Abstract
The SOUL protein is known to induce apoptosis by provoking the mitochondrial permeability transition, and a sequence homologous with the BH3 (Bcl-2 homology 3) domains has recently been identified in the protein, thus making it a potential new member of the BH3-only protein family. In the present study, we provide NMR, SPR (surface plasmon resonance) and crystallographic evidence that a peptide spanning residues 147–172 in SOUL interacts with the anti-apoptotic protein Bcl-xL. We have crystallized SOUL alone and the complex of its BH3 domain peptide with Bcl-xL, and solved their three-dimensional structures. The SOUL monomer is a single domain organized as a distorted β-barrel with eight anti-parallel strands and two α-helices. The BH3 domain extends across 15 residues at the end of the second helix and eight amino acids in the chain following it. There are important structural differences in the BH3 domain in the intact SOUL molecule and the same sequence bound to Bcl-xL.
Keywords: apoptosis, Bcl-xL, Bcl-2 homology 3 domain (BH3 domain), crystal structure, NMR, SOUL, surface plasmon resonance
Abbreviations: BH, Bcl-2 homology; HEBP, haem-binding protein; HSQC, heteronuclear single-quantum coherence; MPT, mitochondrial permeability transition; rmsd, root mean square deviation; RZPD, Deutsches Ressouroenzentrum für Genomforschung; SPR, surface plasmon resonance
INTRODUCTION
The Bcl-2 family are a group of evolutionarily conserved proteins that interact to maintain a balance between newly forming and old, damaged or superfluous dying cells. [1]. They play a central role in the regulation of apoptosis, or programmed cell death, a process that in multicellular organisms leads to the controlled death of unneeded or unwanted cells. A crucial event in apoptosis is the MPT (mitochondrial permeability transition), a drastic increase in the permeability of the mitochondrial inner membrane to low-molecular-mass solutes [2]. Through the regulation of apoptosis, the Bcl-2 proteins have an important function in embryogenesis [3], tissue remodelling [4] and the immune response [5]. Their abnormal behaviour is linked to many diseases, such as autoimmunity [6], neurodegenerative disorders [7] and cancer [8].
The effect of the Bcl-2 proteins on the apoptotic process is due to the presence of one or more conserved regions of amino acid sequences, known as BH (Bcl-2 homology) domains, named BH1, BH2, BH3 and BH4 [9,10]. The proteins of this family that contain only the BH3 domain are pro-apoptotic and function as initial sensors of apoptotic signals resulting from various cellular processes, whereas the pro-survival Bcl-2 family members, such as Bcl-2 or Bcl-xL, wield their effect by binding and sequestering their pro-apoptotic counterparts [11]. Peptides spanning the sequence of BH3 domains appear to exert the physiological activity of the intact proteins and their complexes with anti-apoptotic members of the Bcl-2 family have received considerable attention since this interaction is believed to explain the effect at the molecular level. In particular, the complexes of peptides with the sequences of the BH3 domains of Bad, Bim, Bak, Bid and Beclin 1 with Bcl-xL have been examined by X-ray crystallography and NMR, and the conserved crucial interactions between the peptides and the protein have been identified [12,13].
Cancer cells frequently overexpress the anti-apoptotic members of the Bcl-2 family, and small molecules that incorporate the structural features of the BH3 domains necessary for binding to these anti-apoptotic proteins have been synthesized and are being tested as specific cancer cell killers [14].
SOUL was first identified at the transcriptional level by suppression subtractive hybridization in the chicken retina and pineal gland, and its gene was named ckSoul because of the high transcript levels found in the pineal gland, the organ René Descartes hypothesized was the location of the soul [15]. A few years before this report, human SOUL had been isolated and characterized from saline extracts of human term placentas and had been called PP23 (placental protein 23) [16]. More recently, the protein has also been identified in human amniotic fluid [17]. It has subsequently been shown that the gene coding for this protein is very widely distributed in evolution and it has been characterized in many other species, including the popular model organism of plant biology Arabidopsis thaliana. On the basis of its sequence similarity with the mouse gene p22HBP, which codes for HEBP1 (haem-binding protein 1) or p22HBP, SOUL has also been called the alternative name of HEBP2 [18]. Recombinant mouse SOUL was reported to be a dimer in the absence of haem and to hexamerize upon haem binding, with a dissociation constant in the nanomolar range [19].
A very important observation is that SOUL can induce the MPT, a condition that leads to mitochondrial swelling and cell death [20]. More recently, analysis of the human SOUL sequence revealed the presence of a putative BH3 domain of the Bcl-2 protein family whose deletion abolished the apoptotic effects of SOUL [21].
In the present paper, we provide experimental evidence that SOUL is a BH3-only protein as the sequence spanning amino acids 147–172 interacts with the anti-apoptotic protein Bcl-xL. In addition, we have crystallized both intact SOUL and the complex of a peptide that contains its BH3 domain with Bcl-xL, and solved their three-dimensional structures to 1.6 and 2.0 Å (1 Å=0.1 nm) resolution respectively. The Bcl-xL–SOUL BH3 domain interactions are particularly interesting, since the domain adopts a different structure when bound to Bcl-xL.
EXPERIMENTAL
Protein expression and purification
The cDNA coding for human SOUL (IMAGE ID 3445763), obtained from RZPD (Deutsches Ressourcenzentrum für Genomforschung), was amplified by PCR using primers designed to introduce restriction sites for BamHI and HindIII endonucleases and a sequence coding for a digestion site for thrombin in the C-terminal end in the amplified fragment. After purification, the fragment and the expression vector pQE50 (Qiagen) were digested with the restriction enzymes mentioned above and incubated with ligase to insert the cDNA into the vector respecting the reading frame. BL21 C41 strain Escherichia coli cells were transformed with the resulting vector, grown at 37 °C and protein synthesis was induced overnight at 20 °C with 0.5 mM IPTG (isopropyl β-D-thiogalactopyranoside). Under these conditions of subcloning in pQE50, the expressed intracellular domain is fused to a histidine tag at its C-terminus. The presence of the tag allowed the affinity purification of the fused protein by passing the bacterial extracts through a nickel–Sepharose column. The column was equilibrated with 20 mM Tris/HCl (pH 7.5), 0.5 M NaCl, 10 mM imidazole and 0.02% sodium azide, and the bound protein was eluted with a linear gradient of imidazole from 10 to 500 mM. After the affinity column separation, the tag was removed by thrombin digestion and the protein was purified further by gel filtration on a Superdex G-200 column equilibrated with 20 mM Tris/HCl (pH 7.5), 0.15 M NaCl and 0.02% sodium azide and by hydrophobic interaction chromatography (Lipidex1000).
Recombinant human Bcl-xL (IMAGE ID 2823498; RZPD) was prepared in a similar way. A truncated form lacking the flexible loop spanning amino acids 27–82 and the last 24 amino acids, which are the transmembrane domain, was inserted into the pET15b vector which introduces an N-terminal histidine tag and a thrombin digestion sequence. The purification protocol followed that of SOUL.
Complete removal of the tag was assessed by Western blot analysis using an HRP (horseradish peroxidase)-conjugated anti-His antibody (Sigma–Aldrich). The purified protein was a single band by SDS/PAGE in both cases.
