Abstract
Trinucleotide repeats (TNRs) are a blessing and a curse. In coding regions, where they are enriched, short repeats offer the potential for continuous, rapid length variation with linked incremental changes in the activity of the encoded protein, a valuable source of variation for evolution. But at the upper end of these benign and beneficial lengths, trinucleotide repeats become very unstable, with a dangerous bias toward continual expansion, which can lead to neurological diseases in humans. The mechanisms of expansion are varied and the links to disease are complex. Where they have been delineated, however, they have often revealed unexpected, fundamental aspects of the underlying cell biology. Nowhere is this more apparent than in recent studies, which indicate that expanded CAG repeats can form toxic sites in the genome, which can, upon interaction with normal components of DNA metabolism, trigger cell death. Here we discuss the phenomenon of TNR-induced DNA toxicity, with special emphasis on the role of transcription. Transcription-induced DNA toxicity may have profound biological consequences, with particular relevance to repeat-associated neurodegenerative diseases.
Key words: DNA toxicity, convergent transcription, trinucleotide repeats, apoptosis, cell cycle, ATR pathway
Introduction
Microsatellite sequences—runs of one to eight nucleotides repeated in tandem—comprise about 3% of the human genome and are unevenly distributed throughout chromosomes.1,2 Clustered microsatellite repeats, for example, form the major sequence elements of chromosomal centromeres and telomeres, while microsatellites of various lengths and numbers of repeats are scattered along the chromosome arms.3,4 A common feature of these repetitive sequences is their instability; they gain or lose repeat units much more rapidly than unique sequences acquire mutations, with rates that depend on the nucleotide organization of the repeats, the length of the repeat unit, the number of repeated units, and the purity of the repeat tract.5 Most events of repeat instability do not have detectable consequences in humans, but repeats that are located in transcription units can alter gene function.
Microsatellites in general tend to be excluded from the coding regions of genes because their instability plays havoc with the reading frame, but trinucleotide repeats (TNRs), whose numbers do not affect the reading frame, tend to be overrepresented in exons.3 The relative abundance of TNRs in coding regions and their selective enrichment in genes for transcription factors and other regulatory proteins suggest that TNRs provide a positive evolutionary benefit.4,6,7 Similar evolutionary advantages may extend to repeats located in promoters and untranslated portions of transcripts, where repeat variation may fine tune gene expression.8 These benefits, however, come with a cost.
At more than 20 loci in the human genome, the lengthening of a TNR tract beyond some threshold causes a neurodegenerative or neuromuscular disorder.9–11 Long TNRs tend to become unstable during their transmission to offspring, giving rise to progeny with longer repeat tracts (expansions) or shorter ones (contractions). The typical bias toward expansions leads to progressively more severe disease symptoms in the progeny in each subsequent generation. In addition, as affected individuals age, the continuing expansion-biased repeat instability in somatic tissues, especially brain, may accelerate neuron dysfunction and death, hastening the onset and progression of disease symptoms.12 The mechanisms of repeat instability in germline and somatic tissues and the basis for cell death have been the subjects of intense study over the past two decades.9,13–16
Early experiments in cells and model organisms identified DNA replication, recombination and DNA repair, as potential contributors, singly or in combination, to the repeat instability observed in humans.14,17 Each of these DNA metabolic processes exposes single strands of repeats, which can form secondary structures such as hairpins and slipped-strand duplexes18,19 that may constitute the key, common event leading to changes in repeat-tract length. More recently, transcription through a repeat has also been shown to stimulate repeat instability, presumably by exposing single strand to allow secondary structure formation at repeats and thereby engaging DNA repair processes to generate changes in repeat-tract length.20–23
Among these processes, transcription occupies a unique place. Most TNR-associated disease genes are ubiquitously expressed in mammals, with expression levels that vary among tissues and at different developmental stages.21 Unlike replication, for example, transcription occurs in both proliferating and terminally differentiated cells, both of which display repeat instability.14,15,24 And TNR disease genes are much more frequently transcribed than they are replicated, repaired or recombined. Added to these features, the puzzling phenomenon of antisense transcription, with its inherent possibilities for head-on collisions between converging sense and antisense RNA polymerase II (RNAPII) complexes, has been detected in most TNR-disease genes.25,26 In model systems, convergent transcription not only synergistically enhances repeat instability, but can also trigger cell cycle arrest and cell death.27
By analogy with ‘protein toxicity’ and ‘RNA toxicity,’ which are terms used to describe suspected causes of cell dysfunction and death in some TNR diseases, we have referred to repeat-associated convergent transcription-induced cell death as ‘DNA toxicity.’ We propose this term to indicate the capacity of a DNA sequence, upon interaction with a normal DNA metabolic process like transcription, to trigger cell death. Here, we discuss the basis for DNA toxicity and its potential relevance to our understanding of repeat-associated diseases in humans.
