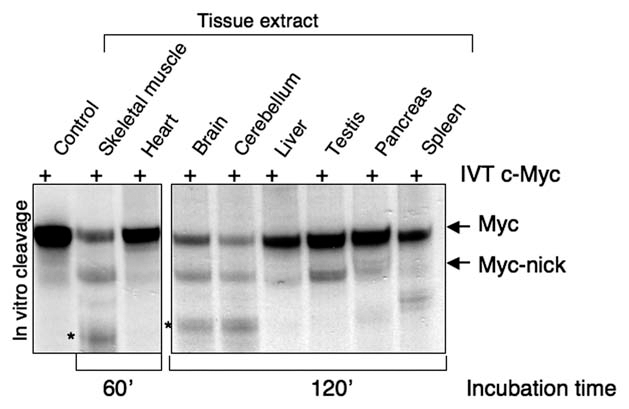
Skeletal muscle, brain and cerebellar extracts display Myc cleavage activity. To assess the cleavage of c-Myc in different tissues, 1 µl of radiolabeled IVT c-Myc was incubated with 30 µg of mouse tissue extracts for the indicated time points (see reference 21 for protocol details). After incubation, the samples were processed for autoradiography to visualize radioactive c-Myc. Tissue extracts were prepared from snap frozen adult mouse tissues in the absence of protease inhibitors. The control lane contains 1 µl of radiolabeled IVT c-Myc used as an input in every reaction. The asterisks indicate a calpain cleavage product of Myc-nick often generated in vitro.
What is the function of Myc-nick during differentiation?
Our work suggests that Myc-nick plays an active role in muscle differentiation by promoting changes in the cytoskeleton that result from increased levels of acetylated α-tubulin. In many cell types, differentiation is accompanied by microtubule stabilization and an increase in tubulin modifications,58 predominantly acetylation of α-tubulin. This modification correlates with neuronal,59,60 muscle,61 and keratinocyte62 differentiation. Previous studies had shown that the N terminus of c-Myc interacts with tubulins.63–65 We found that Mycnick forms a complex with α-tubulin and the acetyltransferase GCN5 to induce α-tubulin acetylation and thereby promote changes in cell morphology. This is analogous to the role played by full-length Myc in transcriptional activation, where Myc forms a complex with acetyltransferases such as GCN5 and TIP60 to acetylate histone tails.66,67 Recently, several acetyltransferases were found to promote α-tubulin acetylation in different cellular contexts. These include ELP3,59 ARD1,68 NAT 10,69 and three members of the GNAT (Gcn5-Related N acetyltransferase) family of acetyltransferases: GCN5,21 MEC17,70 and α-TAT1.71 Two enzymes have been shown to deacetylate α-tubulin, HDAC6 and Sirt2;72–74 however, we have found the induction of α-tubulin acetylation by Myc-nick to be independent of HDACs. Acetylated α-tubulin accumulates in stable microtubules, often found in microtubule based specialized structures such as primary cilium, axon and mitotic spindle. Acetylation of α-tubulin has recently emerged as an important regulator of microtubule function. For a recent review on tubulin acetylation, regulation and function see Perdiz et al., 2010.75
In addition to muscle differentiation, the regulation of Myc function by calpain cleavage is likely to play a role in the differentiation of other cell types. For example, we recently found that Myc-nick is highly expressed in brain and cerebellum, tissues that also display high calpain activity towards Myc in vitro (Fig. 2) and require tubulin acetylation in order to differentiate.59 In these tissues, Myc-nick may be involved in mediating at least some of the functions attributed to the myc gene in terminal differentiation and tissue maintenance. Furthermore, during keratinocyte differentiation calpains are activated28 and there is an increase in tubulin acetylation,62 suggesting a correlation with Myc-nick production and function. In a keratinocyte model for the autoimmune disease Pemphigus vulgaris, a smaller form of Myc, with a size corresponding to that of Myc-nick, was detected predominantly in the cytoplasm.76 Perhaps the positive role attributed to Myc during keratinocyte differentiation49 is carried out, at least in part, by Myc-nick. In this regard, it is worth noting that a previous study showed that blocking cellular deacetylases with TSA could mimic the effects of Myc in promoting keratinocyte differentiation.77
The evidence outlined above provides considerable support for the idea that protein acetylation may be one of the major functions of Myc during differentiation. In addition to α-tubulin acetylation, we surmise that Myc-nick may be involved in the acetylation of other cytoplasmic proteins that regulate terminal differentiation and we are currently searching for cytoplasmic acetylation targets of Myc-nick. Moreover, we found that the interaction between Myc-nick and the acetyltransferase GCN5 is only partially responsible for Myc-nick-induced muscle differentiation, indicating that Myc-nick may play additional roles in the cytoplasm of differentiating cells. We note, however, that while Myc-nick function may be an important component in Myc-induced terminal differentiation, Myc-nick does not promote proliferation, which is a transcriptional function performed by full-length Myc. Therefore, systems that require cell proliferation to determine lineage commitment may require both full-length Myc and Myc-nick in order for lineage progression to occur.
