Abstract
Chk1 is a critical effector of DNA damage checkpoints necessary for the maintenance of chromosome integrity during cell cycle progression. Here we report, that Chk1 co-localized with the nucleolar marker, fibrillarin in response to radiation-induced DNA damage in human cells. Interestingly, in vitro studies using GST pull down assays identified the dual-specificity serine/threonine nucleolar phosphatase Cdc14B as a Chk1 substrate. Furthermore, Chk1, but not a kinase-dead Chk1 control, was shown to phosphorylate Cdc14B using an in vitro kinase assay. Co-immunoprecipitation experiments using exogenous Cdc14B transfected into human cells confirmed the interaction of Cdc14B and Chk1 during cell cycle. In addition, reduction of Chk1 levels via siRNA or UCN-01 treatment demonstrated that Chk1 activation following DNA damage was required for Cdc14B export from the nucleolus. these studies have revealed a novel interplay between Chk1 kinase and Cdc14B phosphatase involving radiation-induced nucleolar shuttling to facilitate error-free cell cycle progression and prevent genomic instability.
Key words: Chk1, nucleoli, DNA damage, Cdc14B, genomic instabiliy, cell cycle
Introduction
Chk1 is an essential serine/threonine kinase required for normal mammalian development.1 We and others have shown that reduction of Chk1 levels in mice results in significant increase of apoptosis, spontaneous DNA damage foci, mammary gland defects, defective T-cell development, defective erythropoiesis and anemia.2–5 During cell cycle progression, Chk1 is a key player in firing origins of replication and monitoring replication fork progression during S phase.6 Recent studies also have shown that Chk1 is essential for spindle checkpoint, chromosome segregation and cytokinesis required to execute normal cell division.7,8 Thus, reduction of Chk1 levels causes various developmental defects in mammals, due to mis-regulation of cell cycle events leading to a loss of chromosomal integrity during cell division.
Orthologs of Chk1 kinase have been identified in all eukaryotes,9 which share a highly conserved N-terminal kinase domain of ∼250 amino acids and a C-terminal regulatory domain of ∼200 residues with an ill-defined function.10 Chk1 regulation during the cell cycle is complex and involves many upstream candidates. In S. pombe and S. cerevisiae, carboxy-terminal ATR/ATM kinase consensus (S/TQ) sites are phosphorylated following DNA damage. This results in increased Chk1 activity required for mediating the checkpoint response.11 In higher eukaryotes, the C terminus of Chk1 kinase has been suggested to play an inhibitory role through its interaction with the kinase domain. Accordingly, phosphorylation of C-terminal residues results in the loss of this inhibition.12 In addition to DNA damage-mediated Chk1 activation, several other proteins such as Claspin and Brca1 are necessary for complete activation of Chk1 kinase.13,14 Activated Chk1 recognizes its target substrates through a consensus sequence motif [R-X-X-S/T].15 An extensively studied and well characterized group of Chk1 substrates are the positive cell cycle regulators, Cdc25 phosphatases.10 In the presence of DNA damage, Chk1 phosphorylates Cdc25C phosphatase on serines embedded in the 14-3-3 recognition sites. This results in the binding of 14-3-3 and nuclear exclusion of 14-3-3 bound Cdc25C, leading to cell cycle arrest and checkpoint activation during G2/M phase to facilitate DNA repair.16,17 Similarly, Chk1 is also required for chromatin remodeling and repair in damaged cells via its phosphorylation of TLK1 at Ser743 to regulate the chromatin assembly factor ASF1A during S phase of the cell cycle.18,19 Recently, additional studies have identified many other critical cell cycle regulators as Chk1 substrates such as TLK1, BubR1, Aurora B, Plk1 in the presence and absence of DNA damage to facilitate cell cycle progression in a timely and error-free manner.8,20
During the cell cycle, multiple phosphorylation and dephosphorylation events regulate the localization, as well as the activity of various proteins within the compartmentalized cell for spatial-temporal regulation of various interconnected signaling pathways. For example, sub-nuclear shuttling of catalytically active human telomerase is induced by the cell cycle stage, transformation and DNA damage. Another well studied example is the nucleolar tumor suppressor protein p14ARF, which induces nucleoplasmic p53 via its binding partners B23 and topoisomerase 1 in response to oncogene activation or DNA damage.21–23 In eukaryotes, Chk1 is primarily thought to be a nucleo-cytoplasmic protein and contains a multipartite unusually long nuclear localization signal (NLS) in its regulatory C-terminal domain.12,17 In mammalian cells, Chk1 also localizes to the centrosome to protect centrosomal CDC2 kinase from inappropriate activation by cytoplasmic CDC25B and inappropriate mitotic entry.24 Interestingly, a recent study demonstrated a two-step mechanism of Chk1 phosphorylation at both Ser317 and Ser 345 required for proper centrosomal localization of Chk1 in the presence and absence of DNA damage.25 Moreover, we have shown that phospho-Chk1 Ser317 localizes to the perichromosomal layer, mid-zone and midbody during mitosis and cytokinesis, respectively.7,26 Inhibition of Chk1 levels in normal mitotic cells results in chromosome mis-segregation and binucleation. Similarly, Zachos et al. has reported the localization of GFP-Chk1 to the midzone and midbody during mitosis.8 This suggests that the sub-cellular translocation of Chk1 throughout cell cycle progression is required for not only checkpoint regulation but also for spatial-temporal regulation during cell cycle progression.