Antisense Transcription in Human Cells
Recent analyses of transcription in human cells have shown that the majority of the genome is traversed by RNAPII28,29 and that a surprisingly high proportion (up to about 30%) of human genes is transcribed in both the sense and antisense directions.30–32 Comparisons of the human sense and antisense transcriptomes have shown that most of the time, but not always, the level of a sense transcript is higher than the level of its corresponding antisense transcript.26 Measurements of antisense transcripts, however, likely underestimate the rates of antisense transcription since sense transcripts tend to be more stable in cells than antisense transcripts. Although antisense RNA was originally considered “junk,” and regulatory sequences for controlling antisense transcription remain largely undefined, antisense transcription may offer novel ways to regulate sense transcription.33–35 It has been proposed, for example, that antisense transcription may modulate the efficiency of sense transcription via head-on collisions with RNAPII, may alter sense transcription by recruiting chromatin remodeling factors, may mask RNA processing sites on sense transcripts to generate alternative RNA products, or that it may allow formation of double-stranded RNA with sense transcripts, which could be processed to form regulatory microRNA molecules.36
Whatever its biological function, antisense transcription may have unintended consequences in the context of long repeat tracts in TNR disease genes. Antisense transcripts have been identified along with sense transcripts at five TNR disease genes in vivo.13,25 In addition, antisense transcriptome analyses identified the antisense transcripts of at least ten other TNR disease genes in multiple human cell lines.26 Thus, it seems likely that simultaneous sense and antisense transcription—convergent transcription—occurs naturally in many TNR disease genes. Whether convergent transcription plays a role in either the instability of repeat tracts or the pathogenesis of TNR diseases has not yet been investigated in human patients or in model organisms; however, studies in human cells indicate that convergent transcription has the potential to dramatically increase repeat instability and to trigger cell death via apoptosis.27
Convergent Transcription Promotes Repeat Instability
The frequent occurrence of sense and antisense transcription in TNR disease genes prompted our interest in testing the effects of antisense and convergent transcription on repeat instability. Using a selective system for assaying repeat contraction, we had originally demonstrated that turning on sense transcription increased CAG repeat contractions 15 fold.20 Subsequent studies by others showed that transcription also stimulates GAA repeat instability in human cells and CAG instability in the Drosophila germline.37,38 To test the effects of antisense transcription, we modified an HPRT minigene to carry a CAG95 repeat tract embedded in its only intron, and arranged for sense and antisense transcription to be driven by distinct inducible promoters that respond to different inducers. In this model system, the long CAG repeat interferes with splicing and prevents HPRT expression. Contraction of the repeat below a threshold of about 39 units permits correct splicing and restores HPRT expression, which can be selected for.
A key difference between sense and antisense transcription is that the two single strands in the repeat region have different properties, with CTG repeats capable of forming more stable hairpins than CAG repeats. In the HPRT minigene, the repeat tract is arranged so that the CAG strand is the nontemplate strand, and thus exposed during sense transcription, while the CTG strand is exposed during antisense transcription. Nevertheless, antisense transcription was found to promote repeat instability with an efficiency similar to sense transcription.20,37 Quite unexpectedly, however, induction of convergent transcription synergistically destabilized the repeats, yielding a significantly higher frequency of contractions than the sum of that caused by sense transcription and antisense transcription alone.27
These observations have been extended in a recent study that tested the effects of transcription through a very large CAG repeat tract consisting of 800 units.39 In this case, transcription was either constitutive or could be turned on by the Cremediated removal of a strong termination stop signal. Repeat lengths were analyzed in the absence of selection, using small-pool PCR methods, which allows both contractions and expansions to be assessed. Individually, sense and antisense transcription both substantially stimulated repeat instability. Moreover, convergent transcription also synergistically enhanced repeat instability. Remarkably, analysis of the altered alleles revealed about equal frequencies of contractions and expansions, with several discrete changes of more than 200 repeats. Together, these two studies support a strong role for transcription, especially convergent transcription, in the repeat instability that is observed in human patients.
