Abstract
Objective
The protein deacetylase SirT1 positively regulates cartilage-specific gene expression, while the proinflammatory cytokine tumor necrosis factor α (TNFα) negatively regulates these same genes. This study was undertaken to test the hypothesis that SirT1 is adversely affected by TNFα, resulting in altered gene expression.
Methods
Cartilage-specific gene expression, SirT1 activity, and results of chromatin immunoprecipitation analysis at the α2(I) collagen enhancer site were determined in RNA, protein extracts, and nuclei of human osteoarthritic chondrocytes left untreated or treated with TNFα. Protein extracts from human chondrocytes transfected with epitope-tagged SirT1 that had been left untreated or had been treated with TNFα were analyzed by immunoblotting with SirT1 and epitope-specific antibodies. The 75-kd SirT1-reactive protein present in TNFα-treated extracts was identified by mass spectroscopy, and its amino-terminal cleavage site was identified via Edman sequencing. SirT1 activity was assayed following an in vitro cathepsin B cleavage reaction. Cathepsin B small interfering RNA (siRNA) was transfected into chondrocytes left untreated or treated with TNFα.
Results
TNFα-treated chondrocytes had impaired SirT1 enzymatic activity and displayed 2 forms of the enzyme: a full-length 110-kd protein and a smaller 75-kd fragment. The 75-kd SirT1 fragment was found to lack the carboxy-terminus. Cathepsin B was identified as the TNFα-responsive protease that cleaves SirT1 at residue 533. Reducing cathepsin B levels via siRNA following TNFα exposure blocked the generation of the 75-kd SirT1 fragment.
Conclusion
These data indicate that TNFα, a cytokine that mediates joint inflammation in arthritis, induces cathepsin B–mediated cleavage of SirT1, resulting in reduced SirT1 activity. This reduced SirT1 activity correlates with the reduced cartilage-specific gene expression evident in these TNFα-treated cells.
Osteoarthritis (OA) is the most common degenerative disease affecting articular cartilage and is characterized by disrupted cartilage extracellular matrix (ECM) homeostasis, ultimately resulting in loss of cartilage without effective replacement. OA is caused in part by exposure of chondrocytes to inflammatory cytokines, such as interleukin-1β (IL-1β) and tumor necrosis factor α (TNFα) (1–4). IL-1β and TNFα have long been known to induce matrix metalloproteinase (MMP) expression in chondrocytes, thereby leading to ECM degradation and cartilage breakdown (3–6). The action of the inflammatory cytokines disrupts the delicate balance between ECM synthesis and degradation in articular cartilage and leads to the destruction of cartilage and the onset of arthritis.
Since OA is usually evident in the fourth to fifth decade of life, it is considered an age-associated disease (1,2). It is therefore likely that gene products regulating lifespan and aging would have an impact on OA. One such protein is SirT1, a lysine deacetylase that is responsible for lifespan extension under conditions of caloric restriction (7–9). SirT1 is an NAD-dependent protein deacetylase that targets both chromatin (histones) and nonchromatin proteins. While SirT1 has been shown to play an important role in a variety of age-related diseases, such as diabetes, cancer, osteoporosis, and neurodegeneration (9–12), little is known of the role it plays in either cartilage biology or OA. Recently, it was demonstrated that SirT1 enhances cartilage-specific ECM gene expression (13). SirT1 appears to accomplish this function, at least for α2(I) collagen, by enhancing SOX9-mediated transcription via the recruitment of a number of transcription activators (i.e., histone acetyltransferases) to the promoter and enhancer sites (13). It has also recently been demonstrated that SirT1 blocks apoptosis in chondrocytes, and that it accomplishes this by multiple mechanisms (14,15). Additionally, evidence indicates that SirT1 levels are reduced in chondrocytes from OA cartilage compared to normal cartilage, suggesting that the altered pattern of gene expression and apoptosis evident in OA is correlated with a reduction in SirT1 levels (13,14).
While OA is generally not considered to be an inflammatory disease, it is nevertheless influenced by inflammatory cytokines, such as IL-1β and TNFα (2,4,16). Interestingly, SirT1 demonstrates a broad anti-inflammatory function in a variety of tissues (8,9). SirT1 likely accomplishes this in part by the deacetylation of the p65 subunit of NF-κB, blocking its ability to bind DNA, thereby inhibiting transcription of proinflammatory genes (17). While it appears that SirT1 can inhibit inflammation, there is no evidence to date suggesting that the opposite is true, that mediators of inflammation can interfere with SirT1 function. In the present study, we explored the idea that an inflammatory cytokine modulates the activity of SirT1. We found that in cells treated with a nonapoptotic dose of TNFα, SirT1 undergoes a cathepsin B–mediated cleavage event, generating a stable 75-kd fragment with reduced enzymatic activity. This novel modification to SirT1 is discussed in terms of the pathologic conditions of arthritis.
MATERIALS AND METHODS
Cell culture and transfections
Human chondrocytes were isolated from the knees of patients with OA who were undergoing total knee arthroplasty. (Tissues were supplied by NDRI.) Isolation was carried out according to the method of Derfoul et al (18). All experiments were initiated on low-passage chondrocytes (P0 or P1), which were grown to confluence. After reaching confluence, cells were incubated with Opti-MEM media (Invitrogen) supplemented with 50 ng/ml of TNFα and 1 ng/ml of IL-1β (R&D Systems) for 24 hours. Untreated cells were used as controls. Where indicated, the following protease inhibitors (Calbiochem) were used in conjunction with TNFα: ALLN (10 μg/ml) (19), calpain inhibitor III (2 μg/ml), calpeptin (1.8 μg/ml), EST (10 μg/ml), and PD150606 (12.5 μg/ml). Human chondrocytes were transfected with FuGene 6 (Roche) in the presence of Opti-MEM.
Plasmids for transient transfections
The pcDNA-FLAG-human SirT1 (pcFL-hSirT1) was a kind gift from Professor Danny Reinberg (New York University, New York, NY).
Intracellular NAD levels and SirT1 activity assays
An NAD/NADH quantification kit (catalog no. k337-100; BioVision) was used according to the recommendations of the manufacturer and as previously described (13). Data are presented as NAD per microgram of total protein.
To determine SirT1 activity, SirT1 was initially immunoprecipitated with either a SirT1 antibody (Upstate Biotechnology) or FLAG antibody (Sigma) where indicated and analyzed for enzymatic activity using a Biomol kit (catalog no. AK500), according to the recommendations of the manufacturer. Values were divided by total protein for equivalent samples. Immunoprecipitates were also analyzed by immunoblotting to assure equivalent levels of SirT1 enzyme.
