Abstract
The ancient p53 paralogs p63 and p73 regulate specific tissue formation, cell survival and cell death via their TA and ΔN isoforms. Targeted disruption of the p73 locus leads to severe defects in the development of the central nervous system (CNS), and p73 has recently been shown to be an essential regulator of neural stem cell maintenance and differentiation in both embryonal and adult neurogenesis. In contrast, global p63−/− mice lack skin and limbs. Moreover, p63 is detectable in embryonic cortex. It has previously been proposed to also play critical pro-death and pro-survival roles in neural precursors of the developing sympathetic and central nervous system, respectively, based on experimental overexpression and siRNA-mediated knockdown of p63.
Here we perform an extensive analysis of the developing central nervous system in global p63−/− mice and their wildtype littermates. Brain and spinal cord of embryos and newborn mice were assessed in vivo for neuroanatomy, histology, apoptosis, proliferation, stemness and differentiation, and in vitro for self-renewal and maturation in neurosphere assays. None of these analyses revealed a detectable phenotype in p63−/− mice. Hence, despite the profound impact of p63 on the development of stratified epithelia and limbs, p63 is completely dispensable for proper development of the central nervous system. Thus, despite their strong homology, the non-overlapping tissue specificity of p63 and p73 functions appears more pronounced than previously anticipated.
Key words: p63, p73, neurons, apoptosis, proliferation, stem cell, differentiation, central nervous system
Introduction
The p63 and p73 gene products were first identified based on their structural similarity to the p53 tumor suppressor.1,2 Moreover, the structural homology between p63 and p73 is even higher than with p53. Unlike p53, however, each of its cousins is required for specific aspects of organ and tissue development. Mice with a targeted deletion of all isoforms of p63 are born without skin and show absent or severely truncated limbs.3 Accordingly, various point mutations and small deletions in the DNA-binding and C-terminal domains of human p63 result in the EEC/AEC syndromes characterized by defects in skin, skin appendages and malformations of hands and feet.4 In contrast, mice lacking all isoforms of p73 display severe defects in the development of the central nervous system (CNS) with 100% penetrance.5 They are characterized by progressive cortical hypoplasia with ensuing ex vacuo hydrocephalus, present already in one third of animals at birth,6 hippocampal dysgenesis with loss or truncation of the lower blade of the neurogenic dentate gyrus,6 thinning of the neurogenic subventricular zone in the forbrain and loss of the Cajal-Retzius neurons.7 Importantly, as recently established, this defect is largely due to p73's role as an essential regulator of neural stem cell self-renewal and maintenance in both embryonal and adult CNS neurogenesis.6,8,9 Moreover, as shown by isoform-specific knockout mice, ΔNp73 can specifically act as a pro-survival factor of differentiated mature postmitotic neurons in discrete regions of the brain.10,11 Furthermore, p73−/− mice exhibit increased apoptosis of sympathetic neurons of the autonomic nervous system in the superior cervical ganglion.12
p63 and p73 are closely related, and the corresponding genes appear to have separated by gene duplication during evolution.13 Two classes of p63 with six different isoforms due to alternative promoters and C-terminal splicing have been described. The transcriptional activation domain-containing TAp63 isoforms can act pro-apoptotically, while the ΔNp63 isoforms can act anti-apoptotically, similarly to the TA and ΔN isoforms of p73. This raises the question whether p63 has a similar central role in neural development as does p73. Indeed, in the case of sympathetic neurons of extracranial ganglia, a specific truncation of the α/β region of the C-terminal tail of the p63 gene (the hypomorphic Brdm2 allele) resulted in decreased developmental apoptosis in vivo, generating superior cervical ganglia with increased numbers of neurons.14 Likewise, cultured sympathetic wild-type neurons, which are reported to predominantly express TAp63, were driven into apoptosis upon high ectopic overexpression of TAp63γ. Conversely, ectopic overexpression of ΔNp63α, β or γ each rescued sympathetic neurons from NGF withdrawal-induced cell death. Thus, in developing sympathetic neurons an essential pro-apoptotic role was ascribed to the transactivating TAp63γ isoform,14 perhaps balancing the anti-apoptotic pro-survival function of N-terminally truncated ΔNp63 and/or ΔNp73 isoforms, which are coexpressed in mature neurons.2,7,12,14
From these findings it was proposed that p63 is a general and essential developmental regulator of all neuronal cells, particularly in the CNS.15–18 In support of this notion, a recent experimental study describes an essential anti-apoptotic pro-survival function of ΔNp63 in neural precursor cells in the developing CNS of mouse embryos.19 However, this study relied entirely on a strategy of shRNA-mediated knockdown of p63 in wildtype neurons. It used a small hairpin RNA to common exons of p63 for in utero electroporation into the brains of E13 wild-type embryos and transfection into cultured cortical precursors to achieve a reduction of p63 levels in neuronal cells. In this system that targets all p63 isoforms, it was reported that GFP-positive cells (used as marker for transfected cells), specifically stem and differentiating cells that migrate to the upper layers of the cortex, were drastically reduced in numbers when p63shRNA was co-transfected compared to control shRNA. This reduction in cell numbers was getting more profound over time from E13/14 to E16/17, suggesting that cell survival decreased in response to removal of p63.19 However, a potential pitfall of this experimental system is that it relies on transient transfections into neurons and the delicate developing brain. This constitutes a considerable stress, and thus may not represent the proper cell conditions and/or interfere with normal development of the CNS. Indeed, these results were challenged by the observation from us and others that the brain of p63−/− mice did not display gross anatomical malformations at birth.7
These disparate results raise the need to reassess the precise role of p63 in neural cell fate in a clean in vivo system that is not experimentally manipulated, i.e., in p63−/− mice. Here we analyze in detail whether the ablation of all isoforms of p63 affects CNS development. We assess neuroanatomy, tissue histology and quantify morphologic parameters and markers for apoptosis and proliferation, neural stemness and differentiation. This is flanked by in vitro examination of primary neurospheres derived from p63-proficient or p63-deficient brains. In all cases, the absence of p63 affected neither CNS development nor the fate and numbers of neural cells. We conclude that p63 is not required for any of the steps that lead to the formation of a normal brain and spinal cord.