Covergent Transcription Induces Apoptosis
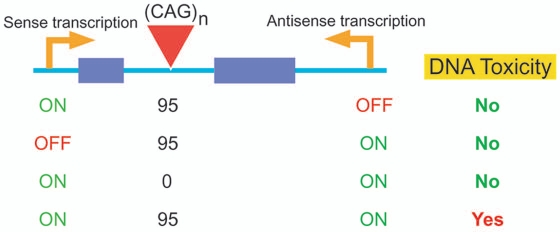
One entirely unexpected consequence of convergent transcription through a long CAG repeat was that it quickly arrested the cell cycle and subsequently stimulated massive cell death by apoptosis.27 Within a few hours to a couple of days after induction of convergent transcription, cells stopped proliferating, with significant alterations in the distributions of cells within the cell cycle.27 In this initial period of cell cycle arrest, no apoptosis was observed. After three days of convergent transcription, however, cells began to die at a dramatic rate, with only 20–40% of cells remaining alive after five days. The extent of cell death depended on the length of the CAG repeat tract and the induced level of convergent transcription. No apoptosis was observed after induction of sense or antisense transcription alone through a CAG95 tract, or induction of convergent transcription through a transcription unit without a CAG tract (Fig. 1). Nor was apoptosis observed if sense and antisense transcription were induced through CAG tracts at different sites in the genome, arguing against an indirect effect mediated, for example, by double-stranded RNA: a line of reasoning confirmed by the lack of effect of knockdown of components of the machinery responsible for generating cellular microRNAs.27 These experiments document for the first time that a DNA sequence—in this case a long CAG repeat tract—can trigger cell death when it interacts with a normal DNA metabolic process: convergent transcription. We refer to this phenomenon as DNA toxicity.
Figure 1.
DNA toxicity at CAG repeat tracts. Sense transcription, antisense transcription, or both were induced across a CAG repeat tract in the intron of the HPRT minigene. Significant cell death—DNA toxicity—was observed only with convergent transcription through a long CAG tract.
The effects of transcription on repeat instability in bacteria and yeast have been interpreted in terms of interactions between transcription and replication.40–42 Such an interaction appears not to be the basis for convergent transcription-induced DNA toxicity, because convergent transcription also induced apoptosis in nonproliferating cells.27 Indeed, in non-proliferating cells apoptosis occurred more rapidly and to a greater extent, with only 10–25% of cells surviving 5 days after induction of convergent transcription. These observations indicate that CAG repeat-dependent DNA toxicity is independent of replication in human cells, which may be especially relevant to the death of terminally differentiated cells in human patients with TNR diseases.
Recent studies in yeast have shown that expanded CAG repeats of 70 or 155 units have the potential to activate DNA damage checkpoints, interfere with normal cell cycle progression, and compromise cell proliferation and survival (C.H. Freudenreich, In press). A strain of yeast carrying an artificial chromosome with an expanded CAG repeat formed microcolonies that were only 50–67% as large as the colonies of yeast with an artificial chromosome that lacked a CAG repeat. This effect on cell growth, which was mediated by a DNA damage response, appeared to be due to frequent and prolonged cell cycle arrests, with significant alterations in the distributions of cells in the phases of the cell cycle. Although transcription through the repeats was detectable, presumably as a read-through from an adjacent gene, the authors interpreted their results as more likely due to checkpoint responses occurring during DNA replication. This repeat-associated DNA toxicity in yeast may be related to early observations in bacteria, where it was shown that long CAG repeats on plasmid DNA interfere with bacterial growth.43 Collectively, these studies indicate that long CAG repeats can be toxic to the cell upon interaction with normal transcriptional or replicative processes.
Convergent Transcription Elicits a DNA Damage Response
Several lines of evidence indicate that convergent transcription through a CAG repeat tract triggers apoptosis via a DNA damage response pathway.27 First, induction of convergent transcription leads to activation of ATR (ATM and Rad3-related)—a major transducer protein kinase of the damage response—via phosphorylation at serine 428; ATR-dependent activation of cell cycle checkpoint kinase 1 (CHK1) via phosphorylation at serine 345; and ATR-dependent activation of p53 by phosphorylation at serine 15. The second major transducer kinase, ATM (ataxia-telangiectasia mutated), is activated with slower kinetics than ATR, via a pathway that does not depend on ATR; nor is ATM responsible for the observed phosphorylation of CHK1 and p53.27 Second, components of the ATR pathway, including ATR itself, ATRIP and TOPBP1, are recruited to the CAG repeat tract after induction of convergent transcription, as determined by ChIP analysis.27 Third, chemical and siRNA-mediated inactivation of components of the ATR pathway increases the fraction of the cell population that die when convergent transcription is induced.27 Thus, activation of the ATR pathway normally acts to suppress apoptosis, presumably by stimulating repair of the DNA structures that initiated the response.