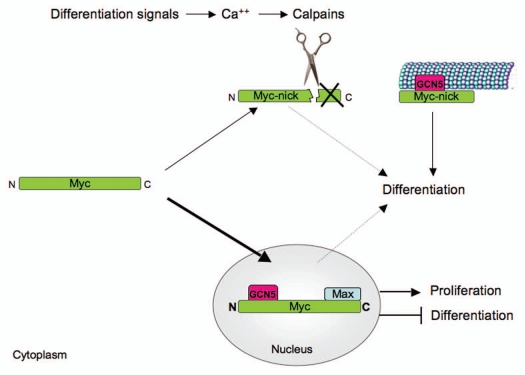
Figure 3 depicts our model for the regulation of Myc by calpain cleavage and for the role of Myc-nick during muscle differentiation. During proliferation, Myc protein is translocated into the nucleus where it activates genes involved in proliferation and represses genes involved in differentiation. In the presence of differentiation signals, there is an increase in intracellular calcium, calpains are activated and Myc is cleaved. This cleavage removes the C-terminus of Myc and produces a transcriptionally inactive form of the protein (Myc-nick). The cleavage of Myc by calpains may cooperate with the transcriptional downregulation of Myc to reduce the pool of transcriptionally active Myc. In the absence of Myc in the nucleus, proliferation genes are downregulated, while genes involved in differentiation are derepressed allowing differentiation to occur. On the other hand, the cleavage of Myc by calpains produces Myc-nick that, in turn, regulates cell morphology and contributes to terminal differentiation by modulating the microtubule cytoskeleton.
Figure 3.
Model for Myc regulation and function during terminal differentiation. When cells are proliferating, Myc protein is efficiently translocated into the nucleus where it activates genes involved in proliferation and represses genes involved in differentiation. In the presence of differentiation signals that promote an increase in intracellular calcium, calpains are activated and cleave Myc removing its C-terminal region (responsible for binding to DNA and to Max). The cleavage of Myc by calpains cooperates with the transcriptional downregulation of Myc to reduce the levels of nuclear Myc. In the absence of Myc in the nucleus, proliferation genes are downregulated while genes involved in differentiation are derepressed allowing differentiation to occur. The cleavage of Myc by calpains also produces Myc-nick that is localized to the cytoplasm and recruits the acetyltransferase GCN5 to microtubules to promote α-tubulin acetylation, thereby regulating cell morphology and contributing to terminal differentiation. The dotted arrows indicate other potential roles, currently under investigation, for full-length Myc and of Myc-nick in terminal differentiation.
Moonlighting Mediated by Calpain Cleavage
Moonlighting, or the ability to perform multiple distinct functions, has been demonstrated to be a property of a surprisingly wide range of proteins including β-actin,78 Ezh2,79 and p53.80,81 Multiple factors such as changes in localization, post-translational modifications and partial proteolytic cleavage can trigger changes in protein function. Cleavage by calpains is well known to cause partial cleavage rather than total protein degradation and may be responsible for increasing the functional diversity of their substrates. For most substrates, the molecular role of the segments produced by calpain cleavage is unknown.
Nevertheless, there are a few examples in the literature where the function of the proteolytic fragment produced by calpain cleavage is well understood. For example, the cleavage of ATG5 converts this cytosolic factor required for the formation of autophagosomes into a mitochondrial proapoptotic factor.82 Moreover, the cleavage of the N terminus of β-catenin that occurs in hippocampal neurons produces a stable form of β-catenin that is insensitive to GSK3β phosphorylation and cannot be degraded by the proteasome. This form of β-catenin preferentially translocates into the nucleus and regulates neuronal plasticity.83 We have now found that Myc is converted into Myc-nick by calpain cleavage switching Myc from a nuclear transcription factor to a cytoplasmic factor that regulates microtubule dynamics. Mycnick is widely expressed and represents a constitutive form of Myc that may play additional functions in the cytoplasm depending on the biological setting. The functional switch caused by calpain cleavage is likely to regulate a large number of substrates, including additional transcription factors, and may emerge as an important permanent post-translational modification that modulates multiple biological processes.
