Abstract
Pluripotent stem cells (PSC s) are maintained by a complex regulatory network orchestrated by transcription factors, epigenetic modifiers and non-coding RNA s. Central to this regulatory network is the Myc family of transcription factors. Defining roles for Myc in PSC s has been problematic, but recently, a number of reports have provided insight in this area. An emerging picture now places Myc as a key regulator of the cell cycle, genomic maintenance and general metabolic activity in PSC s through its ability to directly regulate large numbers of target genes and more indirectly through control of microRNA s. One of Myc's main roles is to repress the activity of genes required for differentiation such as the endoderm master regulator, GATA 6. The general mechanism by which Myc activates target genes is well understood but a remaining major challenge is to understand how it represses gene activity. Here we discuss potential mechanisms for how Myc establishes and maintains the pluripotent state and incorporate proteomics data that supports a model where Myc acts as part of a regulatory network with epigenetic modifiers.
Key words: myc, pluripotency, self-renewal, reprogramming, proteomics
Background
Embryonic stem cells (ESCs) are derived from pluripotent cells within the inner cell mass of blastocyst-stage embryos and like their in vivo counterparts, can differentiate into the three embryonic germ layers. Blastocysts also contain two extraembryonic cell types; primitive endoderm which provides signaling support to the developing pluripotent epiblast and the trophectodermal layer which contributes to the placenta.1–4 ESCs can be maintained for extended periods of time in culture as a stable, self-renewing population but without the appropriate balance of extrinsic signals, they lose pluripotency and differentiate into extra-embryonic cell types or germ layer lineages.5–9 Perhaps the most important extrinsic signaling molecules required for maintenance of murine ESCs (mESCs) are the interleukin-6 family member cytokines, such as leukemia inhibitory factor (LIF).10,11 A major role for LIF in mESCs is to maintain the expression of Myc transcription factors.12 c-, N- and L-myc are a family of sequence-specific, basic helix-loop-helix transcription factors with well-established roles in embryonic development and cancer.13–17 The discovery ∼6 years ago linking Myc to LIFs role in maintaining mESC self-renewal12 has stimulated much interest and insight into pluripotent cell biology. This report will summarize recent advances in this area and describes a model based on new data for how Myc participates in mechanisms relating to pluripotency and reprogramming.
Following the initial report that Myc is critical for self-renewal,12 Yamanaka and colleagues demonstrated that Myc is also important for efficient reprogramming to an induced pluripotent cell (iPSC) state.18,19
Myc Target Genes in Pluripotent Cells
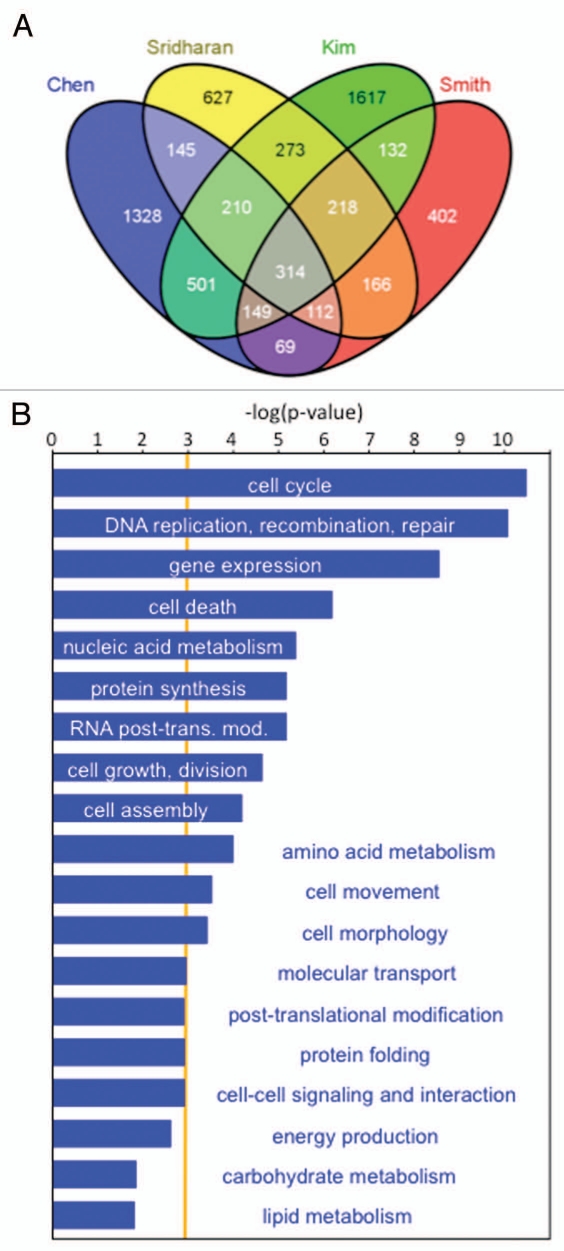
To determine how Myc regulates pluripotency, several ChIP-Chip and ChIP-Seq studies have been performed, resulting in the identification of a large number of in vivo target genes.20–25 Although over 8,000 target genes have been identified by these studies, only ∼19 are common to all six. When different pair-wise comparisons of published Myc target genes are made, the overlap ranges from 6 to 52% (Table 1). Despite the low degree of overlap in Myc targets between different studies, many have been validated independently by ChIP-qPCR assays. Overall, these results indicate that individual studies directed towards global in vivo target gene identification