Results
Normal development of brain and spinal cord in p63−/− embryos.
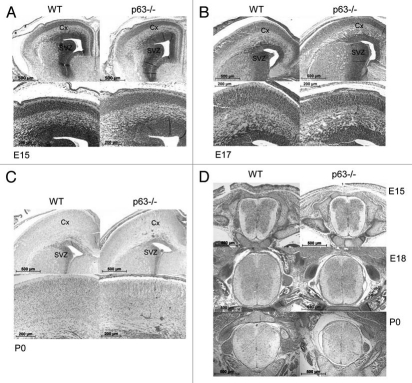
Neurogenesis is the process by which newborn neurons are created from neural stem cells (NSC). In mouse development it starts around day E11 and is completed at birth. To assess the requirement of p63 in neurogenesis, we first did a careful histologic analysis of the CNS from p63−/− mice lacking all isoforms and their wild-type (WT) littermates between stages E15 and P0 [n (E15) = 4, n (E17/18) = 2, n (P0) = 3 mice per genotype]. No anatomic differences were noted upon gross inspection of embryonic and newborn brains. Brains and spinal cords were fixed and paraffin embedded and corresponding coronal (brain) and transverse spinal sections were stained with H&E. No difference in brain histology was seen, neither within the cortical and subcortical layers nor in the neurogenic subventricular zone, which houses neural stem and progenitor cells that migrate up to the cortex (Fig. 1A—C). Likewise, the architecture of the spinal cord did not differ between WT and p63−/− mice (Fig. 1D). The identity of the p63KO mice used in this study was verified by the phenotype (lack of skin) and genotype (PCR) (Sup. Fig. 1A and B). Thus, we conclude that p63 plays no apparent role in the development of the central nervous system. This finding is in complete agreement with an independent concomitant recent report from the Meyer lab7 but not with the idea that p63 is required for survival of neural precursors in the brain.19
Figure 1.
p63−/− mice display normal histology of the brain and spinal cord. (A–C) Corresponding coronal sections of p63−/− and wild-type (WT) littermate mouse brains at stage E15, E17 and P0. Mayer's hematoxylin and eosin (H&E) staining. No structural differences were observed between the two genotypes. SVZ, subventricular zone; Cx, Cortex. (D) Transverse sections of the cervical spinal cord from p63−/− and WT littermate mice. H&E staining. Again p63−/− mice show normal development.
The absence of p63 in the developing central nervous system does not increase apoptosis.
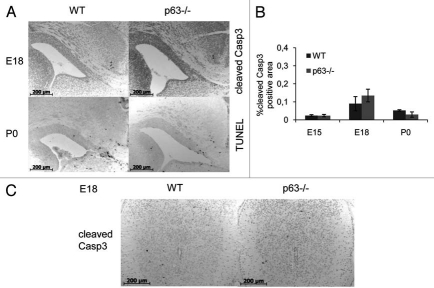
p63 was previously reported to modulate the rate of spontaneous apoptosis in neuronal cells, both in extracranial sympathetic ganglia14 and in cerebral neurons.19 We therefore assessed the number of spontaneous apoptotic cells in brain and spinal cord of WT and littermate p63−/− mice at stages E15, E18 and P0 with two different assays. A small fraction of cells stained positively for activated cleaved caspase 3 and by TUNEL. However, the proportion of positive cells did not vary between mice regardless of p63 status and assay (Fig. 2A and B). Similar findings were obtained in the spinal cord (Fig. 2C). Together, this strongly argues that the developmental apoptotic rate in the central nervous system is not affected by p63.
Figure 2.
The developing central nervous system of p63−/− mice does not exhibit increased apoptosis. (A) Corresponding coronal brain sections from p63−/− and WT littermate embryos were assayed by immunohistochemistry to detect cleaved Caspase 3 (E18), or stained by TUNEL (P0) to visualize spontaneously apoptotic cells. (B) Quantification of cleaved Caspase 3 levels in the ventricular and subventricular zones (VZ/SVZ) and cortical plate (not shown) of p63−/− embryos versus WT littermates at the indicated stages. The percentage of positively stained areas were quantified in brain sections of 2–5 animals per genotype and embryonic stage. At least three coronal sections per animal were analyzed per data point. Error bars represent the standard deviation (SD) of the mean. No significant difference was found between the genotypes (Student's t-test). (C) Immunohistochemical analysis of cleaved Caspase 3 in spinal cord at stage E18 revealed no difference in the number of apoptotic cells.