UV–visible spectra were recorded with a UV/Vis Unicam spectrometer. An aliquot of 250 μM haemin dissolved in DMSO was diluted with 20 mM Tris/HCl (pH 7.5) and 0.15 M NaCl to give a final haemin concentration of 10 μM. The concentration of the haemin solution was determined as described previously [22]. Two other samples were prepared by adding, in addition to haemin, appropriate aliquots of SOUL and BSA dissolved in 20 mM Tris/HCl (pH 7.5) and 0.15 M NaCl to bring their final concentration to 100 μM. These samples thus contained a ratio of ten times the molar concentration of SOUL and BSA with respect to the haemin concentration. The three samples were incubated for 30 min at room temperature (25 °C) and their UV–visible spectra were recorded.
NMR measurements
For the production of 15N-labelled human Bcl-xL lacking only the C-terminal transmembrane domain, host cells were grown in M9 minimal medium using 15NH4Cl as sole nitrogen source. HSQC (heteronuclear single-quantum coherence) NMR spectra were recorded on a Bruker Avance spectrometer operating at 600.13 MHz, equipped with a cryoprobe. The labelled protein, dissolved in 20 mM Tris/HCl (pH 7.5) and 0.15 M NaCl (in 10% 2H2O) and at a concentration of 85 μM, was titrated with the SOUL BH3 peptide dissolved in the same buffer at 600 μM. Nine additions were made so that, after correcting for the precise peptide concentration and taking into account dilutions, the BH3 peptide/protein molar ratio was 0.07, 0.17, 0.26, 0.35, 0.52. 0.69, 1.38, 2.77 and 3.83. After each of the additions, the sample was incubated at 20 °C for approximately 5 min and a one-dimensional 15N-decoupled 1H spectrum and a two-dimensional 1H-15N HSQC spectrum were recorded at the same temperature. Standard sequence schemes with pulsed-field gradients were used to achieve the suppression of the solvent signal.
The dissociation constant was calculated from the shifts in two peaks in fast exchange in the methyl region of the decoupled 1H spectrum fitting the data with the equation:
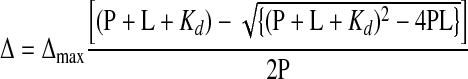 |
where Δ is the chemical shift change, P is the protein concentration, L is the total ligand concentration, and Kd is the dissociation constant. Appropriate corrections for the dilution of protein and ligand were made after the addition of each aliquot during the titration.
SPR (surface plasmon resonance) studies
Bcl-xL was immobilized on a COOH1 research-grade sensor chip (Nomadics) by amine-coupling chemistry using the manufacturer's protocols, and the SOUL BH3 domain peptide was used as the analyte. SPR measurements were carried out in HBS buffer [10 mM Hepes (pH 7.4), 150 mM NaCl, 3 mM EDTA and 0.005% P-20 surfactant] at 20 °C using a SensiQ Pioneer instrument (ICx Technologies). Data were analysed with the Qdat evaluation analysis software.
Crystallization, X-ray data collection, structure solution and refinement
Purified native SOUL was used at approximately 20 mg/ml for the initial screen of crystallization conditions. Molecular Dimensions Structure Screens were used at 20 °C with the hanging-drop method, mixing 1 μl of the protein solution with the same volume of the precipitating solution, and equilibrating against a volume of 0.3 ml of the latter in the reservoir. The conditions yielding small crystals were later refined and the sitting-drop method with larger volumes was also tested until crystals that were large enough for data collection were obtained.
The SOUL BH3 peptide spanning amino acids 147–172 was synthesized by TAG Copenhagen. The complex of recombinant human Bcl-xL with the peptide was prepared by mixing the protein at approximately 4 mg/ml with four times the molar ratio of the peptide. The mixture was incubated for about 1 h and then concentrated to approximately 15 mg/ml, and was then used at this concentration for the crystallization experiments.
Two different crystal forms of native SOUL were obtained, both in the presence of 0.2 M 1-butyl-3-methylimidazolium chloride, because it drastically improved the diffraction properties of the crystals. The first crystal form is hexagonal, space group P6122 with a=b=143.9 Å and c=242.1 Å. It contains four molecules in the asymmetric unit (see Table 1), diffracts to approximately 2.85 Å and appears to be closely related to another crystal form reported in the literature [23]. The crystals grow by adding equal volumes of the protein solution and 0.1 M Tris/HCl (pH 8.5) and 2.0 M ammonium sulfate. The second crystal form is orthorhombic, space group C2221, and a=137.7 Å, b=114.7 Å and c=67.4 Å, and grows by mixing equal volumes of the protein solution and 0.85 M NaH2PO4, KH2PO4 and 0.08 M Hepes (pH 7.5). These crystals diffract to a better resolution, approximately 1.6 Å, contain two molecules in the asymmetric unit and are the crystal form that was solved first using the S.I.R. (single isomorphous replacement) method. The hexagonal crystal form was solved later by molecular replacement.
Table 1. Data collection and refinement statistics.
The values in parentheses refer to the highest resolution shells. For the data collection of the orthorhombic form, the highest resolution interval is 1.69–1.60 Å and for the hexagonal form is 3.00–2.85 Å, whereas for the co-crystals of the BH3 domain with Bcl-xL it is 2.05–1.95 Å. The highest resolution shells used in the refinements are: 1.66–1.60 Å, 2.95–2.85 Å and 1.98–1.95 Å for the co-crystals of the BH3 domain. The ligand of the SOUL crystals is phosphate and that of the complex is sulfate.
| Parameter | Human SOUL native | Human SOUL native | Complex human Bcl-xL–human SOUL BH3 peptide |
|---|---|---|---|
| Space group | C2221 | P6122 | P43 |
| a (Å) | 137.72 | 143.92 | 66.83 |
| b (Å) | 114.66 | 143.92 | 66.83 |
| c (Å) | 67.42 | 242.12 | 175.22 |
| α (°) | 90.0 | 90.0 | 90.0 |
| β (°) | 90.0 | 90.0 | 90.0 |
| γ (°) | 90.0 | 120.0 | 90.0 |
| Molecules in the asymmetric unit (n) | 2 | 4 | 4 |
| Resolution range (Å) | 24.9–1.60 | 80.0–2.85 | 53.1–1.95 |
| Observed reflections (n) | 470976 | 723081 | 276588 |
| Independent reflections (n) | 69663 | 35317 | 54709 |
| Multiplicity | 6.8 (6.5) | 20.5 (20.4) | 5.1 (5.2) |
| Rmerge (%)* | 6.5 (38.1) | 8.5 (38.8) | 7.3 (32.0) |
| I/σ | 18.3 (4.6) | 25.7 (8.3) | 17.6 (6.0) |
| Completeness (%) | 98.9 (98.0) | 99.9 (100.0) | 98.0 (96.5) |
| Reflections in refinement (n) | 69648 | 35223 | 54700 |
| Rcryst (%)† | 17.5 | 23.5 | 21.1 |
| Rfree (%) (test set 5%)‡ | 19.7 | 26.8 | 26.3 |
| Protein atoms (n) | 2974 | 5631 | 5132 |
| Ligand atoms (n) | 5 (phosphate) | − | 5 (sulfate) |
| Water molecules (n) | 395 | − | 172 |
| rmsd on bond lengths (Å)§ | 0.008 | 0.008 | 0.009 |
| rmsd on bond angles (°) | 1.250 | 1.169 | 1.124 |
| Planar groups (Å) | 0.006 | 0.006 | 0.004 |
| Chiral volume deviation (Å3) | 0.081 | 0.074 | 0.068 |
| Average B-factor (Å2) | 21.2 | 73.3 | 36.4 |
| Protein atoms | 20.1 | 73.3 | 36.8 |
| Ligand atoms | 26.7 | − | 49.2 |
| Solvent atoms | 29.4 | − | 27.3 |
*Rmerge=ΣhΣi|Iih–〈Ih〉|/ΣhΣi〈Ih〉, where 〈Ih〉 is the mean intensity of the i observations of reflection h.
