Abstract
Emergence of drug resistance is a major threat to public health. Many pathogens have developed resistance to most of the existing antibiotics, and multidrug-resistant and extensively drug resistant strains are extremely difficult to treat. This has resulted in an urgent need for novel drugs. We describe a database called ‘Database of Drug Targets for Resistant Pathogens’ (DDTRP). The database contains information on drugs with reported resistance, their respective targets, metabolic pathways involving these targets, and a list of potential alternate targets for seven pathogens. The database can be accessed freely at http://bmi.icmr.org.in/DDTRP.
Keywords: DDTRP, drug targets, resistant pathogens, database, drug discovery
Background
Several pathogens have developed resistance against many of the currently available drugs and become a global health threat. Since infections caused by resistant pathogens are associated with higher morbidity and mortality than those caused by susceptible pathogens, increasing resistance is a major concern [1]. Also, while drug resistance is on the rise, discovery and development of newer drugs have been on the decline [2]. Thus, there is a pressing need for novel drugs to handle the problem of drug resistance. Genomic sequences of pathogens could serve as valuable tools in the search for newer drug targets. We employed an in silico approach to identify potential drug targets that could be exploited for drug discovery against some pathogens with reported drug resistance, viz. Mycobacterium tuberculosis, M. leprae, Plasmodium falciparum, P. vivax, Staphylococcus aureus, Sterptococcus pneumoniae and Nesseirria gonorrhea, and made the data available in the form of a database called DDTRP (Database of Drug Targets for Resistant Pathogens). The database can be accessed at http://bmi.icmr.org.in/DDTRP.
Methodology
From the list of currently available drugs for each of the selected pathogens, drugs reported to be associated with resistance due to mutation(s) in their respective target genes were selected and all enzymes involved in each of the targeted metabolic pathways were obtained from KEGG [3]. The protein sequences of these enzymes were compared with that of the human proteome using BLAST [4]. Proteins that had no orthologs in human at an e-value of 0.005 were identified as potential drug targets and made available in the DDTRP. Each target is cross-linked to other useful online resources. The database was built using XAMPP. The flowchart describing the development of the DDTRP is given in Figure 1.
Figure 1.
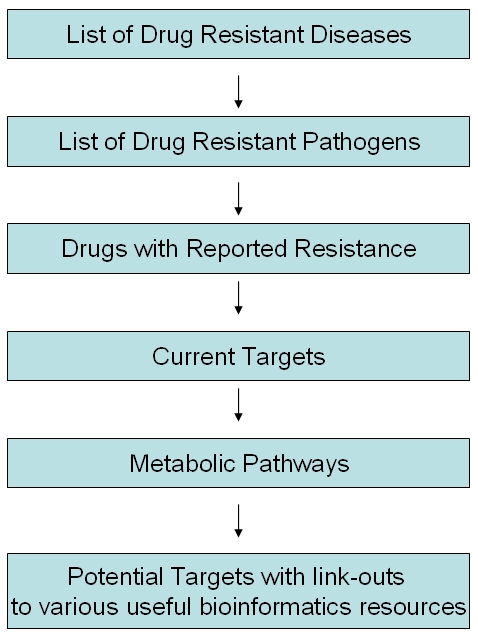
Flow of data in DDTRP
Results and Discussion
Eighty six potential targets were identified for M. tuberculosis, 16 for M. leprae, 47 from Staphylococcus aureus, 43 for Streptococcus pneumoniae, 16 for Plasmodium falciparum, 11 for Plasmodium vivax and 46 for Neisseria gonorrhoeae. Raman et al., (2008) [5] used the method of predicting potential drug targets in M. tuberculosis by comparing its whole genome with that of human proteome. Other investigators like Anishetty et al., (2007) [6] and Agüero et al., (2008) [7] have undertaken various efforts to identify and prioritize targets for tropical diseases including tuberculosis. Our strategy differs from that of others in that we have chosen as our initial dataset proteins from resistance associated metabolic pathways for the prediction of alternate drug targets. A total of 265 proteins were identified by us as potential drug targets for the seven pathogens. Since these targets are involved in metabolic pathways that are known to be critical for the growth or survival of the pathogen and their interruption could bring about a static or cidal effect on the pathogen, the shortlisted targets could be prioritized for further studies. The contents of the database have been summarized in Table 1 (see Table 1).
Features of the database
The database has four browse options for each pathogen:
currently available drugs with resistance,
targets of current drugs,
metabolic pathways involving each of the current targets, and
potential alternate drug targets.
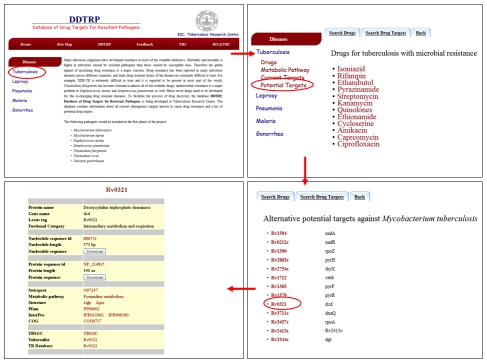
Details for each drug include a brief description of its composition and structure, mechanism of action, target protein, route of administration, dosage, absorption, adverse effects/toxicity and contra-indications. Links have also been provided to other online resources like DrugBank, PharmGKB, Medline Plus, DailyMed and RxList. Current and potential drug targets have been linked to GenBank, Swissprot (UniProt), KEGG, PDB, Pfam, InterPro, COG and other pathogen-specific databases wherever applicable. A provision has been made to browse the database via metabolic pathways. Figure 2 is a screen shot of the data provided for one of the potential targets in the database.
Figure 2.
Information page for a drug target of M. tuberculosis in DDTRP
Future Developments
The database will be periodically updated with drug resistance related information and extended to include pathogens responsible for other emerging and re-emerging infectious diseases.
Conclusion
The Database of Drug Targets for Resistant Pathogens (DDTRP) is a comprehensive database that provides a list of current and alternative drug targets for a group of pathogens of public health significance. This database would be an useful online resource for researchers involved in target identification and drug discovery for various infectious diseases.
Supplementary material
Acknowledgments
We acknowledge the financial support provided by ICMR. We also thank the staff of ICMR-Biomedical Informatics Centre, New Delhi, for administering the database, and Dr. P.R. Narayanan, Former director of TRC, for his kind encouragement and support.
Footnotes
Citation:Sundaramurthi et al, Bioinformation 7(2): 98-101 (2011)
References
- 1.R Isturiz. Int J Antimicrob Agents. 2008;32(4):S201. doi: 10.1016/j.ijantimicag.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 2.MA Fischbach, CT Walsh. Science. 2009;325:1089. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. http://www.genome.jp/kegg/
- 4.SF Altschul, et al. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.K Raman, et al. BMC Syst Biol. 2008;2:109. doi: 10.1186/1752-0509-2-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.S Anishetty, et al. Comput Biol Chem. 2005;29:368. doi: 10.1016/j.compbiolchem.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 7.F Agüero, et al. Nat Rev Drug Discov. 2008;7:900. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.