are not particularly exhaustive and that multiple independent approaches are required to obtain a complete picture. At least some of the variations described can be accounted for by differences in gene expression signatures between different cell lines, differences in the platform technology employed and the bioinformatics analysis used. The routine introduction of deep sequencing-based approaches (Chip-seq and RNA-seq) should circumvent some of these issues. When the criteria used for comparison of data between different studies are relaxed and data from only four studies20–22,25 are included in the analysis, 314 common Myc targets are identified (Fig. 1A). Functional annotation of these target genes shows significant enrichment in processes relating to cell cycle control, DNA replication and repair, gene expression and metabolism (p < 0.001, Fig. 1B). These high-confidence targets therefore form the ‘core’ Myc gene-target network that establishes and maintains PSCs.
Table 1.
Percentage overlap of Myc target genes in mESCs expressed as pair-wise comparisons of the published data sets
| Chen | Kidder | Kim | Lin | Smith | Sridharan | ||
| Chen | 100.00 | 19.94(38.66) | 41.51(34.39) | 12.16(11.54) | 22.77 (41.23) | 27.62(37.82) | 2832a |
| Kidder | 100.00 | 51.61(22.06) | 15.22(7.45) | 33.65(31.43) | 35.50(25.08) | 1459a | |
| Kim | 100.00 | 13.53(15.50) | 23.81(52.05) | 29.73(49.15) | 3417a | ||
| Lin | 100.00 | 6.17(11.78) | 8.36(12.06) | 2980a | |||
| Smith | 100.00 | 51.86(39.23) | 1562a | ||||
| Sridharan | 100.00 | 2065a |
Figure 1.
In vivo target genes for c-myc in mESCs. (A) Venn diagram constructed as in reference 53 showing the proportion of Myc target genes common to four genome-wide ChIP studies.20–22,25 314 target genes are found to be c-myc target genes in all four studies. (B) Functional annotation of target genes found in all four datasets indicates enrichment primarily for regulators of the cell cycle, gene expression, genomic maintenance and metabolic activities. Data were analyzed through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). The orange line represents the cut-off for statistical significance (p-value-0.001). The x-axis denotes—(log) significance where longer bars represent greater enrichment of specified functional categories.
How does Myc Regulate Pluripotency?
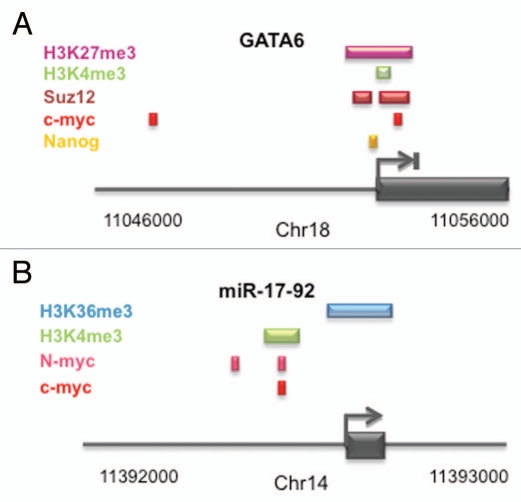
For some time, Myc has been associated with inhibition of cellular differentiation, especially in the context of cancer biology.16,26 Two reports recently showed that simultaneous genetic inactivation of c-MYC and N-MYC in ESCs and iPSCs resulted in rapid loss of PSC colony morphology and pluripotent marker expression.15,25,27 Rather than differentiating randomly into different lineages, we found that c-, N-MYC double knockout (dKO) cells preferentially formed primitive endoderm.25 The mechanism for this involves de-repression of the endoderm master regulator gene, GATA6. These findings indicate that under self-renewing conditions, differentiation towards endoderm is blocked by Myc-dependent repression of GATA6, adding yet another level of complexity to the mechanism by which Myc maintains the pluripotent state. Analysis of the GATA6 locus in mESCs28,29 reveals it to be bivalently marked (H3K4me3 and H3K27me3; Fig. 2A) consistent with it being inactive but poised for activation. Nanog, a known repressor of GATA6,30,31 also binds the regulatory region,32 but it is unclear how and if it works in conjunction with Myc. Since Nanog and Myc repress GATA6, this raises an interesting potential link between two well-established regulators of pluripotency.