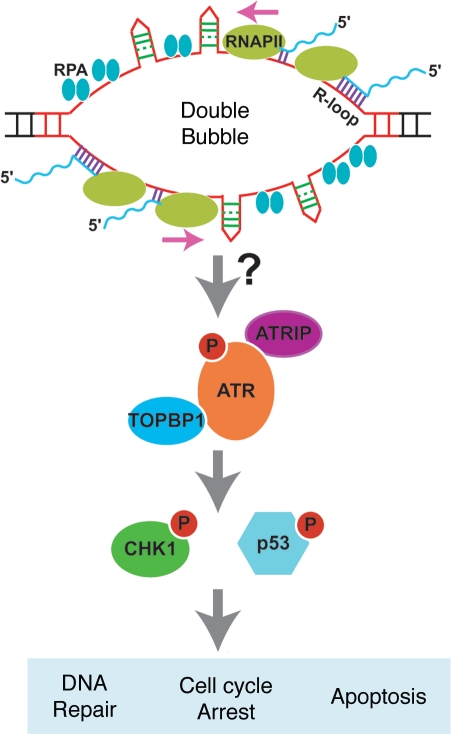
The critical, ATR-activating DNA structures generated by convergent transcription through a CAG repeat tract are unknown, but they were not produced by sense or antisense transcription alone, nor did they occur in the absence of a CAG repeat. These considerations suggest that the problem may arise from the fusion of sense and antisense transcription bubbles, which we will refer to as a double bubble. Figure 2 illustrates several abnormal features of a double bubble that may be important for an ATR response.
Figure 2.
Speculative model for induction of the ATR pathway by convergent transcription through a CAG repeat tract. RNAPII complexes are envisioned to stall at CAG and CTG hairpins on the separated template strand of the double bubble. The hairpins may be stabilized by binding of MSH2/3 (not shown). RPA-coated single strands and R-loops, both of which have been shown to occur during transcription through CAG repeats, are shown as part of the structure of the double bubble. Structural features of the double bubble may be sufficient to activate the ATR pathway, or further processing of the structure may be required (a possibility indicated by the arrow with the question mark).
First, RNAPII complexes may stall at repeat-generated hairpins on both strands. ChIP analysis indicates that RNAPII accumulates at repeat tracts during convergent transcription.27 While it is known that CAG repeats can arrest RNAPII during transcription in vitro,44 CNG hairpins may be converted to more efficient RNAPII roadblocks by binding the mismatch repair (MMR) recognition complex, MSH2/3 (not depicted in Fig. 2), as they do in vitro.45,46 In a similar way, O6-methylguanine can be converted from a nonblocking lesion to one that blocks RNAPII by the binding of MMR proteins.47 The involvement of MSH2/3 in the formation of double bubbles fits with evidence that shows that they also promote transcription-induced CAG repeat instability in human cells.20,22
Second, the single-strand DNA binding protein, replication protein A (RPA) may coat single-strands of DNA on one or both sides of a double bubble (Fig. 2). ChIP analysis showed that RPA accumulates at CAG repeats when convergent transcription is induced.27 RPA might be expected to bind to single-stranded DNA adjacent to hairpins, contributing to the stability of the separated DNA strands. As discussed below, RPA-coated single strands are a critical element of the classic pathway for activating the ATR response.
Finally, RNA-DNA (R-loops) hybrids might be critical to the formation of an ATR-inducing DNA structure (Fig. 2). Studies in bacteria and mammalian cells demonstrated that R-loops form at CAG repeats during sense transcription.48,49 Stable R-loops are thought to form in CNG repeats due to the high thermal stability of rG/dC and rC/dG nucleotide pairs relative to dG/dC pairs.50,51 A recent study using in vitro transcription demonstrated that R-loops can form on DNA strands during convergent transcription through CNG repeat tracts.52 R-loops contribute to repeat instability since their persistence, due to genetic or siRNA mediated deficiency of RNase H, stimulates CAG repeat instability in both bacteria and human cells.48
These proposed features of convergent transcription-induced double bubbles do not link directly to the classic signal for ATR activation, which is RPA-coated single stranded tail protruding from a segment of double-stranded DNA.53 RPA-ssDNA localizes ATR and its binding partner ATRIP to the DNA, while the Rad9-Rad1-Hus1 (9-1-1) complex binds to the adjacent double stranded DNA. Interaction between the ATR-ATRIP and the 9-1-1 complexes allows binding of TOPBP1 (topoisomerase II binding protein 1), which contains the activation domain required for triggering the ATR signaling pathway.
What is missing from the double bubble in Figure 2 is the dsDNA-ssDNA junction. A nick adjacent to a hairpin could create such a junction, but it is unknown whether a hairpin, with its mismatches, could serve at the dsDNA component of the ATR signal. Alternatively, a nick could allow reannealing of adjacent ssDNA to create a suitable dsDNA segment with ssDNA tail. A natural candidate for introducing nicks would be nucleotide excision repair (NER), which is known to be involved in repeat instability induced by transcription through a CAG repeat.23,54 In the absence of its usual dsDNA substrate, NER might operate abortively to nick a double bubble, allowing formation of the classic structure. Finally, it is conceivable that the end of an RNA-DNA hybrid (if RNPII were removed) could serve as the necessary double-stranded junction, with the template DNA strand forming the ssDNA tail.