Chromatin immunoprecipitation (ChIP) analyses
ChIP assays were carried out using an EZ-ChIP kit, according to the recommendations of the manufacturer (Upstate Biotechnology). Polymerase chain reactions (PCRs) were carried out as previously specified (13). Nonspecific IgG was used as a negative control. Input ChIP extracts were used as a positive control and were diluted respectively to obtain a standard curve. Primer information is shown in Supplementary Table 1, available on the Arthritis & Rheumatism web site at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1529-0131.
PCR analyses
Real-time quantitative PCR (qPCR) reactions were carried out using 10 ng of complementary DNA (cDNA) and SYBR Green mixture (Applied Biosystems) with a StepOne real-time PCR system as previously described (13,18). Primer information is shown in Supplementary Table 1.
For analysis of SirT1 alternative splicing, RNA was purified from untreated cells and cells treated with TNFα using an RNeasy kit (Qiagen) and converted to cDNA using a Superscript single-strand kit (Invitrogen). Two hundred nanograms of cDNA per reaction was subjected to one-step reverse transcriptase (RT)–PCR procedure (Invitrogen). The primer sequence (F-CGATTGGGTACCGAGATAAC, R-TGATTTGTTTGATGGATAGTTCAT, flanking positions 416 and 2293 of the cDNA) generated a 1,825-bp product for full-length SirT1. (Results are available online at http://dental.huji.ac.il/newEsite/departments/institute/Supplementary%20Data.pdf.)
Protein isolation and fluorescence-activated cell sorting (FACS) analysis
Whole cell protein extracts and immunoblotting procedures were performed as described elsewhere (13). Primary and secondary antibodies used for immunoblotting, immunoprecipitation, and ChIP analyses are shown in Supplementary Table 2, available on the Arthritis & Rheumatism web site at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1529-0131. FACS analysis for apoptotic cell populations was carried out as specified previously (20).
In vitro cleavage assays
Purified recombinant human SirT1 (5 units at 8 units/μg; Biomol) was incubated with activated purified cathepsin B (0.25 units; Calbiochem), cathepsin K (0.25 units; Enzo Life Sciences), or cathepsin L (0.25 units; R&D Systems) in 5 mM dithiothreitol, 1M Tris HCl (pH 7.5) buffer at 37°C for 30 minutes, 1 hour, 2 hours, and 4 hours. The doses and reaction conditions used for each enzyme were those recommended by the manufacturers for optimal activity. Samples were then processed for immunoblotting or for SirT1 enzyme activity assays. Additional in vitro cleavage assays were conducted after immunoprecipitation of FLAG-tagged SirT1 from transfected chondrocytes and incubation of the immuno-precipitates with active cathepsin B, followed by immunoblotting for SirT1 or assessment of SirT1 enzymatic activity.
In vitro incubation with purified recombinant human SirT1 (5 units at 8 units/μg) and activated cathepsin B (0.25 units), was followed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) purification of the 35-kd carboxy-terminal fragment of SirT1. The 35-kd fragment was excised and subjected to Edman sequencing using an Applied Biosystems 494cLC Procise protein sequencer (Alphalyse).
Tandem mass spectrometry (MS/MS) protein identification
Human chondrocytes were transfected with pcFLHis-hSirT1, and SirT1 was immunoprecipitated with a His antibody. The extracts were then subjected to SDS-PAGE and stained with MagicBlue protein staining (MTR Scientific). The respective 75-kd bands were excised and processed for identification at ProtTech using NanoLC-MS/MS peptide sequencing technology. The mass spectrometry data acquired were used to search the most recent nonredundant Protein Data Bank, which was downloaded from GenBank using proprietary software from ProtTech.
Statistical analysis
Statistical analysis was performed using one-way analysis of variance, assuming confidence levels of 95% (P < 0.05) to be statistically significant. The least significant difference test was carried out to determine the differences between 2 equivalent treatments within a group, assuming confidence levels of 95% (P < 0.05). Data are shown as the mean ± SD. Results of qPCR, ChIP, and SirT1 deacetylation assays and NAD content were calculated from 3 repetitions generated from 3 different samples (n = 9).
RESULTS
Reduced SirT1 activity and formation of a 75-kd SirT1 fragment in human chondrocytes treated with TNFα
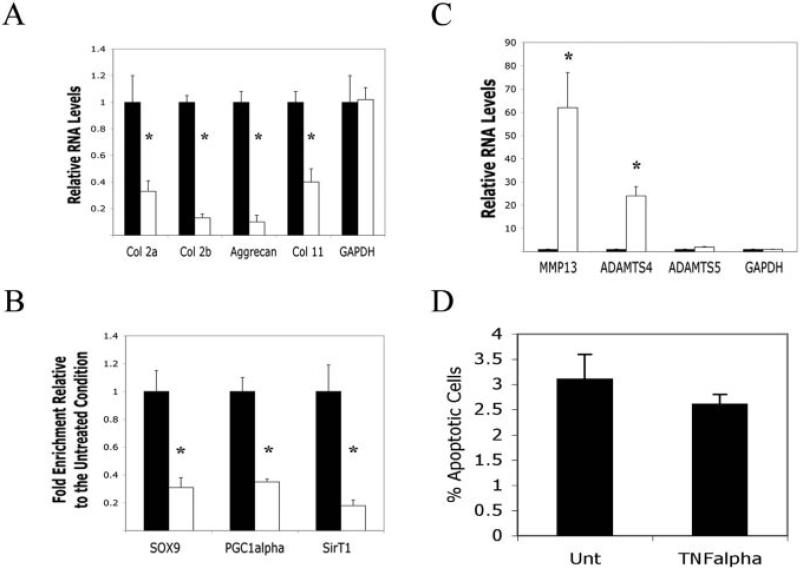
Treatment of human articular chondrocytes with TNFα produced a well-defined change in gene expression (1,4). As shown in Figure 1A, the levels of RNA for a number of cartilage-specific ECM genes were down-regulated by TNFα, which is consistent with the findings of previous studies (1,4). Collagens α2(I), β2(I), and α11(I), and aggrecan showed decreased expression after 24 hours of treatment with TNFα (50 ng/ml). Consistent with a reduction in RNA levels, ChIP analyses of the α2(I) collagen enhancer revealed a reduction in critical activator proteins binding to this sequence in cells treated with TNFα. SOX9, and peroxi-some proliferator–activated receptor γ coactivator 1α (PGC-1α), showed a 3-fold reduction, while SirT1 showed a 5-fold reduction in binding to the enhancer in the presence of TNFα (Figure 1B). Since the association of SOX9, PGC-1α, and SirT1 with the α2(I) collagen enhancer is required for optimal transcription (13,21), their reduction in binding correlated with a reduction in α2(I) collagen messenger RNA (mRNA) levels in cells treated with TNFα.