†Rcryst=Σ||Fobs|–|Fcalc||/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factor amplitudes respectively; summation includes all reflections used in the refinement.
‡Rfree=Σ||Fobs|–|Fcalc||/Σ|Fobs|, evaluated for a randomly chosen subset of 5% of the diffraction data not included in the refinement.
§rmsd from ideal values.
The best crystals of the complex human Bcl-xL–SOUL BH3 peptide grew by mixing equal volumes of the complex solution and 15% PEG [poly(ethylene glycol)] 6000, 0.2 M sodium sulfate and 0.3 M 1-butyl-3-methylimidazolium 2-(2-methoxyethoxy)ethyl sulfate. They are tetragonal, space group P43, with a=b=66.8 Å and c=175.2 Å, and diffract to 2.0 Å resolution.
The diffraction data were collected from crystals frozen at 100 K after a brief immersion in a mixture of 80% of the mother liquor and 20% glycerol. The data set for a gold heavy atom derivative used for phasing were obtained using copper Kα radiation from a Rigaku RU-300 rotating anode X-ray generator with a Mar345 imaging plate area detector. The final data sets used for refinement of this and the other crystal forms were collected at the ID14-2 beamline of the ESRF (European Synchrotron Radiation Facility) in Grenoble (λ=1.001 Å). The data were indexed, integrated and reduced using the programs AUTOMAR, MOSFLM and Scala [24]. The diffraction data statistics of the data sets used for refinement are summarized in Table 1.
Initial phases for the orthorhombic crystals to a 2.3 Å resolution were determined by the single isomorphous replacement method with the derivative data collected at the home source. Two gold sites were located in a difference Patterson map using the program SHELXS [25] and entered as input for the program autoSHARP [26] that was used to locate the minor sites of the derivative, and for density modification and final phasing to 1.8 Å resolution. The electron density map thus produced was of very good quality and could be readily interpreted. The initial model of SOUL was built in this map using the program Coot [27].
Refinement was carried out initially using the program REFMAC [28] and, in a second stage, with the program Phenix.refine [29]. During the process of refinement and model building, the quality of the models was controlled with the program PROCHECK [30]. Solvent molecules were added to the model in the final stages of refinement according to hydrogen-bond criteria and only if their B factors refined to reasonable values and if they improved the Rfree. The model was finally subjected to a final round of TLS refinement.
The structure of the hexagonal crystal form of SOUL was solved using the CCP4 suite of programs for crystallographic computing. The initial phases were calculated by the molecular replacement method as implemented in the program MOLREP [31], with the co-ordinates of the orthorhombic model as the search probe. The automatic search with data up to a resolution of 2.9 Å gave a solution that placed in their correct position three out of the four molecules present in the asymmetric unit. Fixing these co-ordinates, the fourth molecule was found by the same program. The score of this solution was 0.544 and its R factor was 41.6%.
A similar procedure was followed to solve and refine the structure of the complex human Bcl-xL–SOUL BH3 peptide using the co-ordinates of a protomer of Bcl-xL ([32]; PDB code 2YXJ) as the search probe. After the four protomers of Bcl-xL present in the asymmetric unit had been placed in their correct position, it became evident that they dimerized with domain swapping and, at this point, the extra electron density for the four BH3 helices present in the asymmetric unit was also very clear. The final refinement statistics for the models of the three crystal forms are summarized in Table 1.
RESULTS
X-ray structure of human SOUL
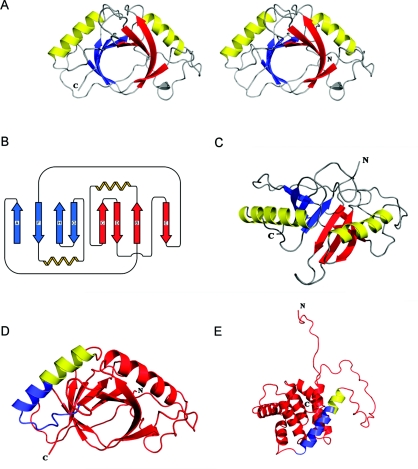
The SOUL monomer is a single domain structure organized as an open distorted β-barrel with eight anti-parallel strands. The barrel is open in the sense that the first and the last strands (A and E) are not in contact in the sheet (Figure 1). Two α-helices connect the second to the third strand and the sixth to the seventh strand. They are both located on one face of the molecule and pack against the curved sheet that forms the barrel. A monomer of SOUL fits into a box with the approximate dimensions 56×47×40 Å. This fold is quite different from that of the canonical member of the BH3-only protein family BID (Figure 1).
Figure 1. Overall structure and folding of SOUL.
(A) Stereodiagram of the SOUL molecule. The eight-stranded β-sheet is shown in two colours, red and blue, to emphasize the presence of two repeated units, each with β-β-α-β-β topology, related by a pseudo-two-fold axis of symmetry. (B) A topological diagram of SOUL. The eight antiparallel β-strands form a distorted β-barrel with the two α-helices arranged on one face. Strands are labelled in the order of their appearance from the N-terminus to the C-terminus using the letters A–H. The eight strands span the following residues: A, 39–43; B, 46–55; C, 89–94; D, 103–110; E, 127–132; F, 135–142; G, 174–178; H, 190–195. The two helices span residues 58–73 and 148–164. (C) Ribbon representation of the SOUL monomer viewed in a direction rotated approximately 90 ° with respect to (A). The two helices are in yellow, and the strands of the β-sheet have been coloured as (A and B) to emphasize the presence of the pseudo-two-fold axis. (D) Ribbon diagram of SOUL with the peptide spanning the BH3 domain represented in light blue for the portion of the chain identified by sequence similarity with known BH3 domains and in yellow for the rest of the second helix of the molecule. (E) Ribbon diagram of the NMR model of BID (lowest energy structure, PDB code 2BID) with the BH3 peptide and domain oriented and colour-coded as in (D). The Figures of the models were prepared using the program PyMOL (http://www.pymol.org).
Examination of the secondary structure (Figure 1B) reveals that the molecule is made up of two repeated units, each with β-β-α-β-β topology related by a pseudo-two-fold axis of symmetry. Two SOUL monomers (A and B) are present in the asymmetric unit of the orthorhombic crystal form (Table 1). The secondary structure assignments of monomer A are, for the β-strands, the following: strand A, residues 39–43, B, residues 46–55, C, residues 89–94, D, residues 103–110, E, residues 127–132, F, residues 135–142, G, residues 174–178 and H residues 190–195. The two α-helices span residues 58–73 and 148–164. In addition, residues 25 and 26 extend the β-sheet and residues 113–116 form an additional helix turn in both monomers. A minor difference between the two monomers was also observed: strand D of molecule B has one residue less at the N-terminal end.
The space within the β-barrel is filled by the side chains of both hydrophobic and hydrophilic residues: Trp48, Met135, Leu137, Leu139 and Trp193, but also Arg41 and Arg132, Asp130, Asp191, Thr90 and Tyr179. Packing of the first helix takes place through hydrophobic residues and the following specific contacts: Trp58 (NE1)–Ser91 (OG), Lys68 (O)–Glu124 (OE), Tyr72 (OH)–Pro120 (O), Tyr72 (O)–Gln77 (OE) and Ile73 (O)–Ile83 (N). Packing of the second helix involves the following specific contacts: Gln154 (OE)–Arg140 (NE) and Leu162 (O)–Lys167 (N).