A new study by Bassermann et al. has defined a novel pathway that is critical for the G2 DNA damage-response checkpoint. In response to DNA damage, mammalian cells in G2 cannot enter mitosis, since they initiate DNA repair. In response to genotoxic stress, a dual-specificity serine/threonine Cdc14B phosphatase translocates from the nucleolus to the nucleoplasm and induces the activation of the ubiquitin ligase APC/CCdh1 and degradation of Plk1. This results in the stabilization of the DNA damage checkpoint activator Claspin and a cell cycle inhibitor Wee1 resulting in an efficient G2 checkpoint. A deubiquitylating enzyme Usp28 facilitates Claspin-mediated activation of Chk1 in response to DNA damage. This study, therefore, has united the fourteen early anaphase release (FEAR)/mitotic exit network (MEN) pathways with the G2/M DNA damage checkpoint pathway.28,29 Furthermore, it established a connection between Cdc14B and Chk1 in G2 DNA damaged cells. Cdc14B is a key player for the FEAR/MEN network and has been extensively studied in budding and fission yeast. In yeast, Cdc14p or Clp1/Flp1 is released during early and late anaphase respectively, to dephosphorylate mitotic cyclin-dependent kinases.30 Cdc14 regulates spindle midzone assembly and function directly through Ase1/hPRC1 and indirectly via the separase-Slk19 complex controlling anaphase B in S. cerevisiae.31 Thus, numerous studies using yeast as a model system have identified a role for Cdc14 in executing anaphase and mitotic exit. In C. elegans, ceCdc14deficient embryos also have central spindle formation defects and mislocalized proteins during anaphase and cytokinesis.32
Mammals have two Cdc14 orthologs, hCdc14A and hCdc14B. The former localizes to the centrosomes and is required for centrosome maturation, sister chromatid segregation and cytokinesis,33,34 whereas, Cdc14B depletion leads to centriole amplification and overexpression prevents unscheduled centriole duplication and S-phase arrest.35 hCdc14B is localized in the nucleolus during interphase, but releases onto the spindle midzone and midbody during anaphase and cytokinesis, respectively.36 In addition, Cho et al. have also demonstrated that Cdc14B is a unique nucleolar phosphatase, which can bundle and stabilize microtubules independent of its phosphatase activity. In a new study by Mocciaro et al. vertebrate cells genetically deleted for Cdc14A or Cdc14B were found to exhibit an increased number of irradiation-induced gamma-H2A.X foci and DNA double-strand breaks as compared to controls, suggesting a role in efficient DNA repair. Taken together these results suggest the existence of a novel regulatory pathway involving Chk1 and Cdc14B as key players required for regulating cell cycle progression.
In this present study, we made the unexpected observation that Chk1 colocalized with the nucleolar marker fibrillarin in a time-dependent manner in response to IR-induced DNA damage. Various biochemical experiments involving GST pull down assays, immunoprecipitation and kinase assays have identified that Chk1 directly interacts with and phosphorylates Cdc14B. Similar to studies by Bassermann et al. we also observed that exogenously transfected Cdc14B translocated from nucleoli to the nucleoplasm in response to IR-induced DNA damage. Interestingly, reducing Chk1 levels in mammalian cells using either siRNA or a chemical inhibitor UCN-01, prevented the translocation of Cdc14B to the nucleoplasm in response to IR-induced DNA damage. These results suggest that Chk1 can directly regulate the nucleolar phosphatase Cdc14B, which has been identified as a key player in the DNA damage response pathway.
Results
Chk1 translocates to the nucleolus in response to irradiation.
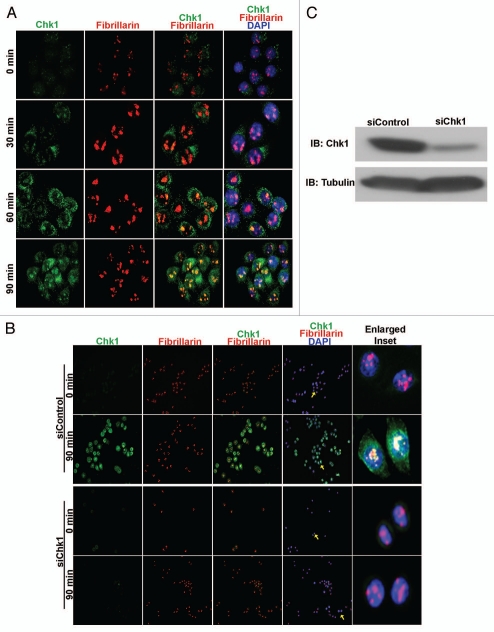
While elucidating the multifunctional role of Chk1 during cell cycle progression,7 we detected Chk1 foci in the nucleolar regions of HeLa cells 30 min post-irradiation at 10 GY. To confirm this observation, we irradiated asynchronous HeLa cell cultures and fixed them at various time points, 0, 30, 60 and 90 min post-irradiation, for immunostaining with the nucleolar marker fibrillarin. Co-immunostaining of irradiated HeLa cells with anti-Chk1 and anti-fibrillarin antibodies revealed co-localization of Chk1 foci with fibrillarin within 30 min. Moreover, an increased number of Chk1 foci was seen within nucleoli of irradiated HeLa cells at 90 min, a time when the maximum co-localization with fibrillarin was observed (Fig. 1A).
Figure 1.
Chk1 translocates to the nucleoli in response to irradiation. (A) Merged images at 40× magnification of irradiated HeLa cells stained with Chk1 (green), fibrillarin (red) and DNA (DAPI-blue) show translocated Chk1 foci in the nucleoli and colocalization with fibrillarin in a time-dependent manner from 0–90 min. (B) Merged images at 20× magnification of irradiated HeLa cells stained with Chk1 (green), fibrillarin (red) and DNA (DAPI-blue) show Chk1 colocalized with fibrillarin at 90 min in siControl-treated HeLa cells and not in siChk1-treated HeLa cells. Enlarged inset shows colocalization of Chk1 with fibrillarin (yellow arrows) at 90 min in irradiated siControl HeLa cells and not in irradiated siChk1 HeLa cells (yellow arrows). (C) Immunoblotting (IB) with Chk1 antibody confirms reduction of Chk1 levels in siChk1-transfected HeLa cells as compared to siControl-transfected HeLa cells.
