Abstract
The non-coding fraction of the human genome, which is approximately 98%, is mainly constituted by repeats. Transpositions, expansions and deletions of these repeat elements contribute to a number of diseases. None of the available databases consolidates information on both tandem and interspersed repeats with the flexibility of FASTA based homology search with reference to disease genes. Repeats in diseases database (RiDs db) is a web accessible relational database, which aids analysis of repeats associated with Mendelian disorders. It is a repository of disease genes, which can be searched by FASTA program or by limitedor free- text keywords. Unlike other databases, RiDs db contains the sequences of these genes with access to corresponding information on both interspersed and tandem repeats contained within them, on a unified platform. Comparative analysis of novel or patient sequences with the reference sequences in RiDs db using FASTA search will indicate change in structure of repeats, if any, with a particular disorder. This database also provides links to orthologs in model organisms such as zebrafish, mouse and Drosophila.
Availability
The database is available for free at http://115.111.90.196/ridsdb/index.php
Keywords: Repeats, Biomedical Informatics, disease, database, homology
Background
The complexity of mammalian genomes is compounded by the presence of large number of repetitive elements whose functions have not yet been fully deciphered. Repeats contribute to more than 50% of the human genome [1]. Depending on their distribution within the genomes, repeat sequences can be divided into tandemly arrayed and interspersed repeats. Aberrant transpositions of these repeats contribute to number of diseases such as cholinesterase deficiency, Ehlers-Danlose Syndrome, Glanzmannthrombasthenia, Lesch-Nyhan syndrome [2], Huntington's disease [3].
Online Mendelian Inheritance in Man (OMIM) [4] database catalogues Mendelian disorders in man and the corresponding genes with description of the diseases in human. TRbase [5] relates tandem repeats to human diseases. Satellog [6] is another database that describes tandem repeats in disease genes with the option of obtaining OMIM IDs. Transpogene [7] covers transposable elements located inside protein coding genes of seven species including human genome with no option for searching either by disease or repeat-related information. None of these contain options for a homology-based search. Here we describe a dynamically searchable database of disease gene reference sequences, with options for text-based and sequencebased searches. The database provides information on the repeat structure within the gene. This information can be retrieved through gene-centric or disease-centric searches or with the help of a query sequence, with FASTA as the search tool.
Methodology
Implementation
RiDs db is a relational database with well-defined schema and has been implemented using the Structured Query Language (SQL) from the MySQL server version 5.1 (www.mysql.com). Tandem repeats were detected using the Tandem Repeats Finder (TRF) program [8] (version 4.01); interspersed repeats were identified with the RepeatMasker program (Version: open-3.2.9), using repeat masker library (RMLib: 20090120). The Disease information was downloaded from the OMIM database [4]. Reference sequences for disease genes were extracted from Ensembl [9] database using BioMart [10] (Homo sapiens genes GRCh37). The default parameters were used for identification of interspersed repeats with rmblast as search engine, DNA source as human. The TRF detection parameters were (match, mismatch and insertion/deletion scores of +2, 7, 7) with a minimal alignment score of 50 as the cut-off for reporting repeats. FASTA [11] program is implemented on local server. FASTA is a DNA/Protein sequence alignment software package, which uses local sequence alignment to identify homologous sequences in databases. The sequence library is created in FASTA format for homology based searching of disease gene sequences.
Database Structure
RiDs db contains disease genes and the corresponding sequences with information on interspersed repeats and tandem repeats contained in them. Four tables (a-d) have been constructed for storing the data in a structured manner using MySQL; The Disease Information Table (a) consists of disease information downloaded from OMIM database [4] with OMIM ID, title, gene name, locus and genetic disorder information. This serves as the central table of RiDs db. The gene symbols were used to extract sequence ID, OMIM ID and sequences from Ensembl database [9] using the BioMart [10] tool. The annotated information was stored in Gene Information Table (b) that is linked with disease information table via OMIM ID as primary/foreign key in the database. The sequences downloaded from Ensembl database were subjected to RepeatMasker analysis for the detection of interspersed repeats. Repeat information was stored in a third table viz. RiDs db Repeat information table (c). The tandem repeats were identified using TRF [8] and the output was stored in Tandem repeats table (d). The tables are linked through the sequence IDs. A library of reference sequences was constructed and stored in FASTA format for sequence based homology search (FASTA search) (Figure 1). Robust PHP scripts were written for efficient processing of data and for making dynamic and interactive Web pages. The main page of RiDs db contains links to various options provided for searching. It can be queried by either free text or limited keyword or by FASTA program. The free or limited text search theme is centralized to
gene centered information,
disease centered information and
Global Search. All queries will guide the user to available disease and repeat information.