Two NMR structures of murine p22HBP have been published [33,34]. The protein has approximately 28% sequence identity with murine SOUL. Comparison of the two SOUL molecules present in the crystallographic asymmetric unit of the orthorhombic crystal form with the two NMR models of murine p22HBP revealed that the four models differ substantially only in rather limited areas (Supplementary Figure S1 at http://www.BiochemJ.org/bj/438/bj4380291add.htm). The zone where the two SOUL molecules in the asymmetric unit differ more from one another are the region before the first strand of the β-sheet and the loop connecting strands C and D. The chains before the first strand are totally exposed to the solvent, whereas in the case of the connection of strands C and D the loop in chain A is in close contact with a symmetry-related molecule, and the equivalent area in molecule B is in contact with the solvent. These differences are thus probably simply a consequence of molecular packing in the crystal. Although there is more variability in the two NMR structures of murine p22HBP, the two models are very similar in the region connecting strands C and D, which is also the region where both are most different from SOUL. This particular region of the SOUL molecule thus appears to be more variable than the rest of the molecule.
The BH3 domain predicted by sequence alignment to be present in SOUL spans residues 158–172, i.e. the last seven residues of the second α-helix of the molecule and eight amino acids in the loop connecting it to strand G. The domain is represented in light blue in Figure 1(D). Additional details on this structure have been included as Supplementary information (at http://www.BiochemJ.org/bj/438/bj4380291add.htm).
Does SOUL bind haemin?
All our attempts to prepare co-crystals of SOUL and haemin failed. When crystallizations were set up, with haemin/SOUL molar ratios of even up to five, the crystals obtained, after screening many different conditions, were invariably those of the apo protein. Soaking the pre-formed crystals did not reveal any electron density other than that of the apo protein. It was thus suspected that the interaction of the two molecules, if present, was not as strong as expected. For this reason, the UV–visible spectrum of haemin was examined alone and in the presence of ten times the molar ratio of SOUL, with BSA used as a control (Supplementary Figure S2A at http://www.BiochemJ.org/bj/438/bj4380291add.htm). Although the sample containing BSA had, due to the ligand–protein interaction, its peak higher and shifted from 390 to 401 nM, as expected for haemin bound with no axial co-ordination to a hydrophobic cavity [35], the sample containing SOUL showed only negligible differences that can be explained by the absorption of the protein present in the sample and cannot be considered as evidence of a SOUL–haemin interaction.
A second control was carried out recording 15N-1H HSQC NMR spectra of 15N-labelled SOUL in the presence of increasing amounts of haemin dissolved in Tris/HCl buffer. Haemin aliquots corresponding to 0.5, 1, 2, 3 and 4 equivalents of SOUL were added to the protein sample and the spectra were recorded at 25 °C (Supplementary Figure S2B). After the addition of up to four equivalents of haemin per protein molecule, the spectrum remained unaltered. Given the time involved to record the different spectra, a kinetic effect can be excluded in this case or at least if there was such an effect it has to be proposed that the reaction is so slow that, even after more than 1 day of observation, no change in the spectrum is detectable at all. These observations thus lead to the conclusion that, with the methods described in the present study, no interaction between haemin and SOUL, that may be considered of physiological relevance, can be observed.
Interaction of the SOUL BH3 peptide with human Bcl-xL
The BH3 domain predicted by sequence alignment to be present in SOUL spans residues 158–172, i.e. the last seven residues of the second α-helix of the molecule and eight residues in the loop connecting it to strand G. BH3 domains are known to be helical and, since we knew that in the structure of SOUL the helix began before, we decided to examine the interaction of a 26-amino-acid-long peptide spanning residues 147–172 with the anti-apoptotic protein Bcl-xL, i.e. covering the entire helix and the region predicted to be part of the BH3 domain by sequence homology. The sequence of the peptide studied is: SAQKNQEQLLTLASILREDGKVFDEK. Figure 1(D) shows the SOUL monomer with the BH3 peptide coloured light blue for the chain predicted to be part of the domain by sequence alignment and yellow for the rest of the peptide corresponding to the N-terminal portion of the second α-helix of SOUL.
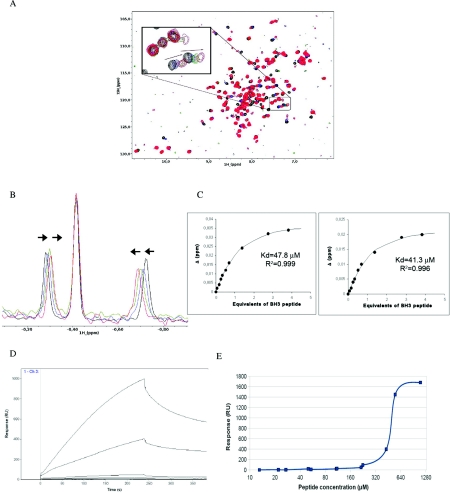
The interaction of the peptide and the protein was examined in solution by one- and two-dimensional NMR and by SPR. Figure 2(A) shows the amidic region of the 15N-1H HSQC NMR spectrum of 15N-labelled human Bcl-xL (lacking the transmembrane domain after amino acid 209) titrated with increasing amounts of the peptide. Note the significant changes in some of the peaks of the protein as the interacting BH3 peptide was added to the solution. Figure 2(B) shows the chemical shift displacements in the two peaks in fast exchange in the methyl region of the 1H spectrum that were used to calculate an approximate dissociation constant of the interaction, and Figure 2(C) shows the fitting of these displacements as a function of equivalents of the BH3 peptide added. The dissociation constant values calculated using the two peaks are in reasonable agreement with one another and are 47.8 and 41.3 μM (see Figure 2C).
Figure 2. Interaction of the SOUL BH3 peptide with human Bcl-xL.
(A) 15N-1H NMR correlation spectra of 15N-labelled human Bcl-xL titrated with increasing amounts of a 26-amino-acid-long peptide spanning residues 147–172 of human SOUL. The peptide sequence is SAQKNQEQLLTLASILREDGKVFDEK. Arrows indicate the direction of peak shifts. Only four spectra are represented in the Figure; the black one is before any addition, the green after the addition of 0.52 equivalents of the peptide, the blue after the addition of 1.38 equivalents and the red after adding 3.83 equivalents of the BH3 domain peptide. (B) Decoupled 1H spectrum showing the shifts in two peaks in fast exchange in the methyl region used to calculate an approximate dissociation constant. The colour of the spectra is the same as in (A) and corresponds to the addition of the same amounts of peptide. (C) Magnitude of the change in the 1H chemical shift of the two selected peaks plotted as a function of the total number of equivalents of BH3 domain peptide added. The best fit curve is shown along with the coefficient of determination (R2) and the calculated dissociation constants. The values determined using the two peaks are 47.8 and 41.3 μM. The equation used to fit the data is given in the Experimental section. (D) SPR studies of the same interaction. The sensorgram shows the binding of the BH3 peptide to truncated immobilized Bcl-xL. Relative units (RU) are plotted as a function of time (in s). (E) A plot of the response as a function of the peptide concentration used to estimate the dissociation constant. The diagram is the result of several experiments and higher peptide concentrations could not be used because the BH3 peptide had a tendency to aggregate.
The SOUL BH3 peptide–Bcl-xL interaction was also studied by SPR. Both intact Bcl-xL and a form lacking amino acids 27–82 (Δ27–82) that is known to bind BH3 domains like the intact protein [36] were immobilized on a sensor chip and the SOUL BH3 domain peptide was used as the analyte. Similar results confirming the interaction were observed for both variants of Bcl-xL. Figure 2(D) shows a sensorgram of the Δ27–82 form of the protein. The dissociation constant estimated with this method is approximately five times the value observed in the NMR experiments. The discrepancy is probably due to errors in the estimate of the peptide concentration used in the experiments. However, we did not detect significant binding between intact SOUL and Bcl-xL (results not shown), which may reflect either a lack of interaction under the conditions tested or more probably the requirement of drastic structural changes in SOUL.