Acknowledgements
We thank Celine Ngouenet for technical assistance and the members of the Eiseman lab for helpful discussions. Work from our laboratory discussed in this review was supported by NIH/NCI grant CA20525 and by a postdoctoral fellowship from EMBO.
References
- 1.Orian A, van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 6.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amati B, Frank SR, Donjerkovic D, Taubert S. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim Biophys Acta. 2001;1471:135–145. doi: 10.1016/s0304-419x(01)00020-8. [DOI] [PubMed] [Google Scholar]
- 9.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc regulates transcriptional pause release. Cell. 141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleine-Kohlbrecher D, Adhikary S, Eilers M. Mechanisms of transcriptional repression by Myc. Curr Top Microbiol Immunol. 2006;302:51–62. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 12.Hann SR, Eisenman RN. Proteins encoded by the human c-myc oncogene: Differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: Cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross-Mesilaty S, Reinstein E, Bercovich B, Tobias KE, Schwartz AL, Kahana C, et al. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc Natl Acad Sci USA. 1998;95:8058–8063. doi: 10.1073/pnas.95.14.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi SH, Wright JB, Gerber SA, Cole MD. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 2010;24:1236–1241. doi: 10.1101/gad.1920310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 18.von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhao X, Heng JI, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popov N, Schulein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(beta-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol. 2010;12:973–981. doi: 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- 21.Conacci-Sorrel M, Ngouenet C, Eisenman RN. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell. 2010;142:480–493. doi: 10.1016/j.cell.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Small GW, Chou TY, Dang CV, Orlowski RZ. Evidence for involvement of calpain in c-Myc proteolysis in vivo. Arch Biochem Biophys. 2002;400:151–161. doi: 10.1016/S0003-9861(02)00005-X. [DOI] [PubMed] [Google Scholar]
- 23.Gonen H, Shkedy D, Barnoy S, Kosower NS, Ciechanover A. On the involvement of calpains in the degradation of the tumor suppressor protein p53. FEBS Lett. 1997;406:17–22. doi: 10.1016/s0014-5793(97)00225-1. [DOI] [PubMed] [Google Scholar]
- 24.Watt F, Molloy PL. Specific cleavage of transcription factors by the thiol protease, m-calpain. Nucleic Acids Res. 1993;21:5092–5100. doi: 10.1093/nar/21.22.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorimachi H, Hata S, Ono Y. Expanding members and roles of the calpain superfamily and their genetically modified animals. Exp Anim. 2010;59:549–566. doi: 10.1538/expanim.59.549. [DOI] [PubMed] [Google Scholar]
- 26.Tompa P, Buzder-Lantos P, Tantos A, Farkas A, Szilagyi A, Banoczi Z, et al. On the sequential determinants of calpain cleavage. J Biol Chem. 2004;279:20775–20785. doi: 10.1074/jbc.M313873200. [DOI] [PubMed] [Google Scholar]
- 27.Kimura Y, Koga H, Araki N, Mugita N, Fujita N, Takeshima H, et al. The involvement of calpain-dependent proteolysis of the tumor suppressor NF2 (merlin) in schwannomas and meningiomas. Nat Med. 1998;4:915–922. doi: 10.1038/nm0898-915. [DOI] [PubMed] [Google Scholar]
- 28.Garach-Jehoshua O, Ravid A, Liberman UA, Reichrath J, Glaser T, Koren R. Upregulation of the calcium-dependent protease, calpain, during keratinocyte differentiation. Br J Dermatol. 1998;139:950–957. doi: 10.1046/j.1365-2133.1998.02548.x. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura M, Mori M, Morishita Y, Mori S, Kawashima S. Specific increase in calcium-activated neutral protease with low calcium sensitivity (m-calpain) in proerythroblastic K562 cell line cells induced to differentiation by phorbol-12-myristate 13-acetate. Exp Cell Res. 1992;200:513–522. doi: 10.1016/0014-4827(92)90203-k. [DOI] [PubMed] [Google Scholar]
- 30.Dedieu S, Poussard S, Mazeres G, Grise F, Dargelos E, Cottin P, et al. Myoblast migration is regulated by calpain through its involvement in cell attachment and cytoskeletal organization. Exp Cell Res. 2004;292:187–200. doi: 10.1016/j.yexcr.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 31.Kumar A, Shafiq S, Wadgaonkar R, Stracher A. The effect of protease inhibitors, leupeptin and E64d, on differentiation of C2C12 myoblasts in tissue culture. Cell Mol Biol. 1992;38:477–483. [PubMed] [Google Scholar]