It is also possible that the features of the double bubble in Figure 2 may be adequate by themselves, since some reports suggest that ATR activation may not be solely dependent on the classic structure.53 For example, constitutive nuclear translocation of the ATR-activation domain of TOPBP1 is enough to activate the ATR pathway, including phosphorylation of CHK1 and p53, in the absence of any obvious DNA damage.55 In addition, in a system that reconstituted the human ATR-mediated checkpoint response to bulky lesions, purified components were used to show that ATR-ATRIP phosphorylates CHK1 in a reaction that requires TOPBP1, is strongly dependent on DNA containing bulky base lesions, but appears to be entirely independent of DNA ends.56,57 Finally, if the classic structure were the only way to activate ATR, it might be expected that the phenotypes generated by the loss of ATR or by the loss of the 9-1-1 complex would be the same; however, the loss of ATR is much more severe than the loss of RAD9 or HUS1.53
Although neither mechanism has been defined, convergent transcription-induced ATR activation resembles the transcriptional stress response, which also involves the ATR- and RPA-dependent activation of CHK1 and p53.58–60 Interference with the progression of the RNAPII complex by treatments such as UV light, actinomycin D and psoralen (which cause pyrimidine dimers, base intercalation and interstrand crosslinks, respectively) activate the ATR pathway.60 But DNA damage is not required since antibodies to the elongating form of RNAPII elicit the same ATR response.60 Thus, it may be that a stalled RNAPII complex in the presence of RPA-coated ssDNA is sufficient to stimulate an ATR response; that is, that the stalled RNAPII complex may serve directly as the sensor for transcriptional stress and triggers the cellular response.54,61 What is remarkable about convergent transcription through a CAG repeat tract is that a single genomic site of transcriptional interference—a single toxic site—is capable of triggering an ATR response that can lead to cell death.
DNA Toxicity and TNR Diseases
The key question, of course, is whether DNA toxicity plays any role in TNR diseases. Most TNR diseases involve the progressive dysfunction and loss of specific differentiated cells—neurons or muscle cells—with the time of symptom onset and rate of disease progression depending strongly on the length of the repeat tract donated by one parent. How the expanded repeat tract leads to cell death is much less clear. If DNA toxicity plays a significant role in TNR diseases, however, then a single expanded allele should be sufficient to manifest the disease symptoms; that is, a disease caused by DNA toxicity should display dominant inheritance. Remarkably, 12 CAG repeat diseases are dominantly inherited62 and the 13th, spinal and bulbar muscular atrophy (SBMA), is X-linked and X inactivation makes its dominance difficult to determine. Currently, the pathogenic mechanisms are grouped into three categories: protein gain of function (“protein toxicity”), RNA mediated (“RNA toxicity”) and unknown.10,63
Protein toxicity.
Nine disorders, including Huntington disease (HD), several spinocerebellar ataxias (SCA1, SCA2, SCA3, SCA6, SCA7 and SCA17), dentatorubral-pallidoluysian atrophy (DRPLA) and SBMA, are caused by expansion of a CAG tract in an exon, where they encode polyglutamine (polyQ) tracts.9 In each case, the extended polyQ tract is thought to alter the properties of the mutated protein, making it toxic and leading to the development of disease symptoms. In HD, which is the most intensively studied, there is robust support for a protein-based toxicity mechanism of disease pathogenesis. It has been shown, for example, that mutation of a specific caspase-6 cleavage site renders the huntingtin protein nontoxic.64 Various post-translational modifications of the protein such as phosphorylation, acetylation and sumoylation may also play roles in the toxicity of the mutant protein.9 Similar strong evidence supports protein toxicity as the mechanism of disease pathogenesis for SCA1.9 Most of diseases in this class are less well studied, but are likely due to the toxicity of the polyglutamine-containing mutant protein. For example, expression of a truncated human huntingtin, corresponding just to exon 1, in a transgenic mouse model is sufficient to cause neurological disorders,65 suggesting an isolated polyQ tract is pathogenic.
RNA toxicity.
Myotonic dystrophy type 1 (DM1) is the single CAG repeat disease caused by dominant RNA toxicity mechanism. Because the repeat tract is oriented in the 3′ UTR of the DMPK gene so that CUG is expressed in the RNA, it is commonly referred to as a CTG repeat disease. Muscular dystrophy type 2 (DM2), which is also dominantly inherited, is caused by the related tetranucleotide repeat, CCTG, which is expanded in the first intron of the ZFN9 gene.63 At these expanded loci, transcription produces long CUG and CCUG tracts in the RNA, which can bind to certain splicing components, leading to an increase in the function of CUG-binding protein 1 and to a reduction in the activity of muscleblind 1.66 As a result of the disturbance in the balance of these splicing factors, transcripts such as those from the chloride channel gene and the insulin receptor gene are spliced abnormally, contributing directly to defined features of the disease pathology.63 Supporting the RNA toxicity hypothesis, a mouse model expressing a CTG250 tract in the 3′-UTR of the human skeletal α-actin gene developed myotonia.67 Interestingly, transgenic mice expressing the DMPK 3′-UTR containing only a CTG5 tract upregulated CUGBP1 and developed DM1-like phenotypes.68 This observation is difficult to reconcile with the RNA toxicity hypothesis, since RNA transcripts with repeat tracts longer than CUG5 are common in human transcriptome.69
Unknown.