X-ray structure of the SOUL BH3 domain peptide complexed with Bcl-xL
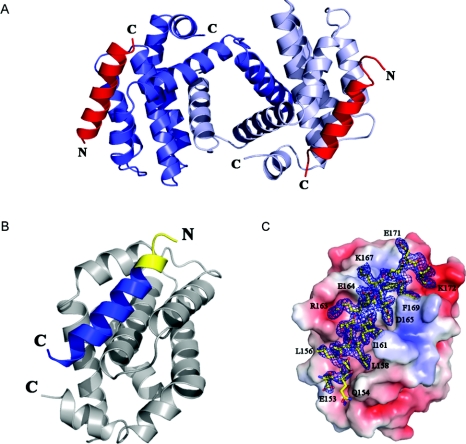
The data collection and refinement statistics of the co-crystals of human Bcl-xL (Δ27–82) with the SOUL BH3 peptide are summarized in Table 1. The crystals are tetragonal, space group P43 and present 50% merohedral twinning. The structure was solved by molecular replacement, initially assuming that the space group was P43212, but the model could not be properly refined. A standard statistical test of the structure factors revealed that the correct space group is P43 with the twinning law h, -k, -l. The law was introduced in the program Phenix.refine [29] and the model was refined to give the statistics listed in Table 1. The asymmetric unit contains four Bcl-xL protomers organized as dimers that exhibit domain swapping, exchanging their N-terminal α1-helix (Figure 3A). This kind of domain swapping has been observed in several co-crystals of BH3 domains and human Bcl-xL (Δ27–82) [13,37–39]. It is considered to be an artifact due to the Δ27–82 deletion, but it does not affect in any way the BH3-domain-binding activity or anti-apoptotic properties of the protein. In fact, the complex in solution of the Beclin 1 BH3 domain and another truncated form of Bcl-xL studied by NMR exhibits the same interactions observed in the crystals [40]. The two dimers present in the asymmetric unit of the crystals of the SOUL BH3 complex are very similar to each other, with an rmsd (root mean square deviation) of 0.607 Å over 314 α carbon atoms of Bcl-xL and the BH3 domains. Their Bcl-xL part is also quite similar to that of the other domain-swapped dimers of human Bcl-xL (Δ27–82) that exchange the N-terminal α1-helix. The rmsd values for the 276 α carbons are 1.845 Å for the Beclin 1 complex ([13]; PDB code 2P1L), 2.425 Å for the BIM L12F mutant peptide complex ([38]; PDB code 3IO8) and 2.216 Å for the helical α/β peptide foldamer complex ([39]; PDB code 3FDM).
Figure 3. Crystal structure of the SOUL BH3 domain peptide complexed with Bcl-xL.
(A) A dimer of human Bcl-xL (Δ27–82) showing swapping of the α1 domains. The two helical SOUL BH3 peptides are represented in red. (B) A protomer of Bcl-xL and the SOUL BH3 peptide in contact with it. The portion of the peptide predicted by sequence homology to be the BH3 domain is in blue, whereas the yellow part is the additional portion of the peptide, the beginning of the second helix of SOUL. (C) Electron density of the SOUL BH3 peptide bound to Bcl-xL oriented as in (B). The molecular surface of Bcl-xL shows the negatively charged residues in red and those positively charged in blue. The 2Fobs−Fc map was contoured at a 1.2 σ level. Selected BH3 peptide amino acids participating in important contacts with Bcl-xL have been labelled in single-letter amino acid notation. The Figure was prepared using the program PyMOL (http://www.pymol.org).
The structures of several BH3 domains in complex with Bcl-xL have been examined. They all reveal that the BH3 sequence forms an amphipathic helix that inserts into a hydrophobic groove on the surface of the anti-apoptotic protein [41–43].
The four SOUL BH3 domain peptides observed in the co-crystals with Bcl-xL present an ordered structure which in every case contains more amino acids at the N-terminus than those predicted by sequence similarity (residues 158–172 of the SOUL sequence). Of the 26-amino-acid-long peptide co-crystallized with Bcl-xL, for only the first three amino acids (Ser147-Ala148-Gln149) there is no clear electron density in any of the four peptides present in the asymmetric unit. In all of the four BH3 domain peptides examined, the helix observed is at least 18 amino acids long (E153QLLTLASILREDGKVFD170). Figure 3(B) represents in two colours the BH3 domain peptide bound to a protomer of Bcl-xL that more closely corresponds to the prediction (chain E). The extra amino acids at the N-terminus are represented in a different colour. Figure 3(C) shows the electron density of the BH3 domain peptide oriented as in Figure 3(B), with the Bcl-xL protomer represented as a space-filling model. In intact SOUL, the last amino acid of the second α-helix is Glu164 and thus eight amino acids with the sequence DGKVFDEK in the loop following the helix change their conformation upon interaction with Bcl-xL to become the last portion of the BH3 helical domain in the complex.
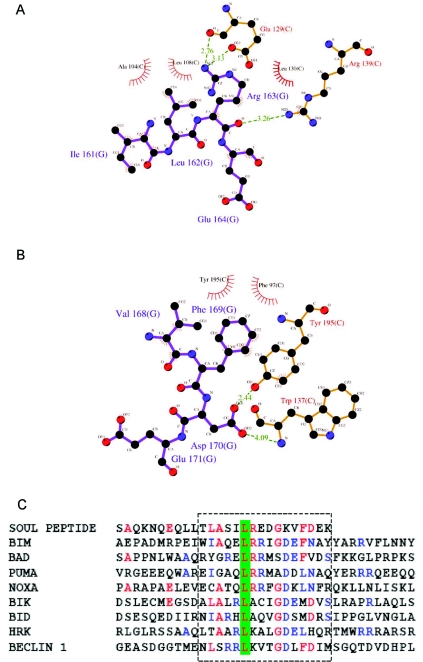
The interactions of the SOUL BH3 domain peptide with Bcl-xL are mostly hydrophobic (Supplementary Table S1 at http://www.BiochemJ.org/bj/438/bj4380291add.htm, and Figure 4), but they also include other more specific contacts, due to charged residues: Gln154 (BH3)–Gln111 (Bcl-xL), Ser160 (BH3)–Glu129 (Bcl-xL), Arg163 (BH3)–Glu129 (Bcl-xL), Asp165 (BH3)–Tyr101 (Bcl-xL) and Asp170 (BH3)–Tyr195 (Bcl-xL). Residues participating in hydrophobic contacts are Leu155, Leu156, Leu158, Leu162 and Phe172 of the BH3 domain peptide and Leu108, Leu112, Leu130, Val126, Val141, Phe97, Phe105, Tyr101, Tyr195 and Trp137 of the Bcl-xL molecule.
Figure 4. SOUL BH3 peptide side chains that interact with Bcl-xL.
(A) Arg163 and other side chains participating in one of the specific contacts of the SOUL BH3 peptide with Bcl-xL. Hydrogen bonds are indicated with green broken lines, whereas the amino acids that make hydrophobic contacts are only indicated but not represented as ball and stick models. (B) The same type of diagram as (A), but with another specific contact in which the key residue is Asp170 of the SOUL BH3 peptide. (C) Sequence alignment of the SOUL BH3 peptide and eight BH3-only proteins. The BH3 domains are boxed and the amino acids that are identical in SOUL and at least two other sequences are in red, whereas those that are identical in at least three sequences are in blue. The leucine residue conserved in all of the sequences is indicated on a green background. (A and B) were prepared using the visualization program LIGPLOT [49].
