Abstract
A chemoenzymatic method was developed for the synthesis of macrocyclic peptides and glycopeptides. Sortase A was found to mediate either head to tail cyclization or oligomerization and then head to tail cyclization of peptides and glycopeptides, depending on the peptide length, to produce 15-mer or higher cyclic peptides and glycopeptides.
Cyclic peptides and glycopeptides show remarkable biological activities, probably due to their inherently relatively rigid and defined conformation and resistance to enzymatic degradation.1 Thus, cyclopeptides and cycloglycopeptides have been widely explored as antibiotics,2 antitumor agents,3 immunoregulative agents,4 and in vaccine development.5 For cyclic peptide and glycopeptide synthesis, chemical methods have been extensively investigated.6–8 However, to achieve effective and regiospecific reactions, orthogonal protecting tactics and often special peptide sequences, such as glycine-/proline-rich peptides, are required. To tackle this synthetic challenge, biological methods, e.g., the split-intein circular ligation of peptide and protein (SICLOPPS) technology,9 have been developed. Even though powerful, these methods are not straightforward for creating large cyclic peptide libraries. Chemoenzymatic approaches using enzyme-mediated peptide cyclization have also been established for the synthesis of cyclopeptides and cycloglycopeptides.10 These methods can be particularly useful owing to the unique properties of enzymatic reactions. However, promiscuous enzymes that can effectively promote peptide cyclization are necessary.
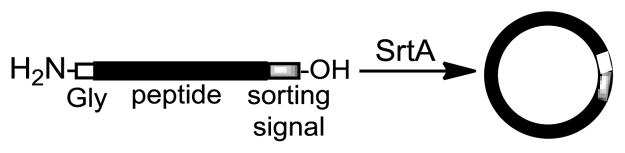
Sortases are a class of transpeptidases found in Gram-positive bacteria, which are responsible for anchoring surface proteins to bacterial cell walls by the so-called “sorting reaction”.11 Each sortase recognizes and reacts with a specific short peptide, known as “sorting signal”, near the C-terminus of the target protein to form a reactive protein-enzyme conjugate and then transfers the acyl group to the N-terminus of an oligoglycine motif of target pepteoglycans.12 For example, the sorting signal of sortase A (SrtA) of Staphylococcus aureus origin is a pentapeptide LPXTG, where X is variable. It has been demonstrated that sortases are quite substrate promiscuous, so sortase-mediated transpeptidation has been proved to be a powerful alternative to native chemical ligation.13–15 Accordingly, SrtA has been utilized to ligate and functionalize peptides and proteins,16–19 anchor proteins to solid surfaces20 and living cells,21–23 etc. Recently, we employed SrtA to effectively couple peptides, glycopeptides, and proteins with glycosylphosphatidylinositols (GPIs) for the synthesis of GPI-linked peptides, glycopeptides, and proteins.24–26 Based on these studies, we envisioned that SrtA might be utilized to perform intramolecular transpeptidation reaction to prepare cyclopeptides (Scheme 1), provided that the substrate peptide contains both the sorting signal and the proper peptide acceptor at its C- and N-termini, respectively. Indeed, Ploegh and co-workers found that SrtA could catalyze protein cyclization to produce moderate to excellent yields of cyclic proteins.27
Scheme 1.