Figure 1.
Repression and activation of gene expression in human chondrocytes treated with tumor necrosis factor α (TNFα). A, Relative RNA levels in chondrocytes left untreated (solid bars) or treated with TNFα (open bars), as determined by quantitative polymerase chain reaction (qPCR) with the indicated human primers. B, Chromatin immunoprecipitation analysis of chondrocytes left untreated (solid bars) or treated with TNFα (open bars), using antibodies to SOX9, peroxisome proliferator–activated receptor γ coactivator 1α (PGC-1α), and SirT1 or a nonspecific IgG as a negative control. Quantitative PCR was performed with primers flanking the enhancer within the first intron of α2(I) collagen. An IgG control showed no PCR product. C, Relative RNA levels in chondrocytes left untreated (solid bars) or treated with TNFα (open bars), as determined by qPCR with the indicated human primers. D, Percentage of apoptotic cells in chondrocytes left untreated (Unt) or treated with TNFα (50 ng/ml) for 24 hours, as determined by flow cytometry. No significant difference between the 2 conditions was noted. Bars show the mean ± SD. * = P < 0.05 versus untreated controls.
In contrast to the cartilage ECM genes, expression of genes encoding the matrix-degrading enzymes MMP-13 and ADAMTS-4 were significantly elevated by TNFα (Figure 1C), consistent with the findings of previous studies (4,16). Thus, TNFα is a divergent regulator of different gene families in human chondrocytes. Interestingly, while chondrocytes responded to TNFα in terms of gene expression, we found that they were resistant to the apoptotic effects of TNFα. We found no increase in chondrocyte death following TNFα treatment (Figure 1D), as determined by FACS analysis.
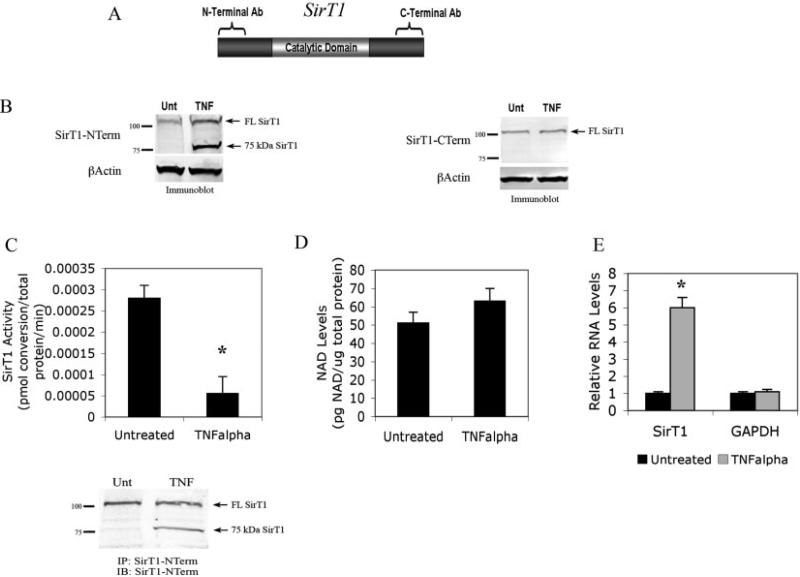
The protein deacetylase SirT1 has recently been demonstrated (13) to enhance expression of cartilage-specific ECM genes, such as α2(I) collagen, aggrecan, and α11(I) collagen, which are shown in Figure 1A. SirT1 and TNFα appear to have opposing effects on cartilage gene expression; this suggests that TNFα may function in part by blocking SirT1 expression or activity. To determine whether this is the case, we assessed levels of endogenous SirT1 by immunoblot analysis in chondrocytes left untreated and chondrocytes treated with TNFα, using 2 commercial anti-SirT1 antibodies (Figure 2A). One antibody reacts with the N-terminal domain of SirT1 (residues 1–130), and the other reacts with the C-terminal end of SirT1 (residues 700–747). Immunoblots using the C-terminal antibody (Figure 2B, right) revealed no change in levels of SirT1. However, immunoblots using the N-terminal reactive antibody (Figure 2B, left) revealed 2 distinctive bands; one was a full-length SirT1 band (110 kd), and the other was a smaller band migrating at 75 kd, which was present only after TNFα treatment. A separate commercially available N-terminal–reactive SirT1 antibody was also used in these immunoblot assays and yielded the same results; namely, a 110-kd SirT1 band and a 75-kd SirT1 band in the TNFα-treated extracts. The 75-kd SirT1 band was also observed following treatment of chondrocytes with IL-1β. (Results are available online at http://dental.huji.ac.il/newEsite/departments/institute/Supplementary%20Data.pdf.)
Figure 2.
Reduction in SirT1 activity in human chondrocytes treated with TNFα. A, Outline of SirT1 and the regions detected by the N-terminal (NTerm)–specific and C-terminal (CTerm)–specific antibodies (Ab). B, Immunoblot of protein extracts from chondrocytes left untreated (Unt) or treated with TNFα with the N-terminal antibody (left) and the C-terminal antibody (right). C, SirT1 activity in chondrocytes left untreated or treated with TNFα after immunoprecipitation with a SirT1 N-terminal antibody. SirT1 was immunoprecipitated from chondrocytes, and the immunoprecipitates were processed for SirT1 activity (top) or for immunoblotting (IB; bottom). Immunoblots in B and C are representative of 6 repetitions. D, Intracellular NAD levels in chondrocyte extracts left untreated or treated with TNFα. E, Relative RNA levels in chondrocytes left untreated or treated with TNFα, determined by qPCR with the indicated human primers. Bars show the mean ± SD. * = P < 0.05 versus untreated controls. FL SirT1 = full-length SirT1 (see Figure 1 for other definitions).
To determine whether this novel 75-kd band representing SirT1 possessed altered enzymatic activity, we monitored endogenous SirT1 activity in extracts derived from chondrocytes treated with TNFα versus those from untreated control chondrocytes. Following immunoprecipitation with a SirT1 N-terminal antibody, TNFα treatment led to a 6-fold decrease in SirT1 activity (Figure 2C, top), indicating that this 75-kd SirT1 band had reduced enzymatic activity (Figure 2C, bottom). Since SirT1 requires NAD for enzymatic activity, we also monitored intracellular NAD levels and found no significant changes between control chondrocytes and TNFα-treated chondrocytes (Figure 2D), indicating that the reduction in activity was not due to a reduction in NAD concentration.