The links are also provided for three model organisms, viz. Mouse [12], Zebrafish [13] and Drosophila [14]. For ease of navigation, links to OMIM, RepeatMasker, TRF, Ensembl and FASTA program are provided.
Figure 1.
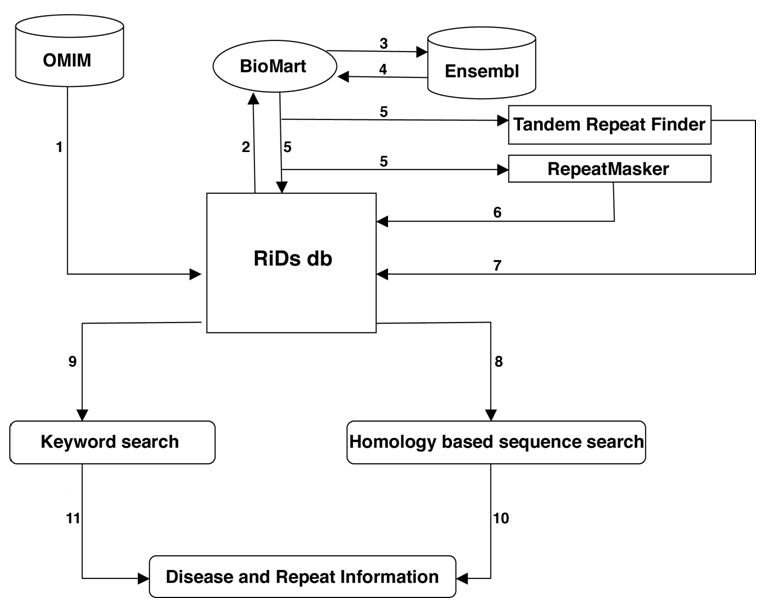
Data flow diagram: The figure describes the input and output for RiDs db. 1) Disease related information from OMIM db; 2-5) disease gene sequences from Ensembl via BioMart subjected to repeat identification using RepeatMasker and TRF; 6, 7) repeat output in RiDs db; 8, 9) modes of search; 10, 11 result.
Conclusion
Repeats are no more considered junk. Abundance of repeats in the human genome indicates probable functions, either for selfpropagation or perhaps to fulfill an unknown requirement at the level of the genome. Therefore to understand these repetitive elements in relation to diseases is important. Repeats in diseases database is a unified platform for studying repeats present in disease genes. Sequence based homology search is a unique feature of this database allowing comparative genomics based query of patient specific as well as novel sequences with reference genes, which will provide more insights into the association of repeats with specific diseases and hence could be a valuable database for biomedical informatics.
Footnotes
Citation:Chaturvedi et al, Bioinformation 7(2): 96-97 (2011)
References
- 1.ES Lander, et al. Nature. 2001;409:860. [Google Scholar]
- 2.PL Deininger, MA Batzer. Mol Genet Metab. 1999;67:183. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 3.M Mitas. Nucleic Acids Res. 1997;25:2245. doi: 10.1093/nar/25.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A Hamosh, et al. Nucleic Acids Res. 2002;30:52. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.T Boby, et al. Bioinformatics. 2005;21:811. [Google Scholar]
- 6.PI Missirlis, et al. BMC Bioinformatics. 2005;6:145. doi: 10.1186/1471-2105-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A Levy, et al. Nucleic Acids Res. 2008;36:D47. doi: 10.1093/nar/gkm949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.G Benson. Nucleic Acids Res. 1999;27:573. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.TJ Hubbard, et al. Nucleic Acids Res. 2009;37:D690. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D Smedley, et al. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WR Pearson, DJ Lipman. Proc Natl Acad Sci U S A. 1988;85:2444. [Google Scholar]
- 12.JA Blake, et al. Nucleic Acids Res. 2009;37:D712. doi: 10.1093/nar/gkn886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.J Sprague, et al. Nucleic Acids Res. 2001;29:87. doi: 10.1093/nar/29.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S Chien, et al. Nucleic Acids Res. 2002;30:149. doi: 10.1093/nar/30.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]