To confirm the specificity of Chk1 antibody and the translocation of Chk1 post-irradiation, we knocked down Chk1 expression using Chk1 siRNA in HeLa cells. Western blot results confirmed a significant reduction in total Chk1 levels with Chk1 siRNA (Fig. 1C). The Chk1 siRNA treated HeLa cells also were subjected to irradiation at 10 GY. The irradiated Chk1 siRNA and control siRNA HeLa cells then were fixed at 0 min and 90 min for co-immunostaining with the anti-Chk1 (G4) and anti-fibrillarin antibodies. Chk1 foci co-localized with fibrillarin were seen at 90 min in control siRNA treated HeLa cells, whereas Chk1 siRNA treated HeLa cells did not contain any detectable Chk1 foci in the nucleoli (Fig. 1B). Thus, these experiments demonstrated that Chk1 translocates to the nucleolus in a time-dependent manner presumably to regulate cell cycle progression in response to IR-induced damage in mammalian cells.
Chk1 interacts with and phosphorylates Cdc14B in vitro.
Since the DNA damage effector Chk1 kinase translocated into nucleoli post-irradiation, we hypothesized that it might regulate the activity of a critical nucleolar protein or proteins involved in the DNA damage response pathway. Recent studies have demonstrated that nucleolar Cdc14B translocates into the nucleoplasm following IR-induced DNA damage to participate in the activation of G2/M checkpoint. Therefore, we performed GST pull down assays to determine whether in vitro translated radiolabeled-Chk1 directly interacts with GST-Cdc14B. We observed that Chk1 interacted directly with GST-Cdc14B, but not with the GST-control (Fig. 2A). Next, to test whether Cdc14B might be one of the potential substrates of Chk1 kinase, we performed kinase assays using either an active Chk1 or a kinase-deficient Chk1 mutant and in vitro translated Cdc14B. Active Chk1, but not the kinase-dead mutant phosphorylated Cdc14B (Fig. 2B). Finally, to confirm these in vitro observations we performed immunoprecipitation experiments using HeLa cells expressing Flag-Cdc14B. Immunoblotting with Flag antibody confirmed that Chk1 interacts with Cdc14B in cycling mammalian cells (Fig. 2C). These results suggest that nucleolar Cdc14B is a new Chk1 substrate possibly involved in the regulation of the IR-induced DNA damage response.
Figure 2.
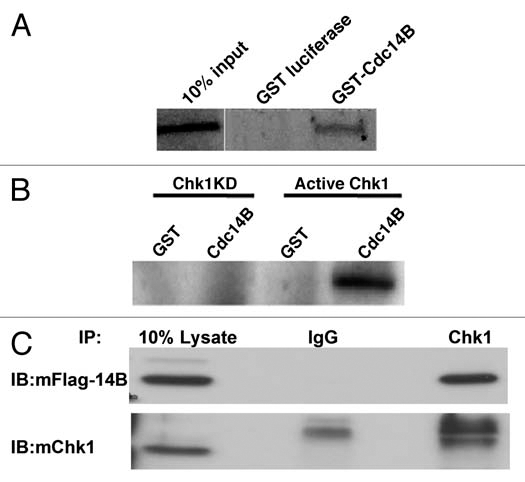
Chk1 interacts with and phosphorylates Cdc14B in vitro. (A) GST pull down assay shows GST-Cdc14B interacts with Chk1 in vitro and not a GST-luciferase control. (B) In vitro kinase assay shows active Chk1, but not kinase dead Chk1 (Chk1KD) phosphorylates Cdc14B. (C) Immunoprecipitation (IP) with polyclonal anti-Chk1 antibody and immunoblotting (IB) with monoclonal anti-Flag antibody (anti-mFlag) show that Chk1 interacts with Flag-Cdc14B in transfected HeLa cells and not the control anti-IgG.
Cdc14B undergoes sub-nuclear shuttling in response to radiation-induced DNA damage.