When considering the total amount of SirT1 in the TNFα-treated extracts represented by both the 110-kd and 75-kd SirT1 bands (Figures 2B and C), it is clear that the total levels increased substantially. To account for this apparent increase in SirT1 protein levels, SirT1 RNA levels were analyzed. As shown in Figure 2E, the levels of SirT1 RNA were increased by 6-fold following TNFα treatment. Thus, TNFα stimulates SirT1 transcription while leading to the formation of a smaller size inactive protein product. Elevated expression of full-length SirT1, following TNFα stimulation, appears to be mediated at least in part by the p65-RelA subunit of NF-κB, which associates with the SirT1 promoter and enhances SirT1 transcription (22).
A TNFα-mediated cleavage site in SirT1 maps to the carboxy-terminal domain
Possible mechanisms for producing a smaller form of SirT1 include alternative splicing and site-specific cleavage. We found no obvious evidence of TNFα-mediated alternative splicing of SirT1 by means of SirT1 mRNA RT-PCR analyses. Only the full-length open-reading frame of SirT1 mRNA was evident with the primers used. (Results are available online at http://dental.huji.ac.il/newEsite/departments/institute/Supplementary%20Data.pdf.) Semiquantitative RT-PCR showed that TNFα elevated the level of SirT1 RNA, which is consistent with the data shown in Figure 2E.
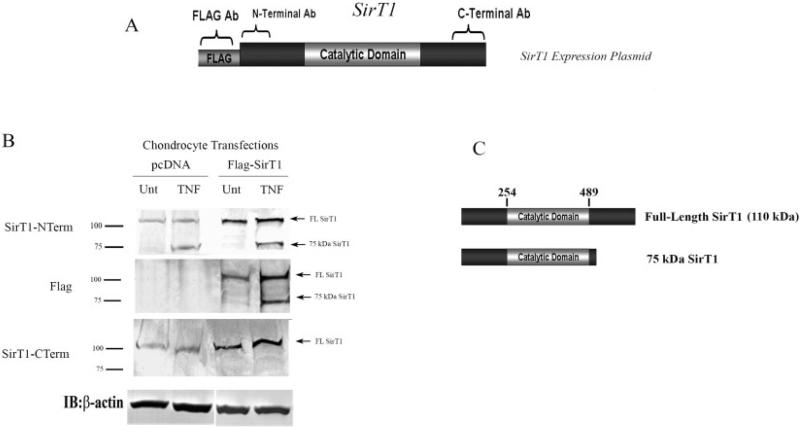
Alternatively, proteolytic cleavage of the C-terminal end of SirT1 was considered as a possible mechanism by which the 75-kd SirT1 fragment was generated. To determine whether this was the case, an N-terminal FLAG-tagged SirT1 expression plasmid (Figure 3A) was used to transfect human chondrocytes. Subsequently, chondrocytes were treated with TNFα, and immunoblotting was performed. As shown in Figure 3B (top), the 75-kd SirT1 band was present in the TNFα-treated chondrocyte extracts and was significantly elevated only in the extracts from the SirT1-transfected cells, when assessed by immunoblotting with the N-terminal–specific SirT1 antibody. Importantly, when the FLAG-specific antibody was used in the immunoblots (Figure 3C, middle), the 75-kd band was evident only in the extracts from TNFα-treated cells. Use of the C-terminal–specific antibody (Figure 3B, bottom) revealed only full-length 110-kd SirT1 protein in the transfected extracts.
Figure 3.
The 75-kd SirT1 fragment is a posttranslationally modified form of SirT1. A, Outline of the FLAG-tagged SirT1 expression plasmid and the regions detected by the N-terminal–specific and C-terminal–specific antibodies. B, Transient transfections of human chondrocytes with the FLAG-tagged SirT1 plasmid or a pcDNA control plasmid. The cells were left untreated (Unt) or were treated with tumor necrosis factor α (TNFα), and protein extracts were analyzed by immunoblotting with the indicated antibodies. Immunoblot is representative of 6 repetitions. C, Cleavage site in SirT1. The area between residues 520 and 540 delineates the cleavage site within the carboxy-terminal domain of SirT1, based on the relative size of the 75-kd fragment. See Figure 2 for other definitions.
To ensure that the 75-kd protein evident in the Western blots was actually SirT1, it was immunoprecipitated and purified, and the band was analyzed by mass spectroscopy. Peptide fragments corresponding to the central domain of SirT1 were confirmed, indicating that the protein was SirT1. (Results are available online at http://dental.huji.ac.il/newEsite/departments/institute/Supplementary%20Data.pdf.) Given the results, it is likely that a TNFα-mediated cleavage site is located within the carboxy-terminus (Figure 3C) between residues 520 and 540.
Generation of the 75-kd fragment of SirT1 by cathepsin-mediated cleavage
We next evaluated whether cathepsins and calpains are involved in TNFα-mediated SirT1 cleavage. Previous evidence indicated that under the influence of TNFα, many lysosomal proteases are capable of translocating to the cytoplasm and nucleus, retaining their activity (23,24).
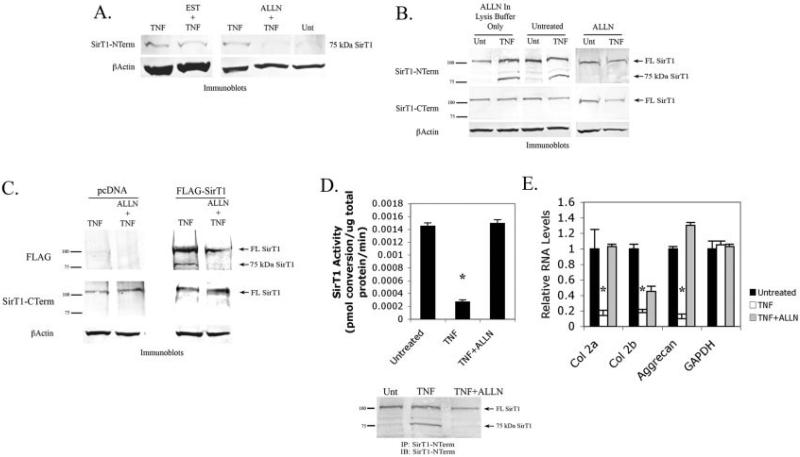
The calpain inhibitor EST was ineffective at blocking the formation of the 75-kd band (Figure 4A). Three additional calpain inhibitors, PD150606, calpeptin, and calpeptin inhibitor III, were also ineffective. (Results are available online at http://dental.huji.ac.il/new Esite/departments/institute/Supplementary%20Data.pdf.) However, ALLN, which blocks both calpains and cathepsins (19), was very effective at blocking the formation of the 75-kd protein (Figure 4A). To determine whether ALLN blocked the action of cathepsin after cell lysis, the inhibitor was added to extracts immediately after lysis. As shown in Figure 4B (top), it was clear that simply adding ALLN to cell extracts did not block the formation of the 75-kd band, while adding ALLN to the culture during the period of TNFα treatment did block its formation. These data indicate that protease activation mediated by TNFα treatment is required for the formation of the 75-kd SirT1 band. Additionally, use of the antibody targeting the carboxy-terminal region of SirT1 showed that levels of full-length SirT1 did not vary significantly in the presence of TNFα and ALLN (Figure 4B, middle), indicating that the protease inhibitor did not affect the full-length protein. That ALLN blocks the ability of TNFα to induce SirT1 mRNA could be explained by its capacity to hinder proteasomal cleavage of IκB (25,26). Cleavage and inactivation of IκB are essential for TNFα-mediated induction of p65-RelA. Thus, in the presence of ALLN, p65-RelA would not be able to induce expression of target genes such as SirT1. Taken together, these data suggest that cathepsins are involved in the TNFα-mediated cleavage of SirT1 in chondrocytes.