The bases for the pathogenesis in SCA8, SCA12 and HD-like 2 (HDL2) are not yet clear. In all three of these diseases, both sense and antisense transcription have been documented in cells.26,70 For SCA8 and HDL2, where antisense transcription was equal to or greater than sense transcription,26 it has been suggested that the pathology may result from expression of a CUG-containing toxic RNA from one strand and a polyglutamine toxic protein from the other (reviewed in ref. 9). An alternative possibility is that sense and antisense transcription through the CAG repeat tract induces DNA toxicity. The situation for SCA12 is less clear; the CAG repeat, which is located in the 5′ UTR and transcribed as CAG, would not be expected to generate either a toxic RNA or protein. Thus, it may also be a candidate for DNA toxicity due to convergent transcription though the repeat tract.
It may prove difficult to tease out the contributions of DNA toxicity to the neurodegeneration in CAG repeat diseases, since in many instances sense and antisense transcription can lead to a toxic protein and toxic RNA. In addition, the overexpression of modified genes, as is often used in TNR studies, may overwhelm cellular coping mechanisms and uncover pathologies that are not present in patients. Notably, the proposed pathogenesis due to toxic RNA and protein do not involve the DNA damage response. However, in an HD mouse model, the DNA damage response, including phosphorylation of ATM at S1981 and of p53 at S15, was found to be activated in various brain regions.71 In fibroblasts from HD or SCA2 patients, an ATM/ATR substrate was phosphorylated at a higher level than normal, implying that the damage response pathways were activated.72 In HD and SCA3 transgenic models, increased levels of p53-S15P was found in neurons of specific brain regions, along with enhanced expression of p53-dependent pro-apoptotic genes Bax and PUMA.73,74 Finally, in a transgenic SCA1 mouse model, p53 was found to promote the progression of neurodegeneration.75 Collectively, these results indicate that DNA damage response pathways are activated in TNR disease models. None of these studies, however, has yet linked damage response activation to convergent transcription through a repeat tract.
Perspectives
TNRs present a fascinating biological conundrum. TNRs offer positive attributes favorable for evolution, but also impose serious health risks for the fraction of the human population who inherit expanded repeats in critical genes. Investigations in both realms have uncovered unexpected complexity, but the dramatic instability of TNRs and the links between expanded TNRs and disease have challenged and informed our understanding of basic cell biology.
Extensive studies in bacteria, yeast, flies, human cells and mice have shown that DNA replication and virtually every aspect of DNA repair—mismatch repair, nucleotide excision repair, base excision repair, single-strand break repair and double strand break repair, including homologous recombination and nonhomologous end joining—can alter the stability of TNRs. As if that weren't enough, transcription through a tract of CAG repeats, in combination with MMR and NER, also destabilizes repeats and convergent transcription enhances repeat instability synergistically, causing both contractions and expansions. This embarrassing diversity of mechanism precludes an easy solution to the problem of what causes repeat expansion in human germline and somatic tissues. And it now seems likely that different mechanisms will be found to operate in different tissues. Nevertheless, our deeper understanding of repeat instability offers the hope of new treatment options to prevent expansion or to promote contraction.
Elucidating the links between expanded CAG repeats and disease pathology has also uncovered surprising biological phenomena, including protein toxicity, RNA toxicity and now DNA toxicity. Further investigations into the ways polyglutamine proteins and CUG- and CCUG-containing RNAs disrupt cellular processes have revealed connections to other cellular processes beyond the function of the affected gene. For example, studies of polyglutamine diseases have identified autophagy as a potential neuroprotective mechanism involved in removing polyglutamine aggregates.76 As the steps that lead from expanded repeat to disease pathogenesis are uncovered, they expose new targets for potential therapeutic approaches. The translation of knowledge into therapy is sorely needed since useful treatments for repeat diseases are almost entirely lacking at present.
The roles of convergent transcription and DNA toxicity in CAG repeat diseases are poorly defined. It is surprising that a single mechanism—convergent transcription through a CAG repeat—can dramatically enhance repeat instability and trigger cell death via apoptosis: two characteristics of CAG repeat diseases in humans. Many questions remain to be addressed, however. What DNA repair proteins or processes, are brought into play by convergent transcription, and how do they account for the synergistic increase in CAG instability over sense or antisense transcription alone? What is the specific structure in double bubbles that activates the ATR pathway? Can it be shown that convergent transcription contributes to CAG repeat instability and cell death in a model organism such as mouse? And what would constitute a proper test of those possibilities? If the history of research in this field is any indication, addressing these questions will likely unravel new biological complexity and improve our understanding of these multifaceted diseases.