- 32.Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, et al. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 33.Ojima K, Kawabata Y, Nakao H, Nakao K, Doi N, Kitamura F, et al. Dynamic distribution of muscle-specific calpain in mice has a key role in physical-stress adaptation and is impaired in muscular dystrophy. J Clin Invest. 2010;120:2672–2683. doi: 10.1172/JCI40658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arthur JS, Elce JS, Hegadorn C, Williams K, Greer PA. Disruption of the murine calpain small subunit gene, Capn4: calpain is essential for embryonic development but not for cell growth and division. Mol Cell Biol. 2000;20:4474–4481. doi: 10.1128/mcb.20.12.4474-4481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kashiwagi A, Schipani E, Fein MJ, Greer PA, Shimada M. Targeted deletion of Capn4 in cells of the chondrocyte lineage impairs chondrocyte proliferation and differentiation. Mol Cell Biol. 2010;30:2799–2810. doi: 10.1128/MCB.00157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 37.Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 38.Leon J, Ferrandiz N, Acosta JC, Delgado MD. Inhibition of cell differentiation: A critical mechanism for MYC-mediated carcinogenesis? Cell Cycle. 2009;8:1148–1157. doi: 10.4161/cc.8.8.8126. [DOI] [PubMed] [Google Scholar]
- 39.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 40.Giuriato S, Ryeom S, Fan AC, Bachireddy P, Lynch RC, Rioth MJ, et al. Sustained regression of tumors upon MYC inactivation requires p53 or thrombospondin-1 to reverse the angiogenic switch. Proc Natl Acad Sci USA. 2006;103:16266–16271. doi: 10.1073/pnas.0608017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakamatsu Y, Watanabe Y, Nakamura H, Kondoh H. Regulation of the neural crest cell fate by N-myc: Promotion of ventral migration and neuronal differentiation. Development. 1997;124:1953–1962. doi: 10.1242/dev.124.10.1953. [DOI] [PubMed] [Google Scholar]
- 44.Wakamatsu Y, Watanabe Y, Shimono A, Kondoh H. Transition of localization of the N-Myc protein from nucleus to cytoplasm in differentiating neurons. Neuron. 1993;10:1–9. doi: 10.1016/0896-6273(93)90236-k. [DOI] [PubMed] [Google Scholar]
- 45.Okano HJ, Park WY, Corradi JP, Darnell RB. The cytoplasmic Purkinje onconeural antigen cdr2 downregulates c-Myc function: Implications for neuronal and tumor cell survival. Genes Dev. 1999;13:2087–2097. doi: 10.1101/gad.13.16.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Toury R, Hauchecorne M, Balmain N. Expression and subcellular localization of the Myc superfamily proteins: c-Myc, Max, Mad1 and Mxi1 in the epiphyseal plate cartilage chondrocytes of growing rats. Cell Mol Biol (Noisy-le-grand) 1997;43:175–188. [PubMed] [Google Scholar]
- 47.Bai MK, Costopoulos JS, Christoforidou BP, Papadimitriou CS. Immunohistochemical detection of the c-myc oncogene product in normal, hyperplastic and carcinomatous endometrium. Oncology. 1994;51:314–319. doi: 10.1159/000227356. [DOI] [PubMed] [Google Scholar]
- 48.Craig RW, Buchan HL, Civin CI, Kastan MB. Altered cytoplasmic/nuclear distribution of the c-myc protein in differentiating ML-1 human myeloid leukemia cells. Cell Growth Differ. 1993;4:349–357. [PubMed] [Google Scholar]
- 49.Gandarillas A, Watt FM. c-Myc promotes differentiation of human epidermal stem cells. Genes Dev. 1997;11:2869–2882. doi: 10.1101/gad.11.21.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frye M, Gardner C, Li ER, Arnold I, Watt FM. Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development. 2003;130:2793–2808. doi: 10.1242/dev.00462. [DOI] [PubMed] [Google Scholar]
- 51.Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Habib T, Park H, Tsang M, de Alboran IM, Nicks A, Wilson L, et al. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J Cell Biol. 2007;179:717–731. doi: 10.1083/jcb.200704173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berta MA, Baker CM, Cottle DL, Watt FM. Dose and context dependent effects of Myc on epidermal stem cell proliferation and differentiation. EMBO Mol Med. 2010;2:16–25. doi: 10.1002/emmm.200900047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gebhardt A, Kosan C, Herkert B, Moroy T, Lutz W, Eilers M, et al. Miz1 is required for hair follicle structure and hair morphogenesis. J Cell Sci. 2007;120:2586–2593. doi: 10.1242/jcs.007104. [DOI] [PubMed] [Google Scholar]