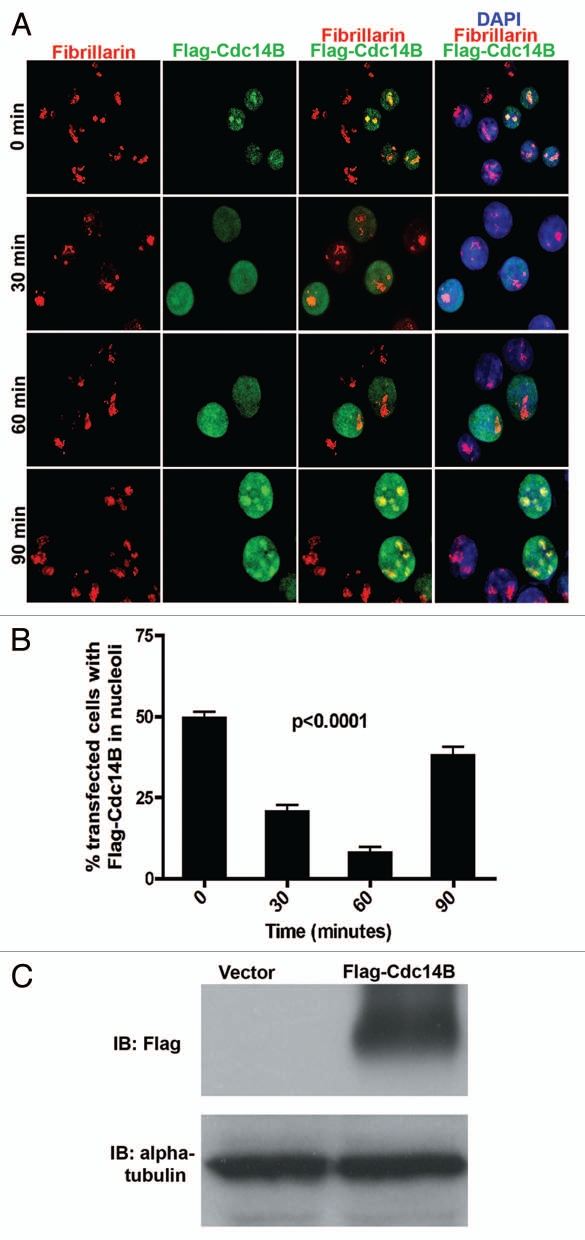
To understand the kinetics of nucleolar to nucleoplasmic translocation of Cdc14B, we transfected asynchronous HeLa cells with exogenous Flag-Cdc14B. Immunoblotting confirmed the expression of Flag-Cdc14B in transfected HeLa cells (Fig. 3C). Forty-eight hours posttransfection the cells were exposed to 10 Gy of radiation to study the spatio-temporal translocation of Flag-Cdc14B from the nucleoli. The irradiated cells were fixed at the same time-points, 0, 30, 60 and 90 min, used for studying Chk1 kinetics in response to irradiation. We co-immunostained the irradiated HeLa cells with anti-fibrillarin and anti-Flag antibodies to follow the sub-nuclear localization patterns of Flag-Cdc14B. At 0 min, Flag-Cdc14B colocalized with fibrillarin in ∼50% of the transfected cells. Whereas, within 30–60 min after radiation treatment, Flag-Cdc14B trans-locates from the nucleoli to the nucleoplasm and forms diffuse foci (Fig. 3A and B). Quantitation of these data confirmed that <25% of transfected cells 30 min post-irradiation show Cdc14B co-localization with fibrillarin (Fig. 3B). Interestingly, 90 min after irradiation, an increase in the Flag-Cdc14B foci co-localizing with fibrillarin in the nucleolus was observed in >40% of the transfected cells (Fig. 3A and B). These results confirm that Cdc14B shuttles between sub-nuclear compartments in response to radiation-induced DNA damage, as previously reported by Bassermann et al. (2009). Therefore, we hypothesized that Chk1 may directly regulate the sub-nuclear translocation patterns of Cdc14B post-irradiation in mammalian cells.
Figure 3.
Sub-nuclear shuttling of Cdc14B in response to radiation-induced DNA damage. (A) Merged images at 40× magnification of irradiated HeLa cells stained for Flag-Cdc14B (green), Fibrillarin (red) and DNA (DAPI-blue) show sub-nuclear shuttling and co-localization of Flag-Cdc14B foci with fibrillarin in a time-dependent manner from 0–90 min. (B) Quantitation indicates a significant decrease in the percentage of transfected HeLa cells showing Flag-Cdc14B foci in the nucleoli at 30 and 60 min post-irradiation, compared to 0 and 90 min. (C) Immunoblotting (IB) with an anti-Flag antibody confirms expression of Flag-Cdc14B in exogenously transfected HeLa cells compared to vector-transfected HeLa cells. Alpha-tubulin was used as loading control.
Reduced Chk1 levels prevent sub-nuclear shuttling of Cdc14B after radiation-induced dna damage.
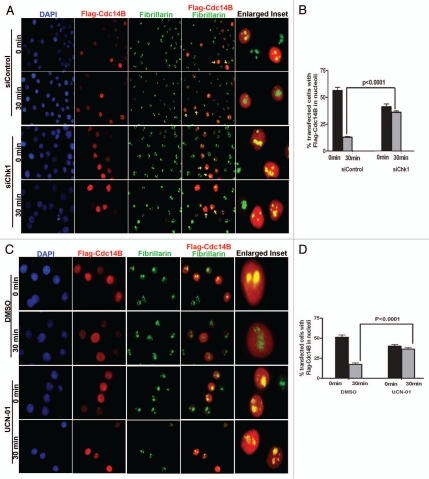
To test this hypothesis, we used two different methods to regulate Chk1 activity. First, we knocked down Chk1 levels using siRNA in Flag-Cdc14B transfected HeLa cells. western blot analysis confirmed a significant decrease in total Chk1 levels in these cells. The Flag-Cdc14B transfected Chk1 siRNA HeLa cells were treated with 10 Gy of radiation and the cells were fixed at 0 and 30 min post-treatment. We then co-immunostained the irradiated cells with anti-Flag and fibrillarin antibodies as before. Interestingly, we observed a significant increase in the percentage of transfected cells showing co-localized Flag-Cdc14B with fibrillarin in the nucleoli at 30 min in Chk1 siRNA HeLa cells as compared to control siRNA transfected HeLa cells. (Fig. 4A and B), thus, supporting our hypothesis that Chk1 might directly control the sub-nuclear translocation of Cdc14B in response to IR-induced DNA damage.
Figure 4.