Figure 4.
Generation of the 75-kd SirT1 band by a cathepsin-mediated cleavage event. A, Immunoblots from protein extracts of chondrocytes left untreated (Unt) or treated with tumor necrosis factor α (TNFα), EST and TNFα, or ALLN and TNFα. B, Immunoblots from protein extracts of chondrocytes left untreated, treated with TNFα, or treated with TNFα and ALLN. C, Transient transfections of chondrocytes with either pcDNA control or FLAG-SirT1. Chondrocytes were left untreated, treated with TNFα, or treated with TNFα and ALLN. Immunoblots from protein extracts of the transfected and treated chondrocytes are shown. D, SirT1 activity in chondrocytes left untreated, treated with TNFα, or treated with TNFα and ALLN after immunoprecipitation with a SirT1 N-terminal antibody. SirT1 was immunoprecipitated from chondrocyte extracts, and the immunoprecipitates were processed for SirT1 activity (top) or for immunoblotting (bottom). Immunoblots in A–D are representative of 6 repetitions. E, Relative RNA levels in chondrocytes left untreated (solid bars), treated with TNFα (open bars), or treated with TNFα and ALLN (shaded bars), determined by quantitative polymerase chain reaction with the indicated human primers. Bars show the mean ± SD. * = P < 0.05 versus untreated controls. See Figure 2 for other definitions.
To support these observations, we further monitored the ability of ALLN to block the cleavage of ectopically expressed SirT1. Human chondrocytes were transfected with pFLAG-SirT1, and the cells were then treated with TNFα in the presence or absence of ALLN. ALLN blocked the formation of the 75-kd band produced by plasmid expression (Figure 4C). Thus, it appears that a cathepsin-like enzyme activated by TNFα acts on both endogenous and ectopically expressed SirT1.
Figure 4D (top) shows that adding ALLN to TNFα-treated cells restored SirT1 enzymatic activity, as well as diminished the 75-kd SirT1 fragment (bottom). Further qPCR analysis indicated that adding ALLN restored chondrocyte-specific gene expression of α2(I) collagen, β2(I) collagen, and aggrecan, even in the presence of TNFα (Figure 4E). These data indicate that blocking cathepsin protease activity during TNFα treatment reduces the repressive effect of this cytokine on cartilage-specific gene expression, SirT1 activity, and formation of the 75-kd SirT1 fragment.
Cleavage of SirT1 by cathepsin B
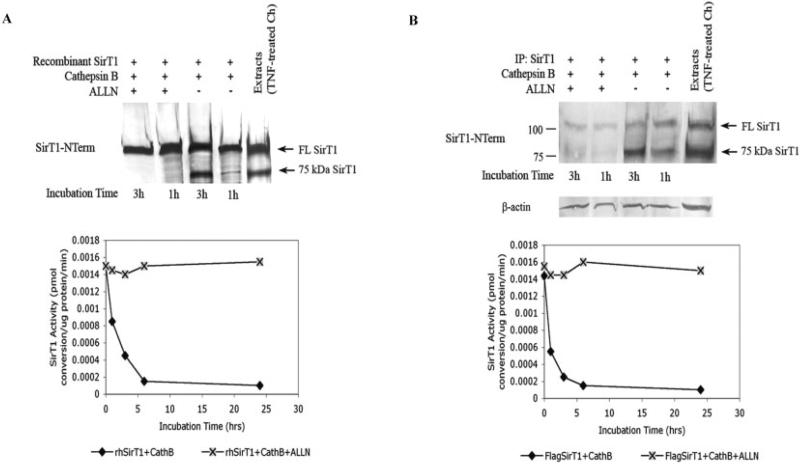
Next, experiments were carried out to determine whether a specific cathepsin was responsible for the cleavage of SirT1 at the carboxy-terminus. Human cathepsin B, cathepsin K, and cathepsin L, along with calpain 1, were tested for their ability to cleave SirT1 in an in vitro reaction. All proteins, including SirT1, were obtained from commercial sources. While cathepsin K, cathepsin L, and calpain 1 could cleave SirT1, they did not produce an appropriate-sized 75-kd fragment (results not shown), indicating that they were unlikely to mediate the TNFα-cleavage of SirT1. Cathepsin B, however, did produce a correctly sized 75-kd fragment. As shown in Figure 5A (top), a 3-hour incubation of pure human SirT1 with cathepsin B yielded a fragment migrating to the correct position, with ~50% cleavage of full-length SirT1, while a 1-hour incubation yielded ~10% cleavage. When SirT1 activity was assessed in these reactions, a 3-hour incubation of SirT1 with cathepsin B reduced the enzyme activity by ~60% (Figure 5A, bottom). Addition of ALLN to the reaction completely blocked cleavage, and the SirT1 activity was not reduced (Figure 5A).
Figure 5.
Cathepsin B (CathB) is responsible for SirT1 cleavage and inactivation. A, Commercially available SirT1, cathepsin B, and ALLN were incubated in a cleavage reaction, as indicated, for 0–25 hours. SirT1 levels, assessed by immunoblot (top), and SirT1 activity (bottom) are shown. rhSirT1 = recombinant human SirT1. B, SirT1 ectopically expressed from transfected chondrocytes (Ch) was immunoprecipitated with a FLAG antibody, and the immunoprecipitates were then incubated in a cleavage reaction with commercially available cathepsin B and ALLN, as indicated, for 0–25 hours. SirT1 levels, assessed by immunoblot (top), and SirT1 activity (bottom) are shown. Immunoblots are representative of 6 repetitions. TNF = tumor necrosis factor (see Figure 2 for other definitions).