Acknowledgements
This work was supported by a grant from the NIH (GM38219) to J.H.W.
References
- 1.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacolla A, Larson JE, Collins JR, Li J, Milosavljevic A, Stenson PD, et al. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 2008;18:1545–1553. doi: 10.1101/gr.078303.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Legendre M, Pochet N, Pak T, Verstrepen KJ. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 2007;17:1787–1796. doi: 10.1101/gr.6554007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mittelman D, Wilson JH. Stress, genomes and evolution. Cell Stress Chaperones. 15:463–466. doi: 10.1007/s12192-010-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albrecht A, Mundlos S. The other trinucleotide repeat: polyalanine expansion disorders. Curr Opin Genet Dev. 2005;15:285–293. doi: 10.1016/j.gde.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 11.Bacolla A, Wells RD. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol Carcinog. 2009;48:273–285. doi: 10.1002/mc.20507. [DOI] [PubMed] [Google Scholar]
- 12.Swami M, Hendricks AE, Gillis T, Massood T, Mysore J, Myers RH, et al. Somatic expansion of the Huntington's disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum Mol Genet. 2009;18:3039–3047. doi: 10.1093/hmg/ddp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y, Hubert L, Jr, Wilson JH. Transcription destabilizes triplet repeats. Mol Carcinog. 2009;48:350–361. doi: 10.1002/mc.20488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 15.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson CE, Edamura KN, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 17.Cleary JD, Pearson CE. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet Genome Res. 2003;100:25–55. doi: 10.1159/000072837. [DOI] [PubMed] [Google Scholar]
- 18.Pearson CE, Wang YH, Griffith JD, Sinden RR. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n. (CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–823. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y, Dion V, Wilson JH. Transcription and triplet repeat instability. In: Wells R, Ashizawa T, editors. Genetic Instability and Neurological Diseases. Amsterdam: Elsevier; 2006. pp. 691–704. [Google Scholar]
- 22.Lin Y, Wilson JH. Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells. DNA Repair. 2009;8:878–885. doi: 10.1016/j.dnarep.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonitel R, Moffitt H, Sathasivam K, Woodman B, Detloff PJ, Faull RL, et al. DNA instability in postmitotic neurons. Proc Natl Acad Sci USA. 2008;105:3467–3472. doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batra R, Charizanis K, Swanson MS. Partners in crime: bidirectional transcription in unstable micro-satellite disease. Hum Mol Genet. 2010;19:77–82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Y, Leng M, Wan M, Wilson JH. Convergent transcription through a long CAG tract destabilizes repeats and induces apoptosis. Mol Cell Biol. 2010;30:4435–4451. doi: 10.1128/MCB.00332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 29.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, et al. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 32.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 33.Morris KV, Vogt PK. Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell Cycle. 2010;9:2542–2545. doi: 10.4161/cc.9.13.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beiter T, Reich E, Williams RW, Simon P. Antisense transcription: a critical look in both directions. Cell Mol Life Sci. 2009;66:94–112. doi: 10.1007/s00018-008-8381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 37.Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 38.Ditch S, Sammarco MC, Banerjee A, Grabczyk E. Progressive GAA.TTC repeat expansion in human cell lines. PLoS Genet. 2009;5:1000704. doi: 10.1371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamori M, Pearson CE, Thornton CA. Bidirectional Transcription Stimulates Expansion and Contraction of Expanded (CTG)*(CAG) Repeats. Hum Mol Genet. 2011;20:580–588. doi: 10.1093/hmg/ddq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowater RP, Jaworski A, Larson JE, Parniewski P, Wells RD. Transcription increases the deletion frequency of long CTG.CAG triplet repeats from plasmids in Escherichia coli. Nucleic Acids Res. 1997;25:2861–2868. doi: 10.1093/nar/25.14.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parniewski P, Bacolla A, Jaworski A, Wells RD. Nucleotide excision repair affects the stability of long transcribed (CTG*CAG) tracts in an orientation-dependent manner in Escherichia coli. Nucleic Acids Res. 1999;27:616–623. doi: 10.1093/nar/27.2.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wierdl M, Greene CN, Datta A, Jinks-Robertson S, Petes TD. Destabilization of simple repetitive DNA sequences by transcription in yeast. Genetics. 1996;143:713–721. doi: 10.1093/genetics/143.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bowater RP, Rosche WA, Jaworski A, Sinden RR, Wells RD. Relationship between Escherichia coli growth and deletions of CTG.CAG triplet repeats in plasmids. J Mol Biol. 1996;264:82–96. doi: 10.1006/jmbi.1996.0625. [DOI] [PubMed] [Google Scholar]