Reduced Chk1 levels prevent sub-nuclear shuttling of Cdc14B after radiation-induced DNA damage. (A) Merged images at 40× magnification of transfected siRNA-treated HeLa cells stained for Flag-Cdc14B (red), fibrillarin (green) and DNA (DAPI-blue) show a reduction in sub-nuclear shuttling and colocalization of Flag-Cdc14B foci with fibrillarin in siChk1 HeLa cells as compared to siControl HeLa cells within 30 min post-radiation. The enlarged inset shows siControl- and siChk1-treated HeLa cells (yellow arrows) in the absence and presence Flag-Cdc14B foci colocalized with fibrillarin in the nucleoli 30 min post-radiation, respectively. (B) Quantitation indicates a significant increase of percentage of transfected HeLa cells showing Flag-Cdc14B foci in the nucleoli at 30 min post-irradiation in siChk1 HeLa cells as compared to siControl HeLa cells. (C) Merged images at 40× magnification of transfected UCN-01 treated HeLa cells stained for Flag-Cdc14B (red), fibrillarin (green) and DNA (DAPI-blue) show the reduction in sub-nuclear shuttling and colocalization of Flag-Cdc14B foci with fibrillarin after UCN-01 treatment as compared to DMSO control within 30 min post-radiation. The enlarged inset shows DMSO and UCN-01 treated HeLa cells (yellow arrows) in the absence and presence of Flag-Cdc14B foci colocalized with fibrillarin in the nucleoli 30 min post-radiation, respectively. (D) Quantitation indicates a significant increase of percentage of transfected HeLa cells showing Flag-Cdc14B foci in the nucleoli at 30 min post-irradiation in UCN-01 treated-HeLa cells, as compared to DMSO treated-HeLa cells.
Alternatively, we used a well studied Chk1 kinase inhibitor UCN-01.39 Similar to the Chk1 siRNA experiments, we treated Flag-Cdc14B transfected HeLa cells with 300 nm UCN-01 for 3 hr and subjected these cells to IR-induced DNA damage at 10 Gy. As a control, we used the DMSO vehicle instead of UCN-01. Once again, the irradiated UCN-01 and DMSO treated Flag-Cdc14B HeLa cells were fixed at 0 and 30 min for co-immunostaining with the anti-Flag and anti-fibrillarin antibodies. We observed a significant increase in the percentage of transfected cells showing co-localized FlagCdc14B with fibrillarin in the nucleoli after 30 min in UCN-01 treated HeLa cells (Fig. 4C and D). Therefore, these results demonstrate that Chk1 regulates Cdc14B translocation between the nucleolus and nucleoplasm in response to IR-induced DNA damage treatment in mammalian cells.
Discussion
The current studies have identified the nucleolus as a previously unrecognized compartment where Chk1 is localized and signals in response to radiation-induced DNA damage to regulate cell cycle progression. The nucleolus is a prominent structure in the cell nucleus with well known functions including ribosomal RNA (rRNA) transcription, pre-rRNA processing and ribosome subunit assembly. It is a dynamic structure that assembles rRNA clusters at mitotic exit and persists throughout interphase, before disassembling again during mitotic entry.40 In the past years, studies have shown that Chk1 is not just shuttling between the nucleus and cytoplasm, but also receives signals at the centrosomes in response to DNA damage.41 DNA damage signals can be generated by external agents such as IR, UV, chemotherapy drugs or during natural cellular processes such as errors during DNA replication and repair.
Sequence analysis of Chk1 proteins from all known species, such as fission yeast42 Drosophila,43 Xenopus17 and human has identified a multiple bipartite NLS in the C-terminal region, which is required for its nuclear localization. Typically, a bipartite NLS consists of two small basic domains separated by a short nine to 12 amino acid sequence.44,45 Similarly, proteins localized to the nucleolus can have nucleolar localizing signals (NoLS), which usually is a stretch of four basic amino acids (RKKR) in the N-terminal half of the protein and is typically linked to a nuclear targeting sequence (KKK).46,47 In case of Chk1, the NLS is present in C terminus10,12 and our in silico analysis did not reveal any well studied NoLS near the NLS of Chk1. Interestingly, recent studies showed that BRCA1 also localized to nucleoli and as nucleoplasmic foci in breast cancer tissues and cell lines using immunohistochemistry. In response to IR-induced radiation BRCA1 was dispersed to various nucleoplasmic sites.48 These unexpected observations open new possibilities for as yet to be discovered nucleolar-nucleoplasmic functions of the DNA damage and repair proteins in the DNA damage response pathway.
As a part of this study, we have uncovered a unique interaction between nucleolar Cdc14B dual-specificity serine/threonine phosphatase and Chk1 kinase. Our in vitro studies have confirmed a direct interaction between these two proteins, while the co-immunoprecipitation results revealed an interaction between Chk1 and Cdc14B in mammalian cells. A recent study has demonstrated that Cdc14B can activate the G2-checkpoint response during IR-induced DNA damage.27 Based upon our observations, it appears that in response to radiation treatment, Chk1 translocates into nucleoli to activate Cdc14B in order to enable Cdc14B export into the nucleoplasm to help mediate this checkpoint response. Even in the context of non-DNA damaged “normal” cells, a fraction of activated Chk1 may translocate into the nucleolus to phosphorylate Cdc14B, which in turn may be required to carry out its functions in the DNA replication-repair pathway. Thus, using siRNA and chemical inhibitor treatment we have shown that reduction of Chk1 levels can lead to accumulation of Cdc14B in the nucleoli in response to DNA damage. A recent study also has demonstrated that disruption of Cdc14B resulted in spontaneous DNA damage foci formation38 similar to that observed following Chk1 inhibition in mammalian cells. Moreover, these Cdc14B-deficient cells also showed delayed DNA repair following DNA damage. Therefore, it is possible that reducing the DNA damage effector Chk1 kinase, may mis-regulate Cdc14B phosphorylation and affect its nucleolar translocation in damaged cells. This in turn may significantly impact Cdc14B's yet to be elucidated DNA repair functions in the sub-nuclear compartment to recover and resume normal cell cycle progression. Taken together with the recent discoveries of Cdc14B functions in mammalian cells, we have demonstrated that Chk1 is one of the kinases that can directly regulate Cdc14B. Based on our results, we propose a working model wherein, Chk1 might be activated by external DNA damage such as IR or during normal DNA replication in a mammalian cell by an unknown kinase. Activated Chk1 then translocates into the nucleolus to possibly phosphorylate Cdc14B and initiate its sub-nuclear shuttling to regulate cell cycle progression in an error-free manner. Thus, DNA damage dependent Chk1-Cdc14B interplay might be required for cell cycle arrest and DNA repair to prevent the intiation of genomic instability.