Similar experiments were performed on SirT1 ectopically expressed in human chondrocytes. The FLAG-tagged SirT1 was purified via immunoprecipitation from untreated cell extracts and incubated with cathepsin B in the presence or absence of ALLN. As shown in Figure 5B (top), a 3-hour incubation yielded a SirT1 fragment migrating to the correct position of 75-kd, with ~80% cleavage of full-length SirT1 (Figure 5B, top), while a 1-hour incubation yielded ~50% cleavage. When SirT1 activity was assessed in these reactions, a 3-hour incubation of SirT1 with cathepsin B reduced the enzyme activity by ~80% (Figure 5B, bottom). Addition of ALLN to the reaction completely blocked cleavage, and the SirT1 activity was not reduced (Figure 5B).
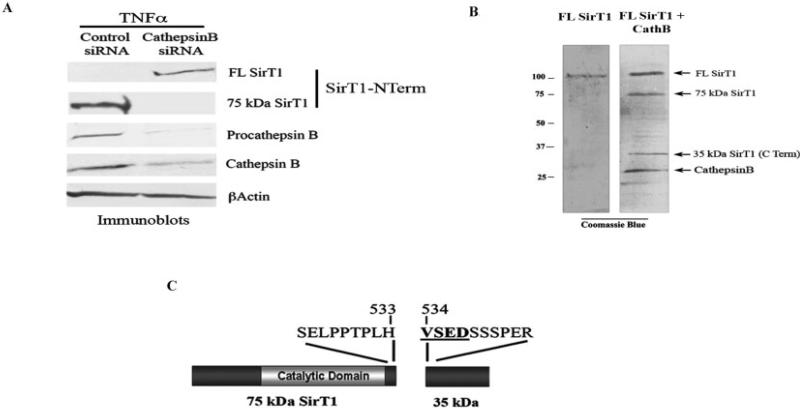
It was hypothesized that a reduction in cathepsin B levels by small interfering RNA (siRNA) approaches would block the formation of the 75-kd SirT1 fragment following TNFα treatment. Chondrocytes were transfected with a cathepsin B siRNA, which resulted in a reduction in both procathepsin B and active cathepsin B (Figure 6A). This reduction in cathepsin B led to a reduction in the 75-kd SirT1 and a corresponding increase in the levels of full-length SirT1 (Figure 6A). These data indicate that cathepsin B is the primary enzyme responsible for TNFα-mediated cleavage of SirT1.
Figure 6.
Cathepsin B (CathB) cleaves SirT1 at amino acid residue 533 and is markedly elevated in osteoarthritis samples. A, Chondrocytes transiently transfected with either control small interfering RNA (siRNA) or cathepsin B siRNA were treated with tumor necrosis factor α (TNFα) for 24 hours, and cell extracts were processed for immunoblotting with the indicated antibodies. B, Commercially available SirT1 and cathepsin B were incubated in a cleavage reaction and then processed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and the gel was stained with Coomassie blue. Cathepsin B migrated to the position of the 27-kd band, as indicated. The 35-kd SirT1 fragment within the gel was excised and processed for N-terminal amino acid sequencing. Immunoblots are representative of 6 repetitions. C, N-terminal amino acid sequencing of the 35-kd SirT1 fragment. See Figure 2 for other definitions.
To identify the cathepsin B–sensitive cleavage site on SirT1, the samples treated as shown in Figure 5A were subjected to SDS-PAGE, and the gel was stained with Coomassie blue (Figure 6B). The 35-kd band corresponding to the complete carboxy-terminal portion of SirT1 resulting from the cathepsin B cleavage was excised (Figure 6B) and subjected to N-terminal sequencing. The resulting sequence shows that the 35-kd band has an amino-terminal sequence beginning at residue 534 (Figure 6C), indicating a cleavage site between residues 533 and 534 of SirT1. This cleavage site corresponds to that identified based on the 75-kd size of the amino-terminal portion of SirT1 (i.e., between residues 520 and 540) (Figure 3C).
DISCUSSION
It is now well accepted that TNFα mediates the joint inflammation evident in many forms of arthritis (1,2). TNFα accomplishes this in part by inducing MMPs and by inhibiting the expression of cartilage-specific genes, such as α2(I) collagen and aggrecan (1) (Figure 1). Thus, to understand how arthritis develops, one must understand the mechanisms by which TNFα and related cytokines exert their effects on gene expression. Herein, we showed that TNFα has a unique function, in that it induces cathepsin B cleavage of SirT1, resulting in a stable but enzymatically inactive SirT1 fragment of 75 kd. Thus, the cleavage and inactivation of SirT1 parallels the down-regulation of cartilage-specific gene expression and the up-regulation of the expression of matrix-degrading enzymes (MMPs) that occur following TNFα treatment of human chondrocytes.
The findings of the present study suggest that mediators of inflammation, such as TNFα, work in opposition to SirT1. Inactivation of SirT1 by TNFα may therefore be necessary for optimal development of the inflammatory response. Importantly, many other proinflammatory cytokines participating in arthritis (i.e., IL-1β, IL-6, etc) have been reported to activate cathepsin B (27,28). Consistent with these findings, in this study, we observed the 75-kd SirT1 fragment in extracts of human chondrocytes exposed to IL-1β, indicating that it may mediate cathepsin B activation, similar to TNFα.
That a mediator of inflammation plays a role in inhibiting SirT1 is noteworthy when the biologic function of this protein deacetylase is considered. SirT1 plays a role in enhancing organism longevity by inhibiting the development of age-related diseases (9–11,29,30). It may be that part of the antiaging function of SirT1 resides in its ability to inhibit inflammation. For example, SirT1 inhibits the inflammation associated with chronic obstructive pulmonary disease (31,32), colitis (33), and hepatic steatosis (34) and can provide a vasoprotective antiinflammatory effect on the vascular endothelium (35) and in adipocytes (36). These antiinflammatory functions of SirT1 may be partly due to its ability to block the mediator of inflammation NF-κB via deacetylation (17,29). When examined in a cartilage context, these functions of SirT1 are consistent with its role in maintaining chondrocyte viability and phenotype as well as cartilage-specific gene expression (13,14). SirT1 could therefore be an important player in reducing the severity of arthritis. Taken together, the data would also suggest that prolonged inflammation may work against any SirT1-mediated enhancement of longevity by blocking SirT1 function.