- 44.Parsons MA, Sinden RR, Izban MG. Transcriptional properties of RNA polymerase II within triplet repeat-containing DNA from the human myotonic dystrophy and fragile X loci. J Biol Chem. 1998;273:26998–27008. doi: 10.1074/jbc.273.41.26998. [DOI] [PubMed] [Google Scholar]
- 45.Owen BA, Yang Z, Lai M, Gajek M, Badger JD, 2nd, Hayes JJ, et al. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 46.Pearson CE, Ewel A, Acharya S, Fishel RA, Sinden RR. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum Mol Genet. 1997;6:1117–1123. doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- 47.Yanamadala S, Ljungman M. Potential role of MLH1 in the induction of p53 and apoptosis by blocking transcription on damaged DNA templates. Mol Cancer Res. 2003;1:747–754. [PubMed] [Google Scholar]
- 48.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci USA. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McIvor EI, Polak U, Napierala M. New insights into repeat instability: Role of RNA.DNA hybrids. RNA Biol. 2010;7:551–558. doi: 10.4161/rna.7.5.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol. 2009;29:3124–3133. doi: 10.1128/MCB.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reddy K, Tam M, Bowater RP, Barber M, Tomlinson M, Nichol Edamura K, et al. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. doi: 10.1093/nar/gkq935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 55.Toledo LI, Murga M, Gutierrez-Martinez P, Soria R, Fernandez-Capetillo O. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev. 2008;22:297–302. doi: 10.1101/gad.452308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi JH, Lindsey-Boltz LA, Sancar A. Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc Natl Acad Sci USA. 2007;104:13301–13306. doi: 10.1073/pnas.0706013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi JH, Lindsey-Boltz LA, Sancar A. Cooperative activation of the ATR checkpoint kinase by TopBP1 and damaged DNA. Nucleic Acids Res. 2009;37:1501–1509. doi: 10.1093/nar/gkn1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ljungman M. Activation of DNA damage signaling. Mutat Res. 2005;577:203–216. doi: 10.1016/j.mrfmmm.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 59.Ljungman M. The transcription stress response. Cell Cycle. 2007;6:2252–2257. doi: 10.4161/cc.6.18.4751. [DOI] [PubMed] [Google Scholar]
- 60.Derheimer FA, O'Hagan HM, Krueger HM, Hanasoge S, Paulsen MT, Ljungman M. RPA and ATR link transcriptional stress to p53. Proc Natl Acad Sci USA. 2007;104:12778–12783. doi: 10.1073/pnas.0705317104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lindsey-Boltz LA, Sancar A. RNA polymerase: the most specific damage recognition protein in cellular responses to DNA damage? Proc Natl Acad Sci USA. 2007;104:13213–13214. doi: 10.1073/pnas.0706316104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 63.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 65.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 66.Osborne RJ, Thornton CA. RNA-dominant diseases. Hum Mol Genet. 2006;15:162–169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 67.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 68.Mahadevan MS, Yadava RS, Yu Q, Balijepalli S, Frenzel-McCardell CD, Bourne TD, et al. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Molla M, Delcher A, Sunyaev S, Cantor C, Kasif S. Triplet repeat length bias and variation in the human transcriptome. Proc Natl Acad Sci USA. 2009;106:17095–17100. doi: 10.1073/pnas.0907112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 71.Illuzzi J, Yerkes S, Parekh-Olmedo H, Kmiec EB. DNA breakage and induction of DNA damage response proteins precede the appearance of visible mutant huntingtin aggregates. J Neurosci Res. 2009;87:733–747. doi: 10.1002/jnr.21881. [DOI] [PubMed] [Google Scholar]
- 72.Giuliano P, De Cristofaro T, Affaitati A, Pizzulo GM, Feliciello A, Criscuolo C, et al. DNA damage induced by polyglutamine-expanded proteins. Hum Mol Genet. 2003;12:2301–2309. doi: 10.1093/hmg/ddg242. [DOI] [PubMed] [Google Scholar]
- 73.Bae BI, Xu H, Igarashi S, Fujimuro M, Agrawal N, Taya Y, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 74.Chou AH, Lin AC, Hong KY, Hu SH, Chen YL, Chen JY, et al. p53 activation mediates polyglutamine-expanded ataxin-3 upregulation of Bax expression in cerebellar and pontine nuclei neurons. Neurochem Int. 2011;58:145–152. doi: 10.1016/j.neuint.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 75.Shahbazian MD, Orr HT, Zoghbi HY. Reduction of Purkinje cell pathology in SCA1 transgenic mice by p53 deletion. Neurobiol Dis. 2001;8:974–981. doi: 10.1006/nbdi.2001.0444. [DOI] [PubMed] [Google Scholar]
- 76.Renna M, Jimenez-Sanchez M, Sarkar S, Rubinsztein DC. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem. 2010;285:11061–11067. doi: 10.1074/jbc.R109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]