These studies raise several questions about a number of undiscovered functions of Chk1 kinase and its localization during interphase. Future studies will be necessary to determine which kinase phosphorylates Chk1 in response to IR-induced DNA damage to facilitate Chk1 translocation into the nucleolus? In the absence of external DNA damage, does Chk1 also shuttle into the nucleolus during normal DNA replication and repair? Are there any new NoLS sequence motifs in the C terminus of Chk1 that enables it to translocate to the nucleolus? Is phosphorylation of Chk1 by ATM or ATR at sites such as Ser 345 and Ser317 on or any other caffeine-sensitive kinase necessary for Chk1 translocation and interaction with Cdc14B in response to DNA damage? What is the functional significance of Chk1 phosphorylation of Cdc14B in mammals? Is it required to regulate phosphatase-independent and -dependent Cdc14B functions during cell cycle progression to prevent genomic instability? Similarly, are there other DNA damage and DNA repair proteins that utilize a similar nucleolar shuttling pathway to regulate cell cycle progression? As more studies uncover the answers to these questions, they should aid in the design of new targeted therapies to treat cancer patients in combination with both chemo- and radiation therapy.
Materials and Methods
Cell culture.
HeLa cells were ordered from ATCC and grown on poly-lysine D coated 12–25 mm glass coverslips (Fisher, Pittsburgh, PA) in DMEM high glucose medium (Invitrogen, La Jolla, CA) supplemented by 10% bovine calf serum (BCS). The cells were maintained in a humid incubator at 37°C in 5% CO2 environment depending on cell type.
Transfection and inhibitors.
HeLa cells were transfected with Chk1 siRNA (Sigma) for 48 hr according to manufacturer's protocol. The transfected cells post-irradiation were fixed with 4% PFA for immunocytochemistry and microscopy. Similarly, HeLa cells were treated with Chk1 inhibitor, UCN-01 (Sigma) at 300 nM as described in Syljuasen et al. The inhibited cells post-irradiation were fixed with 4% PFA for immunocytochemistry and microscopy.
GST pull down assay.
[35S]-methionine-labeled Chk1 was synthesized using TNT quick coupled transcription/translation system (Promega, Madison, WI) with full-length Chk1 cDNA. Recombinant GST-Cdc14B and GST control bound glutathione beads were used to directly bind [35S]-methionine labeled Chk1. The reactions were performed in 0.25 ml phosphate buffer with 0.2% NP40 (pH 7.3) overnight at 4°C. The beads were separated on 10% SDS-PAGE and autoradiography was performed for visualization. [35S]-methionine labeled luciferase was used for control experiments.
In vitro kinase assay.
Recombinant GST and GST-Cdc14B (human) bound to glutathione beads were subjected to thrombin digestion in presence of 0.1 units of thrombin (Novagen, San Diego, CA) in the presence of either 0.6 µg of recombinant GST-Chk1 or kinase-dead GST-Chk1. The digest was performed in 70 mM Tris-HCL (pH 7.5) for 1 hr at 25°C followed by the kinase assay. The kinase reaction was initiated with addition of MgCl2 (10 mM final concentration) DTT (5 mM final concentration) and γ-P32-ATP. The reactions were incubated for 20 min at 30°C followed by 40 min incubation at 25°C. The samples were separated on 10% SDS-PAGE and autoradiography was conducted for visualization.
Co-immunoprecipitation and immunoblotting.
HeLa cells were washed in ice cold PBS three times, lysed using RIPA buffer (1 ml/100 mm dish) for 30 min at 4°C, the adherent cells were scraped and transferred into respective Eppendorf tubes, gently rocked for 15 min at 4°C, centrifuged at 15,000 rpm for 15 min at 4°C and tubes, gently rocked for 15 min at 4°C and the supernatant aliquoted into two new tubes. For immunoprecipitation, samples were precleared and then antibody added to immunoprecipitate the respective proteins and incubated for 45 min at 4°C in a rotary shaker. The complex was spun down and the pellet was transferred to a new tube and washed carefully. Pellets were resuspended in 50 µl of sample buffer with beta-mercaptoethanol, boiled for 2 min, centrifuged in a microfuge for 4 min and resolved using 10% polyacrylamide gels (Bio-Rad, Hercules, CA) using a minigel apparatus, followed by transfer to nitrocellulose membrane for immunoblotting. In addition, HeLa cells (1 × 108) treated with nocodazole (80 ng/ml) were initially subjected to sonication under hypotonic buffer (20 mM phosphate, protease and phosphatase inhibitors) followed by homogenization with (1% Triton-X-100). The supernatants (3.4xmg/ml) obtained by centrifugation were added with 30 µl of either in the presence or absence of free Chk1 antibodies (20 µl). The agarose bound immunoprecipitates were subjected to western blotting using anti-Cdc14B (Zymed, San Francisco, CA) or anti-Chk1 (G4) (Santa Cruz Technology, Santa Cruz, CA) or anti-myc (Abcam, Cambridge, UK) or anti-Flag (Sigma, St. Louis, MO).