The mechanism by which TNFα inhibits SirT1 activity appears to rely on an interesting posttranslational modification. While modifications such as phosphorylation and sumoylation have been reported to alter SirT1 enzymatic activity (37,38), in this study, we demonstrated that SirT1 was subject to site-specific cleavage by cathepsin B, resulting in impaired enzymatic activity. While the cleavage did not appear to occur within the active site of SirT1, enzymatic activity was nevertheless reduced, suggesting that sequences outside the active site are essential for optimal activity. Recent evidence indicates that protein SENP1 associates with the carboxy-terminus of SirT1 and thereby enhances its activity via desumoylation (38). It is therefore possible that cleavage of the carboxy-terminus of SirT1 reduces the ability of factors such as SENP1 to bind and activate SirT1. Additionally, evidence in yeast indicates that SirT1 forms homotrimeric complexes via an interaction domain within the N-terminal part of the protein (39,40). Since the 75-kd fragment retains the N-terminal domain, it is possible that it is able to form these trimeric complexes, yet these complexes may be enzymatically inactive.
The mechanism by which cathepsin B is able to cleave SirT1 is interesting because cathepsins are normally confined to the lysosome. However, under certain conditions, these enzymes are able to translocate to the cytoplasm and nucleus, retaining their activity (23,24). Our unpublished observations indicate that TNFα induces expression of cathepsin B and its translocation from the lysosome to the cytoplasm and nucleus. It has recently been demonstrated, for example, that nuclear histone H3 can be cleaved by cathepsin L even at neutral pH, under conditions of embryonic stem cell differentiation (41). In this study, we showed that TNFα stimulation had the same effect on SirT1, which is typically confined to the nuclear compartment. Baici et al found that chondrocytes derived from OA cartilage possessed elevated levels of active cathepsin B as compared to normal chondrocytes (42,43), which is also consistent withourunpublishedobservations,suggestingthatcathepsin B may play a pathologic role OA development.
In conclusion, our data indicate that the inflammatory cytokine TNFα mediates a proteolytic cleavage of SirT1, producing a stable 75-kd fragment that is incapable of binding chromatin and chromatin-associated coactivators, such as PGC-1α and SOX9. As a result of this cleavage, SirT1 additionally loses a significant level of its enzymatic activity, which may directly deter the expression of cartilage-specific genes. Considering that TNFα alone does not induce apoptosis in chondrocytes, the data would suggest that this fragment of SirT1 serves an alternate biologic function, perhaps even to protect cells against death during the inflammatory response. Current efforts are underway to identify this function.
Supplementary Material
ACKNOWLEDGMENTS
We thank NDRI (Philadelphia, PA) for providing the human cartilage tissue samples. We thank Drs. Evelyn Ralston and Kristina Zaal (NIAMS Light Imaging Section) for their help with the confocal microscopy and Professor Danny Reinberg (New York University, New York, NY) for the generous gift of pcDNA-FLAG-hSirT1.
Supported by the NIH (Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases).
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Dvir-Ginzberg had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Dvir-Ginzberg, Gagarina, Lee, Booth, Gabay, Hall.
Acquisition of data. Dvir-Ginzberg, Gagarina, Lee, Booth, Gabay, Hall.
Analysis and interpretation of data. Dvir-Ginzberg, Gagarina, Lee, Booth, Gabay, Hall.
REFERENCES
- 1.Iannone F, Lapadula G. The pathophysiology of osteoarthritis. Aging Clin Exp Res. 2003;15:364–72. doi: 10.1007/BF03327357. [DOI] [PubMed] [Google Scholar]
- 2.Goldring MB, Goldring SR. Osteoarthritis. J Cell Physiol. 2007;213:626–34. doi: 10.1002/jcp.21258. [DOI] [PubMed] [Google Scholar]
- 3.Davidson RK, Waters JG, Kevorkian L, Darrah C, Cooper A, Donell ST, et al. Expression profiling of metalloproteinases and their inhibitors in synovium and cartilage. Arthritis Res Ther. 2006;8:R124. doi: 10.1186/ar2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrage PS, Mix KS, Brinckerhoff CE. Matrix metalloproteinases: role in arthritis. Front Biosci. 2006;11:529–43. doi: 10.2741/1817. [DOI] [PubMed] [Google Scholar]
- 5.Fernandes JC, Martel-Pelletier J, Pelletier JP. The role of cytokines in osteoarthritis pathophysiology. Biorheology. 2002;39:237–46. [PubMed] [Google Scholar]
- 6.Furuzawa-Carballeda J, Macip-Rodriguez PM, Cabral AR. Osteo-arthritis and rheumatoid arthritis pannus have similar qualitative metabolic characteristics and pro-inflammatory cytokine response. Clin Exp Rheumatol. 2008;26:554–60. [PubMed] [Google Scholar]
- 7.Leibiger IB, Berggren PO. SirT1: a metabolic master switch that modulates lifespan. Nat Med. 2006;12:34–6. doi: 10.1038/nm0106-34. [DOI] [PubMed] [Google Scholar]
- 8.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–25. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 9.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng L, Chen R, Liang F, Tsuchiya H, Murai H, Nakahashi T, et al. Silent information regulator, Sirtuin 1, and age-related diseases. Geriatr Gerontol Int. 2009;9:7–15. doi: 10.1111/j.1447-0594.2008.00504.x. [DOI] [PubMed] [Google Scholar]
- 11.Backesjo CM, Li Y, Lindgren U, Haldosen LA. Activation of SirT1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J Bone Miner Res. 2006;21:993–1002. doi: 10.1359/jbmr.060415. [DOI] [PubMed] [Google Scholar]
- 12.Pfister JA, Ma C, Morrison BE, D'Mello SR. Opposing effects of sirtuins on neuronal survival: SIRT1-mediated neuroprotection is independent of its deacetylase activity. PLoS One. 2008;3:e4090. doi: 10.1371/journal.pone.0004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dvir-Ginzberg M, Gagarina V, Lee EJ, Hall DJ. Regulation of cartilage-specific gene expression in human chondrocytes by Sirt1 and nicotinamide phosphoribosyltransferase. J Biol Chem. 2008;283:36300–10. doi: 10.1074/jbc.M803196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagarina V, Gabay O, Dvir-Ginzberg M, Lee EJ, Brady JK, Quon MJ, et al. SirT1 enhances survival of human osteoarthritic chondrocytes by repressing protein tyrosine phosphatase 1B and activating the insulin-like growth factor receptor pathway. Arthritis Rheum. 2010;62:1383–92. doi: 10.1002/art.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takayama K, Ishida K, Matsushita T, Fujita N, Hayashi S, Sasaki K, et al. SIRT1 regulation of apoptosis of human chondrocytes. Arthritis Rheum. 2009;60:2731–40. doi: 10.1002/art.24864. [DOI] [PubMed] [Google Scholar]