Projected SrtA-mediated synthesis of cyclic peptides
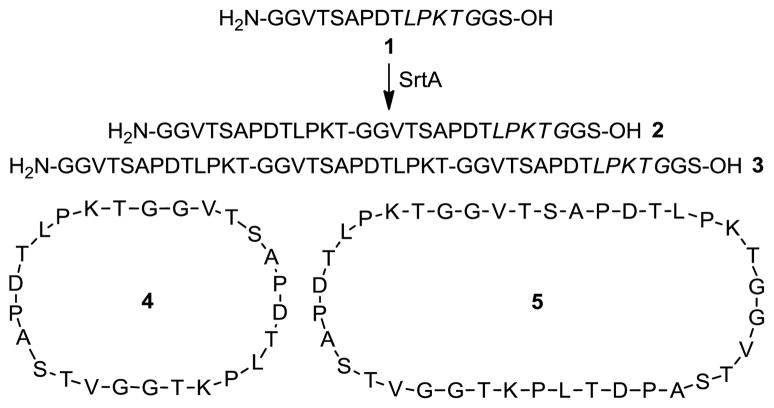
To test the above hypothesis, we prepared a peptide 1 having the sorting signal of SrtA and a diglycine motif at its C- and N-termini, respectively, and examined its reaction with SrtA. As previously reported, the enzymatic reaction was carried out in Tris-HCl buffer containing 0.5 mM of peptide 1 and 20 μM of SrtA,24–26 whereas the reaction was monitored and analyzed by HPLC and MS. After incubation at 37 °C for 20 h, the reaction was quenched and the products were isolated and characterized. We identified 4 major products from this reaction, including the linear dimer 2 (8%), linear trimer 3 (5%), cyclic dimer 4 (53%), and cyclic trimer 5 (10%) (Scheme 2), as well as some unreacted 1 (23%). No direct cyclization product of 1 was observed. All of the products were confirmed by MS.
Scheme 2.
SrtA-mediated oligomerization and clyclization of 1
These results suggest that SrtA first catalyzed peptide head to tail oligomerization to produce 2 and 3 and then promoted their intramolecular head to tail transpeptidation to result in cyclic peptides 4 and 5. That peptide 1 did not cyclize directly suggests that the cyclization reaction can occur only if the peptide reaches certain length. Furthermore, it appears that once the dimmeric and trimeric peptides were generated, their cyclization would prevail under the described conditions, as the linear oligomers 2 and 3, which are the cyclization precursors, were isolated in much lower yields than that of the cyclic counterparts.
After having proved that SrtA could indeed effect peptide cyclization and that this could be a practical method for cyclic peptide synthesis, we then examined how SrtA concentrations might affect the reaction and the product distribution. In these studies, we fixed the concentration of 1 at 0.5 mM, while the concentrations of SrtA were 10 to 60 μM (Table 1). Clearly, the enzyme concentration in the tested range had little influence on the overall reaction yield and product distribution, except for the small decrease and increase of the amount of recovered 1 and the yield of cyclic dimmer 4, respectively. Since the substrate peptide concentration was much higher than the concentration of SrtA, the results seemed surprising, but if we take into consideration that SrtA-catalyzed reactions are reversible,28 because the side product GGS can also function as a peptide acceptor, the results are understandable. Consequently, 1 did not completely convert to the products even in the presence of higher SrtA concentration, and the reactions were in equilibrium to give thermodynamically controlled products.
Table 1.
Product distribution of reactions of 1 in the presence of different concentrations of SrtA
| [SrtA] (μM) | 1a (%)Recovered | 2a (%) | 3a (%) | 4a (%) | 5a (%) |
|---|---|---|---|---|---|
| 10 | 25 | 8 | 5 | 53 | 9 |
| 20 | 23 | 8 | 5 | 53 | 10 |
| 40 | 21 | 7 | 6 | 57 | 9 |
| 60 | 19 | 7 | 7 | 59 | 8 |
% Yields derived from HPLC analysis of reaction mixtures.
We also probed the influence of substrate concentrations on the enzymatic reaction and the product distribution. It is expected that substrate concentrations should have a major influence on the ratios of intermolecular and intramolecular reactions, if both can occur and compete with each other. For these studies, we used relatively high concentration of SrtA (40 μM) in order to shorten the reaction time and facilitate the monitoring, and the substrate peptide concentrations were 0.1 to 10 mM. As disclosed in Table 2, with the increase of substrate concentration, the yields of linear oligomers 2 and 3 and recovered 1 increased, but the yields of cyclic peptides 4 and 5 decreased. The build-up of 1 and linear peptides 2 and 3 in the reaction mixture suggests again that the enzymatic reaction side product GGS may have a major influence on the reaction equilibrium and that with accumulation of GGS subsequent oligomerization and cyclization of 1, 2, and 3 were depressed. Another notable trend revealed in Table 2 is that the reactions gave elevated ratios of total trimeric to total dimeric products with the increase of the concentration of 1, e.g., from 14:70 to 18:40 at 0.1 and 10 mM of 1, respectively. This trend was expected because increased substrate concentrations should improve intermolecular reactions significantly.