The amino acid contributions to the free energy of binding of BH3 peptides to Bcl-xL have been calculated using molecular dynamics simulations coupled with the molecular mechanics/Poisson–Boltzmann surface area method [12]. The Bcl-xL residues that give important contributions are: Phe97, Tyr101, Leu112, Val126, Leu130, Arg139, Tyr185 and Phe146. With the exception of the last residue, they all participate in the interactions with the SOUL BH3 domain peptide. Leu158 and Leu162 of SOUL BH3 correspond to Leu112 and Leu116 of Beclin 1 [13] and to Ile90 and Leu94 of BIM [43], and Phe169 corresponds to Phe123 in Beclin 1 and Phe101 in BIM. Arg163 of the SOUL BH3 domain peptide, one of the residues controlling the specificity of binding, corresponds to Lys117 in Beclin 1 and Arg95 in BIM. These three basic amino acids are hydrogen-bond donors to Glu129 of Bcl-xL.
Two of the specific charged residue contacts of the SOUL BH3 domain peptide and Bcl-xL are represented in Figures 4(A) and 4(B). They involve Arg163 and Asp170 of the BH3 domain peptide. The first interaction is established with Glu129 of Bcl-xL, whereas the second is with Tyr195. Analogous contacts are found in both Beclin 1 and BIM. The sequence of the peptide is aligned to those of several well-known BH3-only proteins in Figure 4(C). Note that some amino acids in the N-terminal region of the peptide are also present in other members of the family.
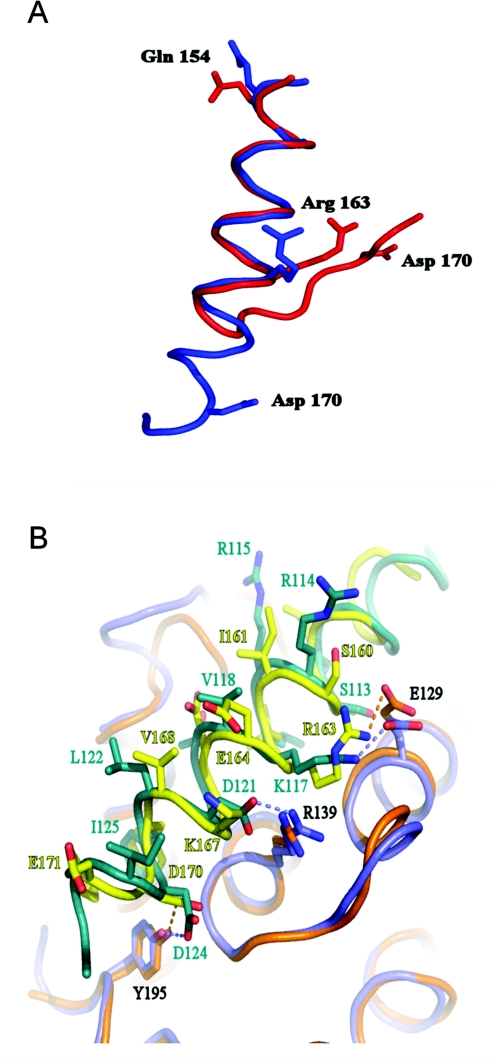
Figure 5(A) superimposes the domain in intact SOUL with the helical conformation bound to Bcl-xL found in the crystals. It is evident that very drastic changes are required to transform the structure of the free domain into the bound one. Figure 5(B) shows that binding of the BH3 domain peptide of SOUL to Bcl-xL (Δ27–82) is quite similar to the binding of another recognized BH3 domain protein Beclin 1. Only one important contact is missing in SOUL, that of an asparagine residue (Asp121 in Beclin 1) with Arg139 of Bcl-xL, which is conserved in all of the BH3 domain peptides studied so far. The absence of this interaction in SOUL might explain the relatively high dissociation constant we have observed for this complex.
Figure 5. Structural changes in the BH3 peptide of SOUL upon binding to Bcl-xL.
(A) Comparison of the peptide bound to Bcl-xL (Δ27–82) (blue) and the same sequence in the intact SOUL molecule (red). Eight amino acids change their conformation to extend the BH3 helix towards its C-terminus. The co-ordinates were superimposed by using the CCP4 suit of programs. Three charged amino acids that give specificity to the interaction are represented as stick models in the two conformations. (B) Superposition of the SOUL BH3 peptide Bcl-xL (Δ27–82) complex (the peptide is in yellow and Bcl-xL is in orange) and the same complex of Bcl-xL (in light blue) and the Beclin 1 BH3 peptide (in green). The PDB code of the model of the Beclin 1 peptide complex is 2P1L [13]. The interactions of the SOUL amino acids Arg163 and Asp170 (shown in Figure 4) are indicated with dotted lines. The aspartate residue is conserved in Beclin 1 and the equivalent of the arginine residue is a lysine. Note that the important contact of Arg139 of Bcl-xL with an aspartate residue in Beclin 1 (conserved in all BH3 domain–Bcl-xL complexes) is missing in the SOUL peptide.
DISCUSSION
The BH3-only members of the Bcl-2 protein family play a central role in the process that leads to programmed cell death or apoptosis. Their effect is due to inhibition through binding of their BH3 domain in the hydrophobic cleft of the anti-apoptotic members of the Bcl-2 family, such as Bcl-2, Bcl-xL or MCL-1. Indeed, one of the criteria to include a protein in this group is a demonstration of its interaction with one of the pro-survival proteins of the Bcl-2 family, in addition to showing that they have a cellular death-inducing activity. On the basis of these criteria, since the discovery of the first member of the family, BIK [44], many other members have been added to the list [9,10]. In many cases, the proteins seem to have additional functions besides their role in inducing cellular death. Although a very large number of complexes of BH3 peptides with pro-survival proteins of the Bcl-2 family are available [12,45], only one NMR structure of an intact BH3-only protein is known, BID [46,47].
Two different mutually non-excluding functions have been attributed to SOUL: haem transport [19] and a role in apoptosis as a BH3-only protein [20,21]. We have used NMR, SPR, UV spectroscopy and crystallography to explore both functions and, in addition, we have determined the three-dimensional structure of the protein. Human SOUL is a monomer with a fold which is quite different from that of the other BH3-only protein whose three-dimensional structure is known, BID, which is similar to Bcl-xL. BID contains eight α-helices: two central hydrophobic helices surrounded by six amphipathic helices with their hydrophilic face exposed to the solvent. The SOUL molecule is similar to murine p22HBP [33,34].
The interaction of human SOUL with haemin was explored by titrating the latter with the former and following the UV spectrum, using BSA as a control. In our experiments, we made sure that the protein used did not contain any traces of the histidine tag used for purification and that no imidazole, residual from the purification in the affinity columns, was present in the samples. We did not find any evidence of an interaction between haemin and SOUL. This result was confirmed by the 15N-1H HSQC NMR spectra of 15N-labelled SOUL in the presence of increasing amounts of haemin. We do not have an explanation as to why our results appear to be at variance with those reported for mouse SOUL [19]. The latter is reported to be a dimer in the absence of haem and a hexamer in the bound state, which is different from both the results of the present study and work on the related p22HBP proteins. It is also worth noting that the histidine residue that Sato et al. [19] found to be essential for haem binding does not seem to be involved in haem binding to p22HBP, which appears to bind this moiety through hydrophobic interactions and not metal co-ordination.