Immunofluorescence and deconvolution microscopy.
For routine immunostaining, cells were permeabilized with 0.5% Triton X-100 for 10 min, blocked with 5% goat serum (Sigma, St. Louis, MO) for 30 min and incubated with primary antibodies anti-Chk1 (G4) or anti-Chk1(FL476) (Santa Cruz Technology, Santa Cruz, CA) and anti-Fibrillarin (Abcam, Cambridge, UK) or anti-Flag (Sigma, St. Louis, MO, USA) overnight at 4°C on a shaker. Cells were subsequently washed three times with 1× PBS, blocked with 5% goat serum (Sigma, St. Louis, MO) for 30 min again and incubated with respective fluorochrome-conjugated secondary antibody for 1 hr. They were then washed five times and counterstained with Vectashield 4,6-diamidino-2-phenylindole (DAPI) to visualize DNA. Images were taken using 20×, 40× and 63á magnifications using either Olympus Fluorescence microscope with Spot imaging software or 1 × 1 or 2 × 2 bin on a Nikon/DeltaVision fluorescence microscope (Applied Precision, Issaquah, WA) as a series of 0.1 or 0.2 µM thick z sections and deconvolution was performed with a Softworx image workstation (10 iterations, algorithm: ratio-conservative).
Statistical analysis.
All the results were confirmed in multiple independent experiments. Data quantitation and analysis was performed by Student′s t-test and expressed as ±SEM using Prism software. p-values of less than 0.05 are considered statistically significant.
Acknowledgements
We would like to thank members of Rosen and Zhang laboratories for their helpful discussions during the entire course of this project. This work is supported by NIH grants CA116097 and CA122623 to P.Z. and NCI R37CA16303 to J.M.R.
References
- 1.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 2.Greenow KR, Clarke AR, Jones RH. Chk1 deficiency in the mouse small intestine results in p53-independent crypt death and subsequent intestinal compensation. Oncogene. 2009;28:1443–1453. doi: 10.1038/onc.2008.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boles NC, Peddibhotla S, Chen AJ, Goodell MA, Rosen JM. Chk1 haploinsufficiency results in anemia and defective erythropoiesis. PLoS One. 5:8581. doi: 10.1371/journal.pone.0008581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 2004;6:45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 5.Zaugg K, Su YW, Reilly PT, Moolani Y, Cheung CC, Hakem R, et al. Cross-talk between Chk1 and Chk2 in double-mutant thymocytes. Proc Natl Acad Sci USA. 2007;104:3805–3810. doi: 10.1073/pnas.0611584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci USA. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peddibhotla S, Lam MH, Gonzalez-Rimbau M, Rosen JM. The DNA damage effector checkpoint kinase 1 is essential for chromosome segregation and cytokinesis. Proc Natl Acad Sci USA. 2009;106:5159–5164. doi: 10.1073/pnas.0806671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC, et al. Chk1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–260. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, et al. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 10.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Sanchez Y. Chk1 in the DNA damage response: conserved roles from yeasts to mammals. DNA Repair (Amst) 2004;3:1025–1032. doi: 10.1016/j.dnarep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Palermo C, Hope JC, Freyer GA, Rao H, Walworth NC. Importance of a C-terminal conserved region of Chk1 for checkpoint function. PLoS One. 2008;3:1427. doi: 10.1371/journal.pone.0001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- 14.Yarden RI, Pardo-Reoyo S, Sgagias M, Cowan KH, Brody LC. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat Genet. 2002;30:285–289. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- 15.O′Neill T, Giarratani L, Chen P, Iyer L, Lee CH, Bobiak M, et al. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J Biol Chem. 2002;277:16102–16115. doi: 10.1074/jbc.M111705200. [DOI] [PubMed] [Google Scholar]
- 16.Dunaway S, Liu HY, Walworth NC. Interaction of 14-3-3 protein with Chk1 affects localization and checkpoint function. J Cell Sci. 2005;118:39–50. doi: 10.1242/jcs.01570. [DOI] [PubMed] [Google Scholar]
- 17.Katsuragi Y, Sagata N. Regulation of Chk1 kinase by autoinhibition and ATR-mediated phosphorylation. Mol Biol Cell. 2004;15:1680–1689. doi: 10.1091/mbc.E03-12-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krause DR, Jonnalagadda JC, Gatei MH, Sillje HH, Zhou BB, Nigg EA, et al. Suppression of Tousled-like kinase activity after DNA damage or replication block requires ATM, NBS1 and Chk1. Oncogene. 2003;22:5927–5937. doi: 10.1038/sj.onc.1206691. [DOI] [PubMed] [Google Scholar]
- 19.Groth A, Lukas J, Nigg EA, Sillje HH, Wernstedt C, Bartek J, et al. Human Tousled like kinases are targeted by an ATM- and Chk1-dependent DNA damage checkpoint. EMBO J. 2003;22:1676–1687. doi: 10.1093/emboj/cdg151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang J, Erikson RL, Liu X. Checkpoint kinase 1 (Chk1) is required for mitotic progression through negative regulation of polo-like kinase 1 (Plk1) Proc Natl Acad Sci USA. 2006;103:11964–11969. doi: 10.1073/pnas.0604987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindstrom MS, Klangby U, Inoue R, Pisa P, Wiman KG, Asker CE. Immunolocalization of human p14(ARF) to the granular component of the interphase nucleolus. Exp Cell Res. 2000;256:400–410. doi: 10.1006/excr.2000.4854. [DOI] [PubMed] [Google Scholar]