Table 2.
Product distributions of SrtA-catalyzed reactions of various concentrations of 1
| [1] (mM) | 1a (%)Recovered | 2a (%) | 3a (%) | 4a (%) | 5a (%) |
|---|---|---|---|---|---|
| 0.10 | 16 | 3 | 5 | 67 | 9 |
| 0.25 | 15 | 3 | 6 | 67 | 8 |
| 0.50 | 21 | 7 | 6 | 57 | 9 |
| 1.00 | 24 | 10 | 7 | 51 | 9 |
| 2.50 | 34 | 17 | 8 | 37 | 8 |
| 5.00 | 35 | 24 | 11 | 24 | 7 |
| 10.00 | 42 | 28 | 13 | 12 | 5 |
Yields derived from HPLC analysis of reaction mixtures.
To determine the minimal size required for a peptide to realize direct head to tail cyclization, we then prepared peptides H2N-G3VTSAPDTLPKTGGS-OH (6), H2N-G4VTSAPDTLPKTGGS-OH (7), and H2N-G5VTSAPDTLPKTGGS-OH (8) that contained 17, 18, 19 amino acids, respectively. Subsequently, the peptides were treated with SrtA under the optimized conditions (0.5 mM peptide and 40 μM SrtA), while these reactions were monitored by HPLC and MALDI-TOF MS. For peptide 6, direct head to tail cyclization product was formed in only 6% yield, and its dimeric cyclic product was formed as the major product (54%) (Table 3). The results were very similar to that of 1. The reaction of peptide 7 offered direct cyclization product as the major product (42%), and the dimeric cyclic product was also observed in a substantial quantity (32%). However, the reaction of peptide 8 gave direct head to tail cyclization product overwhelmingly (79%) with little dimerization or trimerization. These studies clearly demonstrate that, for SrtA-catalyzed peptide cyclization, the minimal size of substrate peptides is an 11-mer, not counting the sorting signal (underlined). However, for an 11-mer peptide, the dimerization reaction can still compete with the cyclization reaction. Thus, to achieve more effective and nearly exclusive cyclization reaction, the substrate peptide should be a 12-mer or longer, which does not count the required sorting signal.
Table 3.
Product distributions of SrtA-catalyzed reactions of 6, 7, and 8
| Substrate peptide | 6 (n=3) | 7 (n=4) | 8 (n=5) |
|---|---|---|---|
| Recovered peptide (%)a | 24 | 18 | 10 |
| Linear dimer (%)a [(GnVTSAPDTLPKT)2GGS] |
7 | 4 | ndb |
| Linear trimer (%)a [(GnVTSAPDTLPKT)3GGS] |
ndb | ndb | ndb |
|
Cyclic monomer (%)a |
6 | 42 | 79 |
| Cyclic dimer (%)a |
54 | 32 | 4 |
| Cyclic trimer (%)a |
9 | 4 | ndb |
% Yields derived from HPLC analysis of reaction mixtures.
nd: not detectable.