Normal proliferation of neural precursors in the VZ/SVZ of the brain and in the spinal cord of p63−/− mice.
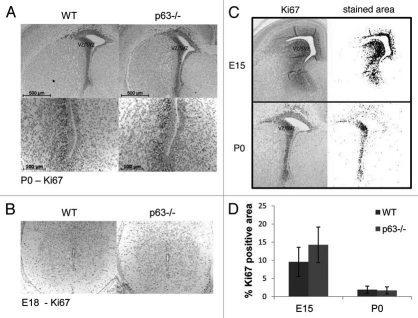
Next, we asked whether p63 influences the rate of proliferation in the neurogenic niche of the CNS. To this end, we immunostained sections of brain and spinal cord of p63−/− and WT littermates at E15 and P0 for Ki67, a marker of DNA replication. However, no difference in number or distribution of Ki67-positive cells within the ventricular and subventricular zone (VZ/SVZ) was found (Fig. 3A and B). This was confirmed by quantitative image analysis (Fig. 3C). No difference was seen in the spinal cord either (Fig. 3D). These results are in agreement with those obtained by in utero electroporation of p63shRNA.19 We conclude that p63 does not affect the proliferation of neural cells in the developing brain.
Figure 3.
Normal proliferation of neural precursors in the VZ/SVZ of the brain and in spinal cord of p63−/− mice. (A and B) Immunohistochemical analysis of brain (A) and spinal cord (B) to visualize the proliferating population of neural precursors. Tissues from p63−/− embryos and WT littermates at the indicated stages were stained for Ki67. VZ/SVZ, ventricular zone, subventricular zone (C and D). (C) For quantitation, a digital mask was derived from images of Ki67-stained VZ/SVZ of corresponding brain sections using the image analysis software Image J.39 (D) Ki67 positive cells in the VZ/SVZ of WT and p63−/− littermates at E15 and P0 were quantified. The percentage of positively stained areas (as determined in C) was quantified in 2–5 animals per genotype per embryonic stage. Four coronal sections per animal were analyzed per data point. Error bars represent the SD of the mean. No significant difference was found between the genotypes (Student's t-test).
p63 does not influence the stemness of neural progenitors.
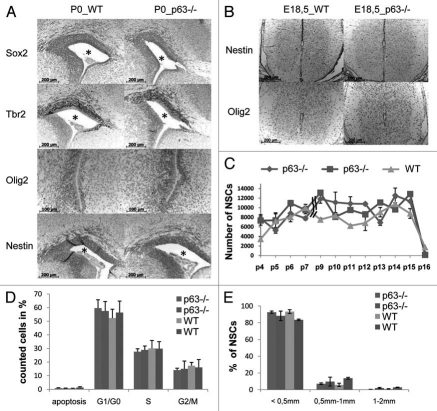
CNS development depends on the availability and commitment of neural stem cells and in mice is complete at the time of birth. We therefore tested whether the absence of p63 affects the abundance of neural stem and progenitor cell populations located in the neurogenic niche around the lateral ventricles. Sections from brain and spinal cord of P0 mice were immunostained with specific early lineage markers to detect particular cell subsets. They included Sox2 transcription factor20) expressed in neural stem cells of the VZ; Tbr2 transcription factor21,22) expressed in the intermediate progenitor cells of the SVZ; Nestin, an intermediate filament of radial glia cells23 and Olig2, a transcription factor of oligodendrocyte progenitors.24 However, no difference in expression pattern or cell distribution could be observed between WT and p63−/− littermates (Fig. 4A). Spinal cord also failed to show any difference (Fig. 4B).
Figure 4.
Normal development of neural and glial progenitors in p63−/− brain and spinal cord. (A and B) Corresponding brain (A) and spinal cord (B) sections from p63−/− mice and WT littermates at the indicated stages were analyzed by immunohistochemistry for markers of neural stem and progenitor cells (Sox2,21 stem cells, located in the VZ; Tbr2,22,23 intermediate progenitor cells, located in the SVZ; Olig2, oligodendrocyte progenitors;25 Nestin, radial glial cells,24 located throughout VZ/SVZ as well as cortex of brain, projections throughout white and grey matter of spinal cord). For Olig2, a close-up is shown to visualize the accumulation of positively stained cells around the inferior horn of the lateral ventricle. The spinal cord shows an even dispersion of Olig2 positive cells within the white and grey matter. Using the same quantitation method as in Figures 2 and 3, no differences could be detected between p63-proficient and -deficient mice in cell number and staining intensities. *Lateral ventricle. (C) In vitro neurosphere assay to quantitate long-term self-renewal of neural stem cells. Primary neural stem cells (NSCs) were derived from the forebrains of E15 littermate embryos with different p63 status. Single cells were seeded in triplicate at clonal density and grown in suspension culture with defined medium. After five days, newly formed neurospheres were counted, dissociated into single cells and reseeded. No significant difference between genotypes is present (Student's t-test). After 15 serial passages, WT and p63−/− cells both stopped proliferating. (D) Flow cytometry analysis of the cell cycle distribution of neural stem and progenitors. Accutase-digested neurospheres were prepared for Propidium iodine staining and analyzed. Values are means ± SEM of five separate experiments. (E) Neurosphere Collagen Assay from E15 forebrains. Freshly isolated cells from two mice per genotype were seeded at clonal density (2,500 cells per sample) in triplicate as in (C). After 16 days, newly formed neurospheres were counted and classified according to size. No significant difference in size distribution was found between genotypes (Student's t-test).