Figure 2.
Epigenetic control of positively and negatively regulated Myc target genes in mESCs. (A) Schematic of the bivalently marked GATA 6 gene29 showing the upstream regulatory region, positional binding of transcriptional regulators and histone modification status in the region.20,25,29 The GATA 6 gene is repressed in pluripotent cells. (B) Schematic of the transcriptionally active murine miR-17-92 cluster and its upstream regulatory region showing binding of c-myc and N-myc together with active histone modifications.20,25,29
Besides a differentiation blockade being a critical part of Myc's role in maintaining pluripotency, several lines of evidence indicate that its ability to control the cell cycle is also important. This is suggested by decreased rates of cell division and a remodeling of the cell cycle following Myc inactivation.25 As mentioned previously, numerous cell cycle regulators including cyclins and cyclin dependent kinases are Myc target genes (Fig. 1B), providing a potential mechanism that could explain cell cycle changes following loss of Myc activity. Similar to the situation in various cancer cells, the mir-17-92 cluster is actively expressed in ESCs and is regulated at the transcriptional level by c- and N-myc.15,20,25 In PSCs, the cluster is marked by the active H3K4me3 and H3K36me3 modifications29 (Fig. 2B). The mir-17-92 locus produces miRNAs that target many cell cycle regulators such as transcripts for E2F transcription factors, cyclins and members of the retinoblastoma (Rb) tumor suppressor family.33,36 Our recent findings indicate that in PSCs, mir-17-92 targets cell cycle regulators such as RB2/p107,25 introducing a post-transcriptional level of cell cycle control regulated by Myc. By extension, during cellular reprogramming it is likely that Myc directs the establishment of an ESC-like cell cycle program by elevating global CDK activity and suppressing tumor suppressor activity.37 To support this, the cell cycle inhibitor p21 inhibits iPSC generation38 and conversely, knockdown of p21 accelerates cell division which leads to faster cellular reprogramming of differentiated cells by Oct4, Sox2, Klf4 and c-myc.39
Identification of Interacting Proteins as a Guide to Myc's Role in Pluripotent Cells
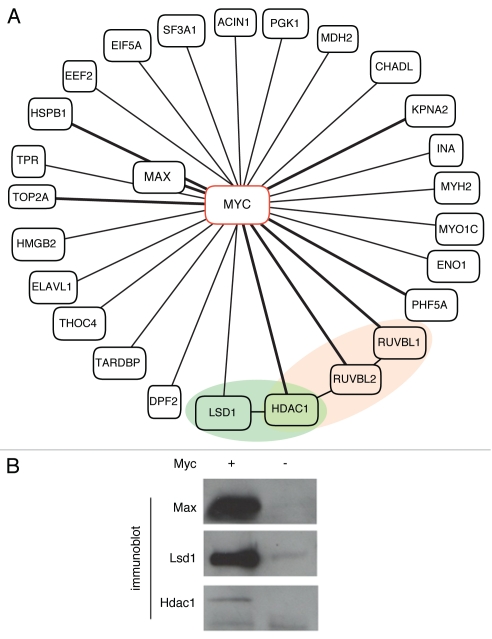
Myc's role as a prolific regulator of gene expression in PSCs can be further understood by defining the types of protein complexes it is assembled into. To characterize Myc-containing complexes in mESCs, we used a mass spectrometry-based proteomics approach. Antibody-coupled magnetic beads were used to isolate Myc complexes from ESC lysates, followed by liquid chromatography-tandem mass spectrometry. Non-specific interacting proteins were identified by subtractive methods using a Myc null cell line.40 The resulting list of 26 proteins that specifically immunoprecipitated with Myc included transcriptional regulators and epigenetic modifiers (Fig. 3A). As expected, Myc's well-characterized binding partner, Max, was identified in immunoprecipitates. Other known interacting proteins identified in this analysis include Ruvbl1, Ruvbl2 (Tip48, Tip49), Hdac1 and Top2a.41,42 The DNA helicases Ruvbl1 and Ruvbl2 are components of multiple chromatin remodeling complexes43,44 and RNAi mediated knockdown of either factor results in loss of the characteristic ESC colony morphology.45 Interestingly, Ruvbl1, Ruvbl2 and Hdac1 are also components of the repressive NURD complex that functions in nucleosome remodeling.46 Previous studies have shown that the NURD complex component MBD3 is required for pluripotency maintenance.47 Recruitment of the repressive histone deacetylase activity associated with NURD46 is therefore one mechanism by which Myc could act to block expression of specific target genes.