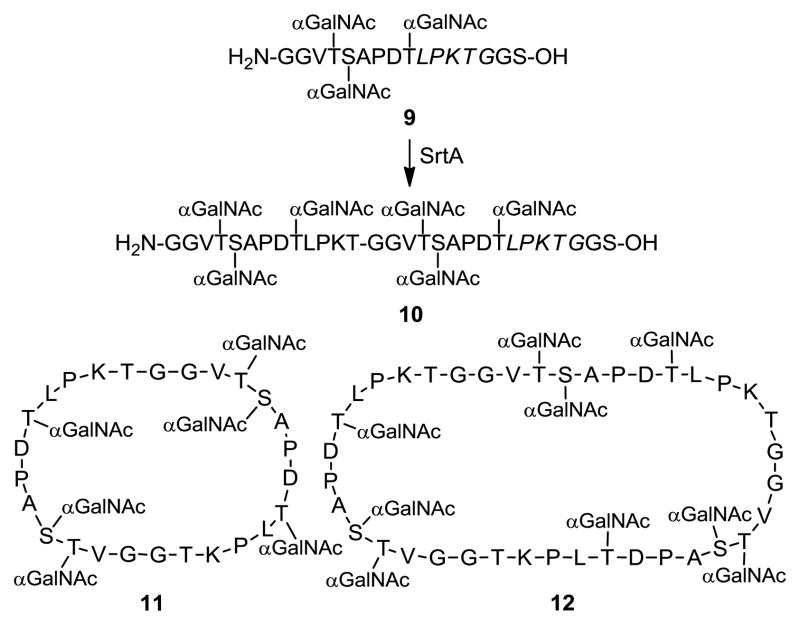
This unprecedented one-pot oligomerization and cyclization procedure was finally employed to perform cyclization of MUC1 glycopeptide 9 (Scheme 3). The reaction was carried out under optimized conditions using 0.25 mM of 9 and 20 μM of SrtA. After reaction at 37 °C for 20 h, the anticipated dimmeric cyclic glycopeptide 11 was obtained in 61% yield, along with a small amount of the dimeric linear product 10 and cyclic trimer 12, in 10% and 8% yields, respectively.
Scheme 3.
SrtA-catalyzed glycopeptide oligomerization and clyclization reactions
In brief, we have demonstrated that SrtA can be utilized to synthesize cyclic peptides and glycopeptides via intramolecular head to tail cyclization reactions of bifunctional peptides and glycopeptides. The peptide size was proved to be important for this application. For 11-mer and longer substrate peptides, not counting the sorting signal at the peptide C-terminus, SrtA promoted direct head to tail cyclization reactions to afford the corresponding 15-mer and larger cyclic peptides predominantly. It was further observed that a 12-mer peptide could cyclize more effectively than an 11-mer to produce a 16-mer cyclic peptide almost exclusively. However, when the substrate peptides were shorter than an 11-mer, SrtA catalyzed an intermolecular head to tail peptide oligomerization first and then an intramolecular head to tail cyclization of the resultant peptides and glycopeptides. This chemoenzymatic method is expected to be widely applicable to prepare macrocyclic peptides and glycopeptides containing 15 or more amino acids, either via SrtA-catalyzed direct cyclization in the case of large peptides/glycopeptides or via SrtA-catalyzed one-pot oligomerization and then cyclization in the case of short peptides/glycopeptides. Compared to existing chemoenzymatic syntheses, the new method is simple and potentially widely useful as SrtA is promiscuous and the only requirement of this synthetic method is that the substrate peptides/glycopeptides must have a short signal peptide and a Gly residue at their C- and N-termini, respectively, which is easily achievable. On the other hand, this requirement has determined that all products of the new synthetic method contain a LPXTG sequence. The impact of this sequence on the biological activity is an interesting issue to explore.
Supplementary Material
Acknowledgments
This work was supported by Wayne State University and in part by NSF (CHE-0554777) and NIH/NIGMS (R01GM090270). Wu thanks Dr. Benjamin Swarts for his help with the automatic peptide synthesizer.
Footnotes
This article is part of the ChemComm “Glycochemistry and glycobiology” web themed issue.