- 22.Rizos H, Darmanian AP, Mann GJ, Kefford RF. Two arginine rich domains in the p14ARF tumour suppressor mediate nucleolar localization. Oncogene. 2000;19:2978–2985. doi: 10.1038/sj.onc.1203629. [DOI] [PubMed] [Google Scholar]
- 23.Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer A, Lukas J, Bartek J. Checking out the centrosome. Cell Cycle. 2004;3:1390–1393. doi: 10.4161/cc.3.11.1252. [DOI] [PubMed] [Google Scholar]
- 25.Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peddibhotla S, Rosen JM. Chking and executing cell division to prevent genomic instability. Cell Cycle. 2009;8:2339–2342. doi: 10.4161/cc.8.15.9169. [DOI] [PubMed] [Google Scholar]
- 27.Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA damage-response checkpoint. Cell. 2008;134:256–267. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coulombe P, Rodier G, Meloche S. Cdc14B/Skp2/p27: a novel cancer axis of evil? Med Sci (Paris) 2009;25:673–675. doi: 10.1051/medsci/2009258-9673. [DOI] [PubMed] [Google Scholar]
- 29.Trautmann S, McCollum D. Cell cycle: new functions for Cdc14 family phosphatases. Curr Biol. 2002;12:733–735. doi: 10.1016/s0960-9822(02)01250-2. [DOI] [PubMed] [Google Scholar]
- 30.Bembenek J, Kang J, Kurischko C, Li B, Raab JR, Belanger KD, et al. Crm1-mediated nuclear export of Cdc14 is required for the completion of cytokinesis in budding yeast. Cell Cycle. 2005;4:961–971. doi: 10.4161/cc.4.7.1798. [DOI] [PubMed] [Google Scholar]
- 31.Khmelinskii A, Lawrence C, Roostalu J, Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J Cell Biol. 2007;177:981–993. doi: 10.1083/jcb.200702145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gruneberg U, Glotzer M, Gartner A, Nigg EA. The CeCDC-14 phosphatase is required for cytokinesis in the Caenorhabditis elegans embryo. J Cell Biol. 2002;158:901–914. doi: 10.1083/jcb.200202054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosso L, Marques AC, Weier M, Lambert N, Lambot MA, Vanderhaeghen P, et al. Birth and rapid subcellular adaptation of a hominoid-specific CDC14 protein. PLoS Biol. 2008;6:140. doi: 10.1371/journal.pbio.0060140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krasinska L, de Bettignies G, Fisher D, Abrieu A, Fesquet D, Morin N. Regulation of multiple cell cycle events by Cdc14 homologues in vertebrates. Exp Cell Res. 2007;313:1225–1239. doi: 10.1016/j.yexcr.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 35.Wu J, Cho HP, Rhee DB, Johnson DK, Dunlap J, Liu Y, et al. Cdc14B depletion leads to centriole amplification, and its overexpression prevents unscheduled centriole duplication. J Cell Biol. 2008;181:475–483. doi: 10.1083/jcb.200710127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nalepa G, Harper JW. Visualization of a highly organized intranuclear network of filaments in living mammalian cells. Cell Motil Cytoskeleton. 2004;59:94–108. doi: 10.1002/cm.20023. [DOI] [PubMed] [Google Scholar]
- 37.Cho HP, Liu Y, Gomez M, Dunlap J, Tyers M, Wang Y. The dual-specificity phosphatase CDC14B bundles and stabilizes microtubules. Mol Cell Biol. 2005;25:4541–4551. doi: 10.1128/MCB.25.11.4541-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mocciaro A, Berdougo E, Zeng K, Black E, Vagnarelli P, Earnshaw W, et al. Vertebrate cells genetically deficient for Cdc14A or Cdc14B retain DNA damage checkpoint proficiency but are impaired in DNA repair. J Cell Biol. 189:631–639. doi: 10.1083/jcb.200910057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, et al. Initial genomics of the human nucleolus. PLoS Genet. 6:1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loffler H, Bochtler T, Fritz B, Tews B, Ho AD, Lukas J, et al. DNA damage-induced accumulation of centrosomal Chk1 contributes to its checkpoint function. Cell Cycle. 2007;6:2541–2548. doi: 10.4161/cc.6.20.4810. [DOI] [PubMed] [Google Scholar]
- 42.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 43.Fogarty P, Campbell SD, Abu-Shumays R, Phalle BS, Yu KR, Uy GL, et al. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr Biol. 1997;7:418–426. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt-Zachmann MS, Nigg EA. Protein localization to the nucleolus: a search for targeting domains in nucleolin. J Cell Sci. 1993;105:799–806. doi: 10.1242/jcs.105.3.799. [DOI] [PubMed] [Google Scholar]
- 45.Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 46.Guo Z, Qian L, Liu R, Dai H, Zhou M, Zheng L, et al. Nucleolar localization and dynamic roles of flap endonuclease 1 in ribosomal DNA replication and damage repair. Mol Cell Biol. 2008;28:4310–4319. doi: 10.1128/MCB.00200-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrysik Z, Bernstein WZ, Deng L, Myer DL, Li YQ, Tischfield JA, et al. The novel mouse Polo-like kinase 5 responds to DNA damage and localizes in the nucleolus. Nucleic Acids Res. 38:2931–2943. doi: 10.1093/nar/gkq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tulchin N, Chambon M, Juan G, Dikman S, Strauchen J, Ornstein L, et al. BRCA1 protein and nucleolin colocalize in breast carcinoma tissue and cancer cell lines. Am J Pathol. 176:1203–1214. doi: 10.2353/ajpath.2010.081063. [DOI] [PMC free article] [PubMed] [Google Scholar]