The interaction of a peptide spanning residues 147–172 of human SOUL with human Bcl-xL was also studied with 15N-1H HSQC NMR spectroscopy using two different forms of human Bcl-xL, the entire molecule lacking only the hydrophobic transmembrane domain after amino acid 209 and another truncated form lacking also amino acids 27–82 (Δ27–82). The results were comparable and indicated in both cases that there is an interaction of the 26-amino-acid SOUL peptide with Bcl-xL with a dissociation constant estimated to be 40–50 μM. These results were confirmed by SPR measurements and by preparing co-crystals of the complex and solving their three-dimensional structure. The new crystal structure revealed swapping of the first helix of the Bcl-xL dimer, a phenomenon associated with the presence of the Δ27–82 truncation. The amino acids participating in the interaction of the BH3 domain and protein were identified. The interactions are mostly hydrophobic, but a significant number of specific charged residue contacts were also found to be present.
When the SPR experiments performed to detect the interaction of the SOUL BH3 domain peptide were repeated using the entire SOUL molecule instead, no interaction was detected between ligand and analyte. This result might be explained by the fact that our findings predict a very drastic conformational change in the protein molecule to allow the portion of polypeptide chain involved in the contacts to adopt the conformation required for the contacts to be established. The last eight amino acids of the BH3 domain are not in a helical conformation in intact SOUL and, in addition, side chains that are important for the interaction point towards the interior of the molecule and are not available on the protein surface. Conformational changes in the anti-apoptotic members of the Bcl-2 family upon interaction with BH3 domains have been described [38], as well as changes in the interacting BH3 domains [41,46]. In addition, it has been shown that Bim, Bad and Bmf have intrinsically unstructured BH3 domains that undergo a localized conformational change upon binding to pro-survival Bcl-2 targets [48]. No important changes were found in Bcl-xL, but the changes in the BH3 domain are remarkable and are sufficient to explain why no interaction is observed with the intact SOUL molecule. These drastic modifications might require conditions that have not yet been found, but that should be explored further given the importance of this interaction in the functionality of the two proteins.
The mechanism of activation of BID, the prototype of BH3-only proteins, involves cleavage by caspase 8 in a region with the sequence LQTDG [46]. The second amino acid in the sequence can be an glutamate residue and the last can be any amino acid other than phenylalanine, glutamate, glutamine, lysine or arginine. This cleavage site is not present in SOUL, although the similar sequence LESDV spans residues 123–127 and is exposed to the solvent in the molecule, in the loop connecting strand D to E. However, an experiment with caspase 8, using human BID as a control, revealed that SOUL is not a substrate of this enzyme (results not shown).
The role of SOUL in inducing apoptosis has been documented, but up to now there has been no information at the molecular level on the mechanism through which this function is accomplished. We have shown that its BH3 domain interacts with a pro-survival member of the Bcl-2 family and thus we have provided new evidence that SOUL behaves like a novel member of the expanding family of BH3-only proteins.
Two very important questions remain unanswered: (i) the nature of the molecular alteration that intact SOUL must undergo for the interaction to take place, and (ii) the precise specificity of the interaction of the SOUL BH3 domain with different members of the Bcl-2 family.
Online data
AUTHOR CONTRIBUTION
Emmanuele Ambrosi produced the SOUL protein, prepared the crystals and solved the structure, and performed the related data analysis; Stefano Capaldi prepared Bcl-xL, crystallized the complex with the peptide and performed the NMR studies; Michele Bovi performed the SPR studies; Gianmaria Saccomani helped in the initial phases of SOUL protein expression, purification and crystallization; Massimiliano Perduca collected and processed the X-ray diffraction data; and Hugo Monaco directed the research and wrote the paper. All authors contributed to revising and improving the final version of the paper prior to publication.
ACKNOWLEDGEMENTS
We are grateful to Michael Assfalg and to Pete Simpson for their help in recording and interpreting the NMR spectra and to the staff of the ESRF (European Synchrotron Radiation Facility) in Grenoble (Proposal MX-805) for assistance during data collection.
FUNDING
This work was supported by the Fondazione Cassa di Risparmio di Verona, Vicenza, Belluno e Ancona. The NMR facility of the University of Verona is funded by the Fondazione Cariverona and the Department of Biotechnology. E.A. is recipient of a scholarship granted by the Fondazione Cassa di Risparmio di Padova e Rovigo, and G.S. was supported by Banca Popolare di Verona e Novara.
References
- 1.Youle R. J., Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 2.Kroemer G., Galluzzi L., Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 3.Metcalfe A. D., Hunter H. R., Bloor D. J., Lieberman B. A., Picton H. M., Leese H. J., Kimber S. J., Brison D. R. Expression of 11 members of the BCL-2 family of apoptosis regulatory molecules during human preimplantation embryo development and fragmentation. Mol. Reprod. Dev. 2004;68:35–50. doi: 10.1002/mrd.20055. [DOI] [PubMed] [Google Scholar]
- 4.Kelekar A., Thompson C. B. Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol. 1998;8:324–330. doi: 10.1016/s0962-8924(98)01321-x. [DOI] [PubMed] [Google Scholar]
- 5.Chao D. T., Korsmeyer S. J. BCL-2 family: regulators of cell death. Annu. Rev. Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 6.Rathmell J. C., Thompson C. B. The central effectors of cell death in the immune system. Annu. Rev. Immunol. 1999;17:781–828. doi: 10.1146/annurev.immunol.17.1.781. [DOI] [PubMed] [Google Scholar]
- 7.Vila M., Przedborski S. Targeting programmed cell death in neurodegenerative diseases. Nat. Rev. Neurosci. 2003;4:365–375. doi: 10.1038/nrn1100. [DOI] [PubMed] [Google Scholar]
- 8.Cotter T. G. Apoptosis and cancer: the genesis of a research field. Nat. Rev. Cancer. 2009;9:501–507. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- 9.Lomonosova E., Chinnadurai G. BH3-only proteins in apoptosis and beyond: an overview. Oncogene. 2008;27(Suppl. 1):S2–S19. doi: 10.1038/onc.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghiotto F., Fais F., Bruno S. BH3-only proteins: the death-puppeteer's wires. Cytometry A. 2010;77:11–21. doi: 10.1002/cyto.a.20819. [DOI] [PubMed] [Google Scholar]
- 11.Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 12.Moroy G., Martin E., Dejaegere A., Stote R. H. Molecular basis for Bcl-2 homology 3 domain recognition in the Bcl-2 protein family: identification of conserved hot spot interactions. J. Biol. Chem. 2009;284:17499–17511. doi: 10.1074/jbc.M805542200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oberstein A., Jeffrey P. D., Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 14.Lessene G., Czabotar P. E., Colman P. M. BCL-2 family antagonists for cancer therapy. Nat. Rev. Drug Discovery. 2008;7:989–1000. doi: 10.1038/nrd2658. [DOI] [PubMed] [Google Scholar]