Figure 3.
The Myc protein interaction network in ESCs. (A) Diagrammatic representation of Myc interacting proteins identified by affinity purification coupled with mass spectrometry. Previously identified protein partners41,50 are joined by bold connectors. Myc interacting protein members implicated in transcriptional repression are shaded. The protein network is based on that generated through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). (B) c-myc assembles into complexes with members of the coRE ST co-repressor complex in pluripotent cells. Co-immunoprecipitation analysis with antibody coupled magnetic beads validates the interaction of c-myc with Lsd1 and Hdac1 in a cell line expressing Myc (AB2.1,6x9e10-c-myc-; +) and not the control Myc null cell line (AB2.1c-myc-/-; −). Max is used as a control.
Another epigenetic regulator with repressive function, Lsd1 (Kdm1a), was also found in Myc-immunoprecipitates (Fig. 3B). Lsd1 functions as a histone demethylase and its action on dimethylated lysine 4 of histone H3 results in gene repression.48 Lsd1 and Hdac1 are components of the CoREST co-repressor complex that cooperates with REST to inhibit neural specification factors in non-neural lineages,49 and Myc has recently been found to interact with Rcor3, another corepressor of Rest.50 Recently, Lsd1 has also been shown to interact with other pluripotency regulators such as Nanog, but perhaps more significant are observations that loss of Lsd1 activity results in increased apoptosis and impaired differentiation of ESCs.51,52 The identification of Lsd1 as a Myc-interacting protein suggests that Myc may repress target genes through a CoREST-dependent mechanism in PSCs.
Another recent proteomic study identified an additional set of Myc interacting proteins in PSCs including Trrap, Ep400, Dmap1, Brd8, Epc1 and Epc2.42 These proteins are members of the NuA4 acetyltransferase complex which appear to be important for maintenance of PSCs.45 Altogether, these proteomic studies indicate that Myc regulates PSCs as a component of different regulatory protein complexes with known roles in gene repression and activation.
Materials and Methods
Myc-interacting proteins were identified using nuclear extracts from mESC lines expressing 9E10-tagged c-myc, or a c-myc null ESC line40 as the control. mESCs were allowed to swell in 10 mM HEPES pH 7.4, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT and protease inhibitors. NP-40 was added to 1.6% final v/v, followed by brief vortexing to disrupt the cellular membrane and centrifugation to pellet nuclei followed by removal of the cytoplasmic supernatant. Nuclear proteins were extracted with 20 mM HEPES pH 7.4, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT and protease inhibitors. Extracts were subjected to immunoprecipitation with tosylactivated magnetic beads (Invitrogen) coupled to affinity-purified 9e10 monoclonal antibody according to the manufacturer's instructions. Complexes were then eluted with glycine (pH 2.5) followed by neutralization with Tris-HCl pH 8.8. In-solution tryptic digests were desalted then analyzed by nLC-MS/MS on a linear ion-trap (LTQ, ThermoFisher) as previously described in reference 54. Data was collated and filtered to obtain a 1% false discovery rate at the protein level using the ProteoIQ software package (BioInquire) and a reverse database to calculate FDR. An immunoprecipation from a cell line not expressing c-myc was used as a negative control and filtered at 5% false discovery (less stringent than the experimental sample). Proteins that were identified by at least two peptides in the experimental sample in at least two of the three replicates but were not identified in the negative control sample were considered high-confidence interacting proteins. Proteins that were identified with three times more coverage in the experimental sample (at 1% FDR) over the negative control (at 5% FDR) generated a list of putative interacting proteins that were further considered.