To further assess stemness of neural cells, we performed in vitro neurosphere assays. Three-dimensional neurospheres consist primarily of Nestin-positive neural stem cells (NSC) and progenitors derived from a single cell.25 Although the neurosphere assay might not completely reflect the in vivo developmental progression, it is a highly informative tool that allows the quantitative study of long-term self-renewal of NSCs. Single cells from E15 brains of p63−/− and WT littermates were grown in defined serum-free suspension cultures to form spheric cell clusters. These clusters were serially passaged every five days by dissociation with accutase, mechanical trituration and resuspension into single cells, followed by clonal neurosphere regeneration. At the end of each passage spheres were counted. However, no significant difference in self-renewal capacity between genotypes was found (Fig. 4C). Moreover, regardless of p63 status, both populations lost their capability to generate neurospheres at passage 16, indicating a depletion of stem cells after the same time span (Fig. 4C). FACS analysis of dissociated neurospheres confirmed an undisturbed cell cycle profile for p63−/− NSCs compared to WT NSCs, further underpinning the absence of a proliferative defect (Fig. 4D). In addition, the size distribution of neurospheres did not differ between genotypes (Fig. 4E). Moreover, p63p73 double knockout mice show an identical brain phenotype to p73 knockout mice. This data further confirms that p63 plays no role in CNS development and does not act as a modifier of p73 function in neurogenesis (Sup. Fig. 2). Thus, we conclude that in contrast to the crucial stem cell role of p63 in stratified epithelia,26 p63 is not required for maintaining the pool of stem and progenitor cells in the central nervous system, supporting our in vivo data.
p63 does not influence the in vivo differentiation and maturation of neural cell lineages during CNS development.
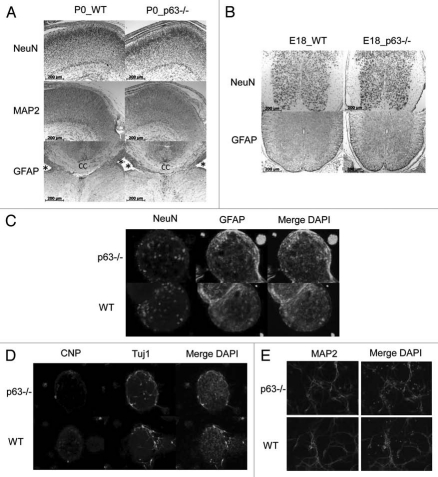
Finally, we determined whether p63 plays a role in the differentiation and maturation of neural stem cells into the three different lineages of neurons, astrocytes and oligodendrocytes. To this end, we used specific differentiation markers. P0 brains were immunostained for NeuN (neuronal nuclei27) and MAP2 (microtubule-associated protein28), both staining differentiated neurons. Astrocytes, which are most prominent in the region surrounding the corpus callosum, were identified by GFAP glial fibrillary acidic protein.29 In each case, the p63 status did not affect the distribution and intensity of these markers in brain (Fig. 5A) and spinal cord (Fig. 5B). Moreover, whole-mount immunofluorescence staining of neurospheres from WT and littermate p63−/− brains confirmed these in vivo findings and revealed that NSCs are able to differentiate into all three lineages in vitro. These included neurons identified by NeuN and Tuj1 (neuronal class III beta-tubulin30) expression, astrocytes by GFAP expression and oligodendrocytes by CNP 2′,3′ cyclic nucleotide 3′ phosphodiesterase expression.31 Again, neither intensity nor distribution of these markers were affected by the p63 status (Fig. 5C and D). Moreover, in differentiation assays of neuronal cell cultures prepared from E18 cortex, which show single neurons and their arborization more clearly, no differences were seen for the neuronal marker MAP2 (Fig. 5E).
Figure 5.
Normal differentiation of neural cell lineages during CNS development in p63−/− mice. (A and B) Brain (A) and spinal cord (B) sections from p63−/− and WT littermate embryos were immunostained for the differentiation markers NeuN (cortical neurons27), MAP2 (neuronal microtubule-associated protein28) and GFAP (astrocytes;29 note the primary location in the area surrounding the corpus callosum). p63−/− mice display normal staining patterns for neurons and astrocytes in brain and spinal cord. *Lateral ventricle; cc, corpus callosum. (C and D) In vitro differentiation of neurospheres derived from E15 forebrains. NSC cells were seeded into differentiation medium. After 5 days neurospheres were stained for markers NeuN (neurons27), GFAP (astrocytes29), CNP (oligodendrocytes31) and Tuj1 (neurons30). Nuclei were counterstained with DAPI. No differences in differentiation patterns were present among genotypes. (E) Primary neuronal cell cultures isolated from the cortex of E18 littermates were immunostained for the neuronal marker MAP2.28 No difference in MAP2 positivity was present among genotypes.
Taken together, we have to conclude that p63 does not influence the number or differentiation of neural progenitors and of newly born post-mitotic cells in the developing CNS in vivo and in vitro.