- 15.Zylka M. J., Reppert S. M. Discovery of a putative heme-binding protein family (SOUL/HBP) by two-tissue suppression subtractive hybridization and database searches. Brain Res. Mol. Brain Res. 1999;74:175–181. doi: 10.1016/s0169-328x(99)00277-6. [DOI] [PubMed] [Google Scholar]
- 16.Bohn H., Winckler W. Isolation and characterization of five new soluble placental tissue proteins (PP22, PP23, PP24, PP25, PP26) Arch. Gynecol. Obstet. 1991;248:111–115. doi: 10.1007/BF02390087. [DOI] [PubMed] [Google Scholar]
- 17.Gianazza E., Wait R., Begum S., Eberini I., Campagnoli M., Labo' S., Galliano M. Mapping the 5–50 fraction of human amniotic fluid by 2-DE and ESI-MS. Proteomics Clin. Appl. 2007;1:167–175. doi: 10.1002/prca.200600543. [DOI] [PubMed] [Google Scholar]
- 18.Jacob Blackmon B., Dailey T. A., Lianchun X., Dailey H. A. Characterization of a human and mouse tetrapyrrole-binding protein. Arch. Biochem. Biophys. 2002;407:196–201. doi: 10.1016/s0003-9861(02)00471-x. [DOI] [PubMed] [Google Scholar]
- 19.Sato E., Sagami I., Uchida T., Sato A., Kitagawa T., Igarashi J., Shimizu T. SOUL in mouse eyes is a new hexameric heme-binding protein with characteristic optical absorption, resonance raman spectral and heme-binding properties. Biochemistry. 2004;43:14189–14198. doi: 10.1021/bi048742i. [DOI] [PubMed] [Google Scholar]
- 20.Szigeti A., Bellyei S., Gasz B., Boronkai A., Hocsak E., Minik O., Bognar Z., Varbiro G., Sumegi B., Gallyas F., Jr Induction of necrotic cell death and mitochondrial permeabilization by heme binding protein 2/SOUL. FEBS Lett. 2006;580:6447–6454. doi: 10.1016/j.febslet.2006.10.067. [DOI] [PubMed] [Google Scholar]
- 21.Szigeti A., Hocsak E., Rapolti E., Racz B., Boronkai A., Pozsgai E., Debreceni B., Bognar Z., Bellyei S., Sumegi B., Gallyas F., Jr Facilitation of mitochondrial outer and inner membrane permeabilization and cell death in oxidative stress by a novel Bcl-2 homology 3 domain protein. J. Biol. Chem. 2010;285:2140–2151. doi: 10.1074/jbc.M109.015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuzelová K., Mrhalová M., Hrkal Z. Kinetics of heme interaction with heme-binding proteins: the effect of heme aggregation state. Biochim. Biophys. Acta. 1997;1336:497–501. doi: 10.1016/s0304-4165(97)00062-7. [DOI] [PubMed] [Google Scholar]
- 23.Freire F., Romão M. J., Macedo A. L., Aveiro S. S., Goodfellow B. J., Carvalho A. L. Preliminary structural characterization of human SOUL, a haem-binding protein. Acta Crystallogr. Sect. F Struct. Biol. Crystal. Commun. 2009;65:723–726. doi: 10.1107/S174430910902291X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–767. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Sheldrick G. M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 26.Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P., Lohkamp B., Scott W. G., Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 31.Vagin A., Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 32.Lee E. F., Czabotar P. E., Smith B. J., Deshayes K., Zobel K., Colman P. M., Fairlie W. D. Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family. Cell Death Differ. 2007;14:1711–1713. doi: 10.1038/sj.cdd.4402178. [DOI] [PubMed] [Google Scholar]
- 33.Dias J. S., Macedo A. L., Ferreira G. C., Peterson F. C., Volkman B. F., Goodfellow B. J. The first structure from the SOUL/HBP family of heme-binding proteins, murine P22HBP. J. Biol. Chem. 2006;281:31553–31561. doi: 10.1074/jbc.M605988200. [DOI] [PubMed] [Google Scholar]
- 34.Gell D. A., Westman B. J., Gorman D., Liew C., Welch J. J., Weiss M. J., Mackay J. P. A novel haem-binding interface in the 22 kDa haem-binding protein p22HBP. J. Mol. Biol. 2006;362:287–297. doi: 10.1016/j.jmb.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Beaven G. H., Chen S. H., D'Albis A., Gratzer W. B. A spectroscopic study of the haemins human-serum-albumin system. Eur. J. Biochem. 1974;41:539–546. doi: 10.1111/j.1432-1033.1974.tb03295.x. [DOI] [PubMed] [Google Scholar]
- 36.Muchmore S. W., Sattler M., Liang H., Meadows R. P., Harlan J. E., Yoon H. S., Nettesheim D., Chang B. S., Thompson C. B., Wong S. L., et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 37.Kvansakul M., Yang H., Fairlie W. D., Czabotar P. E., Fischer S. F., Perugini M. A., Huang D. C., Colman P. M. Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain-swapped dimer that binds a highly selective subset of BH3-containing death ligands. Cell Death Differ. 2008;15:1564–1571. doi: 10.1038/cdd.2008.83. [DOI] [PubMed] [Google Scholar]
- 38.Lee E. F., Czabotar P. E., Yang H., Sleebs B. E., Lessene G., Colman P. M., Smith B. J., Fairlie W. D. Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands. J. Biol. Chem. 2009;284:30508–30517. doi: 10.1074/jbc.M109.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee E. F., Sadowsky J. D., Smith B. J., Czabotar P. E., Peterson-Kaufman K. J., Colman P. M., Gellman S. H., Fairlie W. D. High-resolution structural characterization of a helical α/β-peptide foldamer bound to the anti-apoptotic protein Bcl-xL. Angew. Chem. Int. Ed. Engl. 2009;48:4318–4322. doi: 10.1002/anie.200805761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng W., Huang S., Wu H., Zhang M. Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of beclin-1. J. Mol. Biol. 2007;372:223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- 41.Sattler M., Liang H., Nettesheim D., Meadows R. P., Harlan J. E., Eberstadt M., Yoon H. S., Shuker S. B., Chang B. S., Minn A. J., et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 42.Petros A. M., Nettesheim D. G., Wang Y., Olejniczak E. T., Meadows R. P., Mack J., Swift K., Matayoshi E. D., Zhang H., Thompson C. B., Fesik S. W. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis and biophysical studies. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X., Dai S., Zhu Y., Marrack P., Kappler J. W. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity. 2003;19:341–352. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 44.Boyd J. M., Gallo G. J., Elangovan B., Houghton A. B., Malstrom S., Avery B. J., Ebb R. G., Subramanian T., Chittenden T., Lutz R. J., et al. Bik, a novel death-inducing protein shares a distinct sequence motif with Bcl-2 family proteins and interacts with viral and cellular survival-promoting proteins. Oncogene. 1995;11:1921–1928. [PubMed] [Google Scholar]
- 45.Lama D., Sankararamakrishnan R. Anti-apoptotic Bcl-XL protein in complex with BH3 peptides of pro-apoptotic Bak, Bad and Bim proteins: comparative molecular dynamics simulations. Proteins. 2008;73:492–514. doi: 10.1002/prot.22075. [DOI] [PubMed] [Google Scholar]
- 46.Chou J. J., Li H., Salvesen G. S., Yuan J., Wagner G. Solution structure of BID, an intracellular amplifier of apoptotic signalling. Cell. 1999;96:615–624. doi: 10.1016/s0092-8674(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 47.McDonnell J. M., Fushman D., Milliman C. L., Korsmeyer S. J., Cowburn D. Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists. Cell. 1999;96:625–634. doi: 10.1016/s0092-8674(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 48.Hinds M.G, Smits C., Fredericks-Short R., Risk J.M, Bailey M., Huang D. C., Day C.L. Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ. 2007;14:128–136. doi: 10.1038/sj.cdd.4401934. [DOI] [PubMed] [Google Scholar]
- 49.Wallace A. C., Laskowski R. A., Thornton J. M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.