Conclusions
Although Myc is essential for maintenance of pluripotency, its function in PSCs has until recently been unclear. Recent work has identified a diverse range of in vivo target genes that point towards roles in gene regulation associated with cell cycle control, DNA replication/repair and metabolic control. Importantly, Myc seems to maintain the pluripotent state by blocking differentiation pathways, as shown by its direct effect on the GATA6 gene. Further studies will address the question of whether Myc blocks differentiation along other pathways. The finding of active and inactive target genes identified in PSCs points towards roles for Myc in gene activation and repression. This is consistent with the regulatory complexes Myc assembles into, with activating and repressive activities.
Acknowledgements
This work was supported by grants from the National Institute of Child Health and Human Development (HD049647; S.D.) and the National Institute for General Medical Sciences (GM75334; S.D. and L.W.).
Abbreviations
- PSC
pluripotent stem cell
- ESC
embryonic stem cell
- iPSC
induced pluripotent stem cell
- LIF
leukemia inhibitory factor
- dKO
double knockout
References
- 1.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Rossant J, Tam PPL. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 4.Evans M, Kaufman M. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 5.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 6.Niwa H, Miyazaki J, Smith AG. Quantitative expression of oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 7.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 8.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 9.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007 doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 10.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 11.Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 12.Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 13.Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 14.Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, et al. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- 15.Smith K, Dalton S. Myc transcription factors: Key regulators behind establishment and maintenance of pluripotency. Regen Med. 2010;5:947–959. doi: 10.2217/rme.10.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 17.Kuttler F, Mai S. c-myc, genomic instability and disease. Genome Dyn. 2006;1:171–190. doi: 10.1159/000092507. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kidder BL, Yang J, Palmer S. Stat3 and c-myc genome-wide promoter occupancy in embryonic stem cells. PLoS One. 2008;3:3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin CH, Lin CW, Tanaka H, Fero ML, Eisenman RN. Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-myc. PLoS One. 2009;4:7839. doi: 10.1371/journal.pone.0007839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith KN, Singh AM, Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole MD, Henriksson M. 25 years of the c-myc oncogene. Semin Cancer Biol. 2006;16:241. doi: 10.1016/j.semcancer.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, et al. Myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010;80:9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 29.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ralston A, Rossant J. Genetic regulation of stem cell origins in the mouse embryo. Clin Genet. 2005;68:106–112. doi: 10.1111/j.1399-0004.2005.00478.x. [DOI] [PubMed] [Google Scholar]
- 31.Hough SR, Clements I, Welch PJ, Wiederholt KA. Differentiation of mouse embryonic stem cells after RNA interference-mediated silencing of Oct4 and Nanog. Stem Cells. 2006;24:1467–1475. doi: 10.1634/stemcells.2005-0475. [DOI] [PubMed] [Google Scholar]
- 32.Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for nanog in embryonic stem cells. Stem Cells. 2007;25:2534. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 33.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 35.Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Nat Acad Sci USA. 2008;105:2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh AM, Dalton S. The cell cycle and myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell. 2009;5:141–149. doi: 10.1016/j.stem.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanna J, Saha K, Pando B, Van Zon J, Lengner CJ, Creyghton MP, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baudino TA, McKay C, Pendeville-Samain H, Nilsson JA, Maclean KH, White EL, et al. c-myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002;16:2530–2543. doi: 10.1101/gad.1024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koch HB, Zhang R, Verdoodt B, Bailey A, Zhang CD, 3rd, Menssen A, et al. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle. 2007;6:205–217. doi: 10.4161/cc.6.2.3742. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, et al. A myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, et al. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 44.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodeling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 45.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 47.Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 48.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 49.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 50.Agrawal P, Yu K, Salomon AR, Sedivy JM. Proteomic profiling of Myc-associated proteins. Cell Cycle. 2010;9:4908–4921. doi: 10.4161/cc.9.24.14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2008;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 53.Oliveros JC. VENN. An interactive tool for comparing lists with Venn Diagrams. 2007. bioinfogp.cnb.csic.es/tools/venny/index.html.
- 54.Lim JM, Sherling D, Teo CF, Hausman DB, Lin D, Wells L. Defining the regulated secreted proteome of rodent adipocytes upon the induction of insulin resistance. J Proteome Res. 2008;7:1251–1263. doi: 10.1021/pr7006945. [DOI] [PubMed] [Google Scholar]