Discussion
Here we report that the absence of p63 does not detectably influence neurogenesis in the developing mouse brain and spinal cord. Our conclusion is based on extensive side-by-side analyses of global p63KO mice and their WT littermates, including neuroanatomy and histology from E15 to birth, cell proliferation and survival, stemness of neural progenitors and their ability to undergo proper differentiation in vivo and in vitro. Our data indicate that p63 plays no discernable role in embryonic CNS neurogenesis and the maintenance of newborn postmitotic cells in the perinatal period. The absence of a CNS phenotype in p63KO mice establishes that p63 is not required for brain and spinal cord development. Alternatively, it is possible that the absence of p63 can be completely compensated by other molecular pathways. Of note, an independent concomitant immunohistochemical study on p73KO and p63 KO mice also concludes that p63 is dispensible for CNS development. Global p63−/− mice, which express normal levels of p73, were found to lack any brain malformations and exhibit normal numbers of Cajal-Retzius cells.7
Our findings are inconsistent with the previous notion that p63 is an essential pro-survival regulator of neural development of the CNS. This view was based on an experimental system that relied on in utero electroporation of p63shRNA into wild-type embryonic brains and transfection into cultured neuronal precursors to knock down p63 levels.19 We propose that the transfected cells used in that assay system may have died due to a non-specific stress response that resulted from the transfection procedure and/or co-expression of fluorescent marker proteins. However, while p63 is clearly not required for survival of neural cells in a normally developing brain, it is quite possible that p63 is capable of attenuating such a stress response, which could explain the dramatic in vivo effects seen upon its knockdown.
Our results are also in some contrast with the previous proposal of an essential pro-apoptotic p63 function in developing sympathetic neurons of the autonomic nervous system.14 While sympathetic neurons clearly depend on p73,12 it is possible that they, in contrast to CNS neurons, also differentially depend on p63 to undergo normal developmental apoptosis. This study, however, awaits confirmation in a complete p63 knockout mouse, since the reported experiments used the p63 Brdm2 mouse, which harbors a partial exon 10′–14 deletion with hypomorphic rather than complete knock-out properties.32–34 In any case, given our results, a putative pro-apoptotic function of p63 in sympathetic neurons can clearly not be generalized to neurons and glia of the developing CNS.
Consistent with our results, p63 expression in the developing CNS is scant. By PCR amplification, TAp63 and ΔNp63 isoforms can be detected on the mRNA level in murine embryonic cortex.14 However, these mRNA levels are very low, since in-situ hybridization with a specific p63α probe in E14.5 embryos did not detect any signal in brain and spinal cord, while it readily detected p63 signals in the corresponding stratified epithelia of the same embryos (genepaint.org; Set ID EB555). In sharp contrast and consistent with the phenotype of knockout mice, p73 mRNA is highly expressed in developing CNS and readily detectable by in-situ hybridization (genepaint.org; Set ID EB1922; reviewed in ref. 3). p63 protein expression in the developing mouse CNS is also very scant and limited to a subpopulation of p73-expressing Cajal-Retzius (CR) neurons, albeit expressed at much lower levels than p73. CR cells occur in the marginal zone of the developing cortex (cortical hem), a transient signaling center at the interface of the cortex and the choroid plexus. This zone is the origin of most CR cells that typically express Reelin. Immunohistochemistry of E12.5 brains shows that p73 is highly expressed in all CR cells in the prospective neocortex and that a large proportion of p73-positive CR cells are also positive for p63, however, with lower signal intensity than p73.7 Thus, the proportion of p73 to p63 proteins in CR neurons is reflected in their relative mRNA levels. In addition, p73 protein is expressed in a subset of cycling progenitors of the neurogenic subventricular zone (SVZ) at P5. In contrast, p63 and p53 expression was only seen in a few rare cells whose cycling status remained unknown.7 In human cortex at gestational week eight, p63 protein was also only detected in Reelin-expressing CR neurons covering the developing cortex as faint nuclear staining. Here, p63 was limited to the medial cortex adjacent to the cortical hem, with staining becoming undetectable toward the lateral areas. Again, as in mice, the human p63 signal in CR cells was fainter than the p73 signal, and much less intense than the strong p63 signal in the skin. Both for human and mouse the important question, which p63-isoform, TA or ΔN, is expressed in CR cells could not be answered. At any rate, however, in contrast to p73 p63 expression seems to be a rather accessory feature of CR cells without functional relevance.7
Despite some low level expression of p63 in CR cells, the lack of a detectable p63 function in the developing CNS is in stark contrast to the essential role of p73 in neurogenesis that we and others recently reported in references 6, 8 and 9. Analyzing global p73KO mice with the same criteria used in the p63 study here, we found that p73 is critically required for neural stem cell maintenance in vivo as well as in vitro in neurosphere assays.6 Thus, although both genes are derived from the same ancestor, their products closely related13 and their CNS expression very similar, these studies reveal a fundamental functional difference between p63 and p73 in CNS development. While it cannot be excluded that p73 might be capable to substitute for p63 in this organ, p63 is incapable of replacing p73. This is all the more surprising since p63 and p73 proteins are capable of forming heterotetramers,35 strongly suggesting that they could regulate each other even in a situation where one of them has no function on its own. The fact that p63 has no obvious function in normal brain development but nonetheless is expressed,7 taken together with a pro-survival effect of p63 apparent in electroporated embryonic brains,19 leaves open the possibility that p63 might have some protective role during neurogenesis when it takes place under the specific condition of stress.
p63−/− mice are not viable after birth and therefore a purely developmental model. Thus, it remains unclear whether p63 plays an important role as prosurvival maintenance factor of mature postmitotic neurons in the adult brain or stem cell factor in adult neurogenesis. Moreover, it remains unclear whether p63 plays a role in pathologic conditions of neurodegeneration, cerebral ischemia or aging. These questions will need to be addressed by long-lived conditional p63 knockout models. At least TAp63 isoform-specific KO mice were not reported to display gross cerebral abnormalities.36,37 On the other hand, in contrast to perinatal mouse brain, adult human cortex expresses high levels not only of p73 but also p63 protein in neurons of all neocortical layers, the hippocampal dentate hilus and Ammons' horn,7,38 suggesting a maintenance role for both p63 and p73 in adult brain.