- 16.Hedbom E, Hauselmann HJ. Molecular aspects of pathogenesis in osteoarthritis: the role of inflammation. Cell Mol Life Sci. 2002;59:45–53. doi: 10.1007/s00018-002-8404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derfoul A, Miyoshi AD, Freeman DE, Tuan RS. Glucosamine promotes chondrogenic phenotype in both chondrocytes and mesenchymal stem cells and inhibits MMP-13 expression and matrix degradation. Osteoarthritis Cartilage. 2007;15:646–55. doi: 10.1016/j.joca.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Strachan GD, Rallapalli R, Pucci B, Lafond TP, Hall DJ. A transcriptionally inactive E2F-1 targets the MDM family of proteins for proteolytic degradation. J Biol Chem. 2001;276:45677–85. doi: 10.1074/jbc.M103765200. [DOI] [PubMed] [Google Scholar]
- 20.Gagarina V, Carlberg AL, Pereira-Mouries L, Hall DJ. Cartilage oligomeric matrix protein protects cells against death by elevating members of the IAP family of survival proteins. J Biol Chem. 2008;283:648–59. doi: 10.1074/jbc.M704035200. [DOI] [PubMed] [Google Scholar]
- 21.Kawakami Y, Tsuda M, Takahashi S, Taniguchi N, Esteban CR, Zemmyo M, et al. Transcriptional cooactivator PGC-1α regulates chondrogenesis via association with Sox9. Proc Natl Acad Sci U S A. 2005;102:2414–9. doi: 10.1073/pnas.0407510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Li L, Gao P, Chen H, Zhang R, Wei Y, et al. Involvement of the p65/RelA subunit of NF-κB in TNF-α-induced SIRT1 expression in vascular smooth muscle cells. Biochem Biophys Res Commun. 2010;397:569–75. doi: 10.1016/j.bbrc.2010.05.160. [DOI] [PubMed] [Google Scholar]
- 23.Guicciardi EM, Deussing J, Miyoshi H, Bronk SF, Svingen PA, Peters C, et al. Cathepsin B contributes to TNF-α–mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J Clin Invest. 2000;106:1127–37. doi: 10.1172/JCI9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leist M, Jaattela M. Triggering of apoptosis by cathepsins. Cell Death Differ. 2001;8:324–6. doi: 10.1038/sj.cdd.4400859. [DOI] [PubMed] [Google Scholar]
- 25.Tsubuki S, Saito Y, Tomioka M, Ito H, Kawashima S. Differential inhibition of calpain and proteasome activities by peptidyl aldehydes of di-leucine and tri-leucine. J Biochem. 1996;119:572–6. doi: 10.1093/oxfordjournals.jbchem.a021280. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis T. A ubiquitin ligase complex essential for the NF-κB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 1999;13:505–10. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 27.Aisa MC, Beccari T, Costanzi E, Maggio D. Cathepsin B in osteoblasts. Biochem Biophys Acta. 2003;1621:149–59. doi: 10.1016/s0304-4165(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 28.Chae HJ, Ha KC, Lee GY, Yang SK, Yun KJ, Kim EC, et al. Interleukin-6 and cyclic AMP stimulate release of cathepsin B in human osteoblasts. Immunopharmacol Immunotoxicol. 2007;29:155–72. doi: 10.1080/08923970701511579. [DOI] [PubMed] [Google Scholar]
- 29.Lee JH, Song MY, Song EK, Kim EK, Moon WS, Han MK, et al. Overexpression of SIRT1 protects pancreatic beta-cells against cytokine toxicity by suppressing the nuclear factor-κB signaling pathway. Diabetes. 2009;58:344–51. doi: 10.2337/db07-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Julien C, Tremblay C, Emond V, Lebbadi M, Salem N, Bennett DA, et al. Sirtuin 1 reduction parallels the accumulation of tau in Alzheimer disease. J Neuropathol Exp Neurol. 2009;68:48–58. doi: 10.1097/NEN.0b013e3181922348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang SR, Wright J, Bauter M, Seweryniak K, Kode A, Rahman I. Sirtuin regulates cigarette smoke-induced proinflammatory mediator release via RelA/p65 NF-κB in macrophages in vitro and in rat lungs in vivo: implications for chronic inflammation and aging. Am J Physiol Lung Cell Mol Physiol. 2007;292:L567–76. doi: 10.1152/ajplung.00308.2006. [DOI] [PubMed] [Google Scholar]
- 32.Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I. SirT1, an anti-inflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Resp Crit Care Med. 2008;177:861–70. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh UP, Singh NP, Singh B, Hofseth LJ, Price RL, Hagarkatti M, et al. Resveratrol (trans-3,5,4′-trihydroxystilbene) induces silent mating type information regulation-1 and down-regulates nuclear transcription factor-κB activation to abrogate dextran sulfate sodium-induced colitis. J Pharmacol Exp Ther. 2010;332:829–39. doi: 10.1124/jpet.109.160838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SirT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–38. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Csiszar A, Labinskyy N, Jimenez R, Pinto JT, Ballabh P, Losonczy G, et al. Anti-oxidative and anti-inflammatory vasoprotective effects of caloric restriction in aging: role of circulating factors and SirT1. Mech Ageing Dev. 2009;130:518–27. doi: 10.1016/j.mad.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshizaki T, Milne JC, Imamura T, Schenk S, Sonoda N, Babendure JL, et al. SirT1 exerts anti-inflammatory effects and improves insulin sensitivity in adipocytes. Mol Cell Biol. 2009;29:1363–74. doi: 10.1128/MCB.00705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, et al. Phosphorylation regulates SIRT1 function. PLoS One. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–62. doi: 10.1038/ncb1645. [published erratum appears in Nat Cell Biol 2007;9:1442].
- 39.Zhao K, Chai X, Clements A, Marmorstein R. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat Struct Biol. 2003;10:864–71. doi: 10.1038/nsb978. [DOI] [PubMed] [Google Scholar]
- 40.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–27. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 41.Duncan EM, Muratore-Schroeder TL, Cook RG, Garcia BA, Shabanowitz J, Hunt DF, et al. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem differentiation. Cell. 2008;135:284–94. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baici A, Horler D, Lang A, Merlin C, Kissling R. Cathepsin B in osteoarthritis: zonal variation of enzyme activity in human femoral head cartilage. Ann Rheum Dis. 1995;54:281–8. doi: 10.1136/ard.54.4.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baici A, Lang A, Horler D, Kissling R, Merlin C. Cathepsin B in osteoarthritis: cytochemical and histochemical analysis of human femoral head cartilage. Ann Rheum Dis. 1995;54:289–97. doi: 10.1136/ard.54.4.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.