- 55.Watt FM, Frye M, Benitah SA. MYC in mammalian epidermis: How can an oncogene stimulate differentiation? Nat Rev Cancer. 2008;8:234–242. doi: 10.1038/nrc2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rouah E, Wilson DR, Armstrong DL, Darlington GJ. N-myc amplification and neuronal differentiation in human primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1989;49:1797–1801. [PubMed] [Google Scholar]
- 57.Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in xenopus. Dev Cell. 2003;4:827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 58.Bulinski JC, Gundersen GG. Stabilization of post-translational modification of microtubules during cellular morphogenesis. Bioessays. 1991;13:285–293. doi: 10.1002/bies.950130605. [DOI] [PubMed] [Google Scholar]
- 59.Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 60.Witte H, Neukirchen D, Bradke F. Microtubule stabilization specifies initial neuronal polarization. J Cell Biol. 2008;180:619–632. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gundersen GG, Khawaja S, Bulinski JC. Generation of a stable, posttranslationally modified microtubule array is an early event in myogenic differentiation. J Cell Biol. 1989;109:2275–2288. doi: 10.1083/jcb.109.5.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee WH, Kim JY, Kim YS, Song HJ, Song KJ, Song JW, et al. Upregulation of class II beta-tubulin expression in differentiating keratinocytes. J Invest Dermatol. 2005;124:291–297. doi: 10.1111/j.0022-202X.2004.23506.x. [DOI] [PubMed] [Google Scholar]
- 63.Alexandrova N, Niklinski J, Bliskovsky V, Otterson GA, Blake M, Kaye FJ, et al. The N-terminal domain of c-Myc associates with alpha-tubulin and microtubules in vivo and in vitro. Mol Cell Biol. 1995;15:5188–5195. doi: 10.1128/mcb.15.9.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niklinski J, Claassen G, Meyers C, Gregory MA, Allegra CJ, Kaye FJ, et al. Disruption of Myc-tubulin interaction by hyperphosphorylation of c-Myc during mitosis or by constitutive hyperphosphorylation of mutant c-Myc in Burkitt's lymphoma. Mol Cell Biol. 2000;20:5276–5284. doi: 10.1128/mcb.20.14.5276-5284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gatti G, Maresca G, Natoli M, Florenzano F, Nicolin A, Felsani A, et al. MYC prevents apoptosis and enhances endoreduplication induced by paclitaxel. PLoS One. 2009;4:5442. doi: 10.1371/journal.pone.0005442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, et al. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohkawa N, Sugisaki S, Tokunaga E, Fujitani K, Hayasaka T, Setou M, et al. N-acetyltransferase ARD1-NAT1 regulates neuronal dendritic development. Genes Cells. 2008;13:1171–1183. doi: 10.1111/j.1365-2443.2008.01235.x. [DOI] [PubMed] [Google Scholar]
- 69.Shen Q, Zheng X, McNutt MA, Guang L, Sun Y, Wang J, et al. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp Cell Res. 2009;315:1653–1667. doi: 10.1016/j.yexcr.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 70.Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, et al. MEC-17 is an alphatubulin acetyltransferase. Nature. 2010;467:218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major {alpha}-tubulin K40 acetyltransferase {alpha}TAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 73.Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 75.Perdiz D, Mackeh R, Pous C, Baillet A. The ins and outs of tubulin acetylation: More than just a post-translational modification? Cell Signal. 2010 doi: 10.1016/j.cellsig.2010.10.014. In Press. [DOI] [PubMed] [Google Scholar]
- 76.Williamson L, Raess NA, Caldelari R, Zakher A, de Bruin A, Posthaus H, et al. Pemphigus vulgaris identifies plakoglobin as key suppressor of c-Myc in the skin. EMBO J. 2006;25:3298–3309. doi: 10.1038/sj.emboj.7601224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frye M, Fisher AG, Watt FM. Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS One. 2007;2:763. doi: 10.1371/journal.pone.0000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hofmann WA, de Lanerolle P. Nuclear actin: To polymerize or not to polymerize. J Cell Biol. 2006;172:495–496. doi: 10.1083/jcb.200601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 80.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 81.Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 82.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 83.Abe K, Takeichi M. NMDA-receptor activation induces calpain-mediated beta-catenin cleavages for triggering gene expression. Neuron. 2007;53:387–397. doi: 10.1016/j.neuron.2007.01.016. [DOI] [PubMed] [Google Scholar]