In sum, our study further supports the concept that p63 and p73 functions are each highly specific for different subsets of organogenesis within the body. While p63 drives morphogenesis in striated epithelia, p73 is a key driver of central nervous system organogenesis. Despite its specific expression in CR cells of the developing cortex, p63 is completely dispensable in normal brain and spinal cord development.
Materials and Methods
Mouse strains.
This study was approved by the Göttingen University Animal Care Committee, and use was in accordance with institutional guidelines. p63−/− mice with a targeted deletion of the common Exons 6–8 generated by F. McKeon3 and enriched for the SV129 genetic background for more than seven generations, were maintained through interbreeding the heterozygotes. For staged embryos, the day of the vaginal plug was designated E0. Global p63+/− and global p73+/− mice5 were also interbred to generate p63p73 double knockout mice.
Embryo preparation and histology.
Pregnant females were killed by cervical dislocation. With a Y-cut, the uterus was excised to obtain the embryos. Embryos and newborn P0 mice were decapitated. After removing the skin of the WT embryos to improve fixation and mimic the skinless p63−/− mice, head and body were fixed separately in 4% paraformaldehyde for 24 hrs at room temperature. After three washes in PBS for 1 hr each, the tissue was dehydrated in an ethanol-isopropanol-xylol series and paraffin embedded. Coronal sectioning started at the level of the eyes and continued up to the hippocampus; transverse sectioning of the spinal cord started at the cervical portion. Serial sections (1 µm) were stained with Mayer's Hematoxylin and Eosin. For brain sections, subsequent immunohistochemistry was mainly done at the level of the lateral ventricles and basal ganglia anterior to the hippocampus. Spinal cord sections were obtained at the level of heart and lung.
Immunohistochemistry.
Immunohistochemistry was performed on 1 µm paraffin sections. After antigen retrieval by microwave boiling for 15 min in 10 mM citrate buffer pH 6.0, sections were incubated for 16 hrs at 4°C with the following antibodies: cleaved Caspase 3 (1:200, Cell Signaling, 5A1E, #9664); glial fibrillary acidic protein (1:1,000, GFAP, Abcam, ab7260); Ki67 (1:25, Dako, TEC-3, M7249); microtubule-associated protein (1:400, MAP2ab, Thermo Scientific, AP20, MS-249-S); Nestin (1:50, Millipore, rat-401, #MAB353); NeuN, neuronal nuclei (1:300, Millipore, A60, MAB377); Olig2, oligodendrocyte transcription factor 2 (1:500, R&D systems, AF2418); Sox2 (1:1,000, Chemicon international, AB5603) and Tbr2 (1:1,000, Chemicon, AB9618). After applying a biotinylated secondary antibody (GE Healthcare, 1:200) and Extravidin-Peroxidase (Sigma-Aldrich, E2886, 1:1,000), sections were stained with 3,3′-diaminobenzidine (DAB) and counterstained with hematoxylin. TUNEL-staining on 1 µm paraffin sections was done in a humidified chamber. After permeabilizing the tissue with 50 µg/ml proteinase K (Sigma, P6556), an in-situ end labeling mix (Roche, terminal desoxynucleotide transferase) was applied. For detection of the hybridization product, an anti-digoxigenin antibody coupled to alkaline phosphatase (Roche, cat no 1093274) was used. The sections were stained with 5-Brom-4-Chlor-3-Indolyl-Phosphat/Nitro Blue Tetrazolium chloride (BCIP/NBT; Roth) and counterstained with nuclear fast red (Merck). Images were acquired on a Zeiss Axio Scope MAT with AxioVision Rel. 4.8 software.
Image analysis and quantification.
Image analysis and quantification of immunohistochemistry was achieved by custommade scripts using the ImageJ software39 (National Institutes of Health, rsbweb.nih.gov/ij), and data analysis was performed using the ‘R’ statistical computing language (www.r-project.org). For quantification of cleaved Caspase 3 and Ki67 positive cells, 3–6 coronal brain sections per animal were analyzed by calculating the percentage of the positively stained area above a predefined threshold in relation to the entire area imaged.
Neurosphere assays.
Forebrains were harvested from E15 WT and p63 mice, and dissociated into single cells by trituration and digestion with accutase. Cells were plated in standardized condition at 105 cells/60 mm dishes/5 ml NeuroCult® NSC Basal Medium (Mouse) with proliferation supplements and EGF (all from StemCells Techn. Inc.,), following the manufacturer's instructions. Neurospheres were allowed to form for 5 days (this was considered passage 0), and passaged every 5 days. Passage 3 neurospheres were dissociated with accutase and replated in triplicates at 105 cells/60 mm dishes/5 ml medium to monitor regeneration of new neurospheres from single cells. At the end of each passage, the number of neurospheres and total number of cells were assessed. Subsequently, 105 dissociated cells were replated in triplicates and so on. Results shown are representative of independent litters (n = 3 WT and n = 4 KO).