Electronic Supplementary Information (ESI) available: Experimental procedures and data; NMR and MS spectra of the intermediates and final products. See DOI: 10.1039/b000000x/
Notes and references
- 1.Gibson SE, Lecci C. Angew Chem Int Edit. 2006;45:1364–1377. doi: 10.1002/anie.200503428. [DOI] [PubMed] [Google Scholar]
- 2.Gentilucci L, De Marco R, Cerisoli L. Curr Pharm Design. 2010;16:3185–3203. doi: 10.2174/138161210793292555. [DOI] [PubMed] [Google Scholar]
- 3.Singh EK, Sellers RP, Alexander LD, McAlpine SR. Curr Opin Drug Disc. 2008;11:544–552. [PubMed] [Google Scholar]
- 4.McGeary RP, Fairlie DP. Curr Opin Drug Discov Devel. 1998;1:208–217. [PubMed] [Google Scholar]
- 5.Jeon I, Lee D, Krauss IJ, Danishefsky SJ. J Am Chem Soc. 2009;131:14337–14344. doi: 10.1021/ja9052625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambert JN, Mitchell JP, Roberts KD. J Chem Soc Perk T 1. 2001:471–484. [Google Scholar]
- 7.Davies JS. J Pept Sci. 2003;9:471–501. doi: 10.1002/psc.491. [DOI] [PubMed] [Google Scholar]
- 8.avassoli AN, Todd A, Benkovic, Stephen J. Nucleic Acids and Molecular Biology. 2005;16:293–305. [Google Scholar]
- 9.Cheriyan M, Perler FB. Adv Drug Del Rev. 2009;61:899–907. doi: 10.1016/j.addr.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 10.Grunewald J, Marahiel MA. Microbiol Mol Biol R. 2006;70:121. doi: 10.1128/MMBR.70.1.121-146.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 12.Clancy KW, Melvin JA, McCafferty DG. Biopolymers. 2010;94:385–396. doi: 10.1002/bip.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukiji S, Nagamune T. Chembiochem. 2009;10:787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- 14.Proft T. Biotechnol Lett. 2010;32:1–10. doi: 10.1007/s10529-009-0116-0. [DOI] [PubMed] [Google Scholar]
- 15.Pritz S. Mini-Rev Org Chem. 2008;5:47–52. [Google Scholar]
- 16.Mao HY, Hart SA, Schink A, Pollok BA. Journal of the American Chemical Society. 2004;126:2670–2671. doi: 10.1021/ja039915e. [DOI] [PubMed] [Google Scholar]
- 17.Parthasarathy R, Subramanian S, Boder ET. Bioconjugate Chem. 2007;18:469–476. doi: 10.1021/bc060339w. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto T, Sawamoto S, Sakamoto T, Tanaka T, Fukuda H, Kondo A. J Biotechnol. 2011;152:37–42. doi: 10.1016/j.jbiotec.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Pritz S, Kraetke O, Klose A, Klose J, Rothemund S, Fechner K, Bienert M, Beyermann M. Angew Chem Int Edit. 2008;47:3642–3645. doi: 10.1002/anie.200705718. [DOI] [PubMed] [Google Scholar]
- 20.Tomizaki KY, Usui K, Mihara H. Chembiochem. 2005;6:783–799. doi: 10.1002/cbic.200400232. [DOI] [PubMed] [Google Scholar]
- 21.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Nat Chem Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka T, Yamamoto T, Tsukiji S, Nagamune T. Chembiochem. 2008;9:802–807. doi: 10.1002/cbic.200700614. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto T, Nagamune T. Chem Commun. 2009:1022–1024. doi: 10.1039/b818792d. [DOI] [PubMed] [Google Scholar]
- 24.Guo XQ, Wang QL, Swarts BM, Guo ZW. J Am Chem Soc. 2009;131:9878. doi: 10.1021/ja903231v. [DOI] [PubMed] [Google Scholar]
- 25.Wu ZM, Guo XQ, Wang QL, Swarts BM, Guo ZW. J Am Chem Soc. 2010;132:1567–1571. doi: 10.1021/ja906611x. [DOI] [PubMed] [Google Scholar]
- 26.Wu ZM, Guo XQ, Guo ZW. Chem Commun. 2010;46:5773–5774. doi: 10.1039/c0cc00828a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antos JM, Popp MW, Ernst R, Chew GL, Spooner E, Ploegh HL. J Biol Chem. 2009;284:16028–16036. doi: 10.1074/jbc.M901752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamura Y, Hirakawa H, Yamaguchi S, Nagamune T. Chem Commun. 2011;47:4742–4744. doi: 10.1039/c0cc05334a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.