For Collagen Assays 2,500 cells per sample were seeded as single cells in triplicate into 35 mm culture dishes following the manufacturer's instructions (StemCells Techn. Inc.). Every 7 days the cultures were replenished with the Complete NeuroCult Replenishment Medium. After 16 days, newly formed NSCs in solid collagen were counted and classified according to diameter size.
Differentiation assays.
Cells from freshly dissociated neurospheres from E15 forebrains were seeded onto poly-L-Ornithine/Laminin coated plastic chamber slides in NeuroCult® NSC Basal Medium with differentiation supplements (StemCells Techn. Inc.,) following the manufacturer's instructions. After 5–7 days differentiated cells were fixed in 4% PFA and processed for immunofluorescence.
Primary cell culture.
Murine cortex was dissected from E18 WT and p63−/− brains, meninges removed and the neural tissue dissociated into single cells by trypsinization and trituration. 125,000 cells per well were seeded in Neurobasal Medium (supplemented with Glutamine, apo-Transferrin, Glutathion, Super-Oxide-Dismutase, B27 and antibiotics) on poly-L-Ornithine/Laminin coated 8-well chamber slides. Every 3–4 days 50% of the medium was exchanged. After 10 days cells were fixed in 4% PFA and processed for immunofluorescence.
Immunofluorescence of NSCs/single cells.
Immunofluorescence was performed using standard protocols. Slides with NSCs or single cells were incubated in a humidified chamber with primary antibodies at 4°C over night. Next day mouse/rabbit secondary antibodies coupled to Alexa-488 (green channel) or Alexa-594 (red channel) were applied to the cells for 30 minutes at 37°C. Nuclei were counterstained using 4′,6-diamidin-2-phenylindol (DAPI).The following primary antibodies were used: GFAP, glial fibrillary acidic protein (1:1,000, Abcam, ab7260)NeuN, neuronal nuclei (1:200, Millipore, A60, MAB377) Tuj1, neuronal class III beta-tubulin (1:1,000, Covance, PRB-435P) CNP, 2′,3′-cyclic nucleotide 3′-phosphohydrolase (1:500, Millipore, MAB326) MAP2, microtubule-associated protein (1:400, MAP2a,b, Thermo Scientific, AP20, MS-249-S).
Cell cycle analysis.
Neurospheres were collected and digested with accutase. After cell washing with ice-cold PBS, cells were fixed in 70% ethanol at −20°C at least over night. After cell washing with PBS, cells were stained with propidiumiodide (PI)-solution [50 µg/ml (Sigma), 20 µg/ml RNase in PBS] for 30 min at 37µC. FACS samples were counted with GUAVA EasyCyte Plus cytometer (GUAVA Technologies) and analyzed with ModFit LT (Verity Software House).
Genotyping of p63.
WT and p63−/− embryos could easily be distinguished by their morphological phenotype. Additional genotyping PCR was performed to confirm the p63−/− phenotype and to differentiate between WT and p63+/− mice. The following primers were used:
p63-fwd: 5′-GGT GCT TTG AGG CCC GGA TC-3′ binds in Exon 6 of Trp63 gene
p63-Neo: 5′-GAA AGC GAA GGA GCA AAG CTG-3′ binds in Neo cassette of p63−/− mice
p63-rev: 5′-TTC TCA GAT GGT ACC GCT CC-3′ binds in Intron 6/7 of Trp63 gene
Primers p63-fwd and p63-rev give rise to a fragment of 550 bps, specific for WT.
Primers p63-Neo and p63-rev give rise to a fragment of 450 bps, specific for p63−/−.
Acknowledgements
This work was supported by the German Cancer Aid, the US National Cancer Institute (grant R01CA93853), the EU 6th Framework Program, the German Research Foundation DFG, and the Wilhelm Sander Stiftung. L.H. is supported by the Göttingen Graduate School of Neurosciences and Molecular Biosciences, funded by the German Excellence Initiative. We thank F. McKeon for the p63−/− mice, Doris Bode, C. Hippel and A. Dickmanns for excellent technical assistance, and Wolfgang Brück and Tanja Vogel for helpful discussions.
Supplementary Material
References
- 1.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 2.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 3.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 4.Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, et al. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99:143–153. doi: 10.1016/s0092-8674(00)81646-3. [DOI] [PubMed] [Google Scholar]
- 5.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 6.Talos F, Abraham A, Vaseva AV, Holembowski L, Tsirka SE, Scheel A, et al. p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ. 2010;17:1816–1829. doi: 10.1038/cdd.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez-Acosta NC, Cabrera-Socorro A, Morlans MP, Delgado FJ, Suarez-Sola ML, Sottocornola R, et al. Dynamic expression of the p53-family members p63 and p73 in the mouse and human telencephalon during development and in adulthood. Brain Res. 2010 doi: 10.1016/j.brainres.2010.11.041. [DOI] [PubMed] [Google Scholar]
- 8.Agostini M, Tucci P, Chen H, Knight RA, Bano D, Nicotera P, et al. p73 regulates maintenance of neural stem cell. Biochem Biophys Res Commun. 403:13–17. doi: 10.1016/j.bbrc.2010.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujitani M, Cancino GI, Dugani CB, Weaver IC, Gauthier-Fisher A, Paquin A, et al. TAp73 acts via the bHLH Hey2 to promote long-term maintenance of neural precursors. Curr Biol. 2010;20:2058–2065. doi: 10.1016/j.cub.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 10.Tissir F, Ravni A, Achouri Y, Riethmacher D, Meyer G, Goffinet AM. DeltaNp73 regulates neuronal survival in vivo. Proc Natl Acad Sci USA. 2009;106:16871–16876. doi: 10.1073/pnas.0903191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 24:549–560. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289:304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- 13.Yang A, Kaghad M, Caput D, McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002;18:90–95. doi: 10.1016/s0168-9525(02)02595-7. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs WB, Govoni G, Ho D, Atwal JK, Barnabe-Heider F, Keyes WM, et al. p63 is an essential proapoptotic protein during neural development. Neuron. 2005;48:743–756. doi: 10.1016/j.neuron.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs WB, Kaplan DR, Miller FD. The p53 family in nervous system development and disease. J Neurochem. 2006;97:1571–1584. doi: 10.1111/j.1471-4159.2006.03980.x. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs WB, Walsh GS, Miller FD. Neuronal survival and p73/p63/p53: a family affair. Neuroscientist. 2004;10:443–455. doi: 10.1177/1073858404263456. [DOI] [PubMed] [Google Scholar]
- 17.Miller FD, Kaplan DR. To die or not to die: neurons and p63. Cell Cycle. 2007;6:312–317. doi: 10.4161/cc.6.3.3795. [DOI] [PubMed] [Google Scholar]
- 18.Nicotera P, Melino G. Neurodevelopment on route p63. Neuron. 2005;48:707–709. doi: 10.1016/j.neuron.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 19.Dugani CB, Paquin A, Fujitani M, Kaplan DR, Miller FD. p63 antagonizes p53 to promote the survival of embryonic neural precursor cells. J Neurosci. 2009;29:6710–6721. doi: 10.1523/JNEUROSCI.5878-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pevny LH, Nicolis SK. Sox2 roles in neural stem cells. Int J Biochem Cell Biol. 2010;42:421–424. doi: 10.1016/j.biocel.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, et al. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, et al. Pax6, Tbr2 and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells and post-mitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lendahl U, Zimmerman LB, McKay RD. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 25.Marshall GP, 2nd, Reynolds BA, Laywell ED. Using the neurosphere assay to quantify neural stem cells in vivo. Curr Pharm Biotechnol. 2007;8:141–145. doi: 10.2174/138920107780906559. [DOI] [PubMed] [Google Scholar]
- 26.Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullen RJ, Buck CR, Smith AM. Neu N, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 28.Caceres A, Banker GA, Binder L. Immunocytochemical localization of tubulin and microtubule-associated protein 2 during the development of hippocampal neurons in culture. J Neurosci. 1986;6:714–722. doi: 10.1523/JNEUROSCI.06-03-00714.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodhams PL, Basco E, Hajos F, Csillag A, Balazs R. Radial glia in the developing mouse cerebral cortex and hippocampus. Anat Embryol (Berl) 1981;163:331–343. doi: 10.1007/BF00315709. [DOI] [PubMed] [Google Scholar]
- 30.Ferreira A, Caceres A. Expression of the class III beta-tubulin isotype in developing neurons in culture. J Neurosci Res. 1992;32:516–529. doi: 10.1002/jnr.490320407. [DOI] [PubMed] [Google Scholar]
- 31.Lee J, Gravel M, Zhang R, Thibault P, Braun PE. Process outgrowth in oligodendrocytes is mediated by CNP, a novel microtubule assembly myelin protein. J Cell Biol. 2005;170:661–673. doi: 10.1083/jcb.200411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 33.Talos F, Wolff S, Beyer U, Dobbelstein M, Moll UM. Brdm2—an aberrant hypomorphic p63 allele. Cell Death Differ. 2010;17:184–186. doi: 10.1038/cdd.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolff S, Talos F, Palacios G, Beyer U, Dobbelstein M, Moll UM. The alpha/beta carboxy-terminal domains of p63 are required for skin and limb development. New insights from the Brdm2 mouse which is not a complete p63 knockout but expresses p63 gamma-like proteins. Cell Death Differ. 2009;16:1108–1117. doi: 10.1038/cdd.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coutandin D, Lohr F, Niesen FH, Ikeya T, Weber TA, Schafer B, et al. Conformational stability and activity of p73 require a second helix in the tetramerization domain. Cell Death Differ. 2009;16:1582–1589. doi: 10.1038/cdd.2009.139. [DOI] [PubMed] [Google Scholar]
- 36.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cabrera-Socorro A, Pueyo Morlans M, Suarez Sola ML, Gonzalez Delgado FJ, Castaneyra-Perdomo A, Marin MC, et al. Multiple isoforms of the tumor protein p73 are expressed in the adult human telencephalon and choroid plexus and present in the cerebrospinal fluid. Eur J Neurosci. 2006;23:2109–2118. doi: 10.1111/j.1460-9568.2006.04750.x. [DOI] [PubMed] [Google Scholar]
- 39.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with Image J. Biophotonics International. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.