Abstract
Brown algae are the predominant primary producers in coastal habitats, and like land plants are subject to disease and parasitism. Eurychasma dicksonii is an abundant, and probably cosmopolitan, obligate biotrophic oomycete pathogen of marine brown algae. Oomycetes (or water moulds) are pathogenic or saprophytic non-photosynthetic Stramenopiles, mostly known for causing devastating agricultural and aquacultural diseases. Whilst molecular knowledge is restricted to crop pathogens, pathogenic oomycetes actually infect hosts from most eukaryotic lineages. Molecular evidence indicates that Eu. dicksonii belongs to the most early-branching oomycete clade known so far. Therefore Eu. dicksonii is of considerable interest due to its presumed environmental impact and phylogenetic position. Here we report the first large scale functional molecular data acquired on the most basal oomycete to date. 9873 unigenes, totalling over 3.5Mb of sequence data, were produced from Sanger-sequenced and pyrosequenced EST libraries of infected Ectocarpus siliculosus. 6787 unigenes (70%) were of algal origin, and 3086 (30%) oomycete origin. 57% of Eu. dicksonii sequences had no similarity to published sequence data, indicating that this dataset is largely unique. We were unable to positively identify sequences belonging to the RXLR and CRN groups of oomycete effectors identified in higher oomycetes, however we uncovered other unique pathogenicity factors. These included putative algal cell wall degrading enzymes, cell surface proteins, and cyclophilin-like proteins. A first look at the host response to infection has also revealed movement of the host nucleus to the site of infection as well as expression of genes responsible for strengthening the cell wall, and secretion of proteins such as protease inhibitors. We also found evidence of transcriptional reprogramming of E. siliculosus transposable elements and of a viral gene inserted in the host genome.
Introduction
Like all other living organisms, algae suffer from diseases, which may range from spectacular outbreaks in natural populations to significant losses in multibillion dollar crops such as nori [1]. As aquaculture continues to rise worldwide, and with algae considered as a sustainable biofuel source, pressure is mounting to design efficient disease control methods. More generally, parasites and pathogens are increasingly being considered of equal importance with predators for ecosystem functioning [2]. In aquatic as well as terrestrial environments, altered disease patterns in disturbed environments are blamed for sudden extinctions, regime shifts, and spreading of alien species. Likewise, algal pathogens exert a range of complex, profound, sometimes subtle, and often unexpected impacts in aquatic ecosytems. These range from host generation shifts to changes in biogeochemical cycling and atmosphere chemistry (e.g. [3]).
Despite mounting recognition of their importance, molecular knowledge on algal pathogens is hitherto restricted to viruses (reviewed in [1]). Likewise, the molecular characterisation of algal immune responses is in its infancy [4]. To help address these fundamental and ecological questions, we have developed a laboratory-controlled pathosystem involving the genome model seaweed Ectocarpus siliculosus [5] and the oomycete pathogen Eurychasma dicksonii [6], [7].
Eu. dicksonii is the most common and widespread eukaryotic pathogen of marine algae [8]. It occurs in all cold and temperate seas worldwide [9]. In culture, it infects virtually all species of brown algae tested (more than 40, [6]), including representatives of all major orders of this phylum. As brown algae are the predominant primary producers of temperate and polar rocky shores, and Eu dicksonii occurs in frequent epidemics [10], we infer that this pathogen contributes to shaping natural algal populations, therefore profoundly impacting ecosystem functioning [11].
Importantly, molecular taxonomy unambiguously designates Eu. dicksonii as the most early-branching clade within the oomycete lineage [7], [12]. Oomycetes (or water moulds) are classified within the group of Stramenopiles together with diatoms, golden-brown and brown algae [13]. They are secondarily non-photosynthetic organisms which exhibit either pathogenic or saprophytic lifestyles [14], [15]. In fact, many of the most devastating agricultural and aquacultural pathogens belong to the oomycetes [16], [17], [18], [19]. For example, Phytophthora infestans, the causal agent of potato late blight, is responsible for annual crop losses and pesticide costs exceeding £5 billion worldwide [20]. Many other species cause significant environmental damage, especially when recently introduced (e.g. Phytophthora ramorum). Therefore, the best-studied oomycete species are phylogenetically highly-derived plant pathogens, of which five fully sequenced genomes are hitherto available. However, pathogenic oomycetes are taxonomically much more diverse, and infect a remarkable palette of hosts ranging from marine algae, crustaceans, plants, nematodes, fungi, insects, to fishes and mammals [18]. From a physiological standpoint, Eu. dicksonii is typical of most basal oomycetes, inasmuch as it is an obligate biotroph, exclusively growing and propagating within a living host. After encysting at the surface of an algal cell, infectious spores inject their protoplasm into the host cytoplasm, and develop into a multinucleate coenocytic intracellular thallus. After having progressively filled the infected algal cell, the latter differentiates into a sporangium that releases new infectious zoospores [7]. The extremely broad host range of Eu. dicksonii is unusual among biotrophic pathogens, and may reflect a low level of specialization, suggesting that it may have retained ancestral infection mechanisms.
In summary, Eu. dicksonii is of considerable experimental interest because of its basal phylogenetic position, its presumed environmental impact, and amenability to molecular studies. Here, we set out to investigate the physiology of the E. siliculosus – Eu dicksonii host-pathogen interaction by constructing cDNA libraries of axenic cultures. We aimed to address what pathogenicity determinants and strategies Eu. dicksonii uses to infect E. siliculosus and what are the consequences of infection on algal physiology. Overall, we describe here a very original dataset that underlines both the taxonomical distance between Eu dicksonii and better-studied oomycetes, and the specialised marine lifestyle of this pathogen.
Materials and Methods
Biological material and microscopy
Axenic cultures of the fully sequenced Ectocarpus siliculosus strain CCAP 1310/4 and of Eurychasma dicksonii CCAP 4018/1 were obtained as described by [21]. They were maintained in half strength Provasoli medium [22] at 15°C, under illumination with daylight type fluorescent lamps at a 2–10 mE.m−2.s−1 irradiance and a 12 h photoperiod, as described by [6]. Being an obligate biotroph, Eu. dicksonii was maintained in co-culture with its E. siliculosus host and transferred into fresh medium every second week, together with some new uninfected algal host. The Eu. dicksonii CCAP 4018/1 strain was originally established from a single developing parasitic thallus, which was propagated into a clonal healthy algal host. For microscopy, infected algae were fixed for 45 min with 4% paraformaldehyde in microtubule-stabilising buffer [23], dipped into methanol for 30 s, and transferred into a DAPI solution (10 mg/mL in sterile seawater) for 5 minutes and observed under an epifluoresence microscope equipped with the following filter set: L365 FT 395, LP 420.
Construction of cDNA libraries and sequencing strategy
Fifty E. siliculosus cells each containing a developing intracellular Eu. dicksonii thallus were dissected under the stereomicroscope using a glass Pasteur pipette, and transferred into RNALater®. Total RNAs were extracted, subjected to reverse-transcription using a poly-dT oligonucleotide, PCR-amplified, and cloned directionally into a pBluescript II sk+ vector (Vertis Biotechnologie AG, Germany). After plating, 3000 colonies were robot-picked. Their insert was further sequenced from both ends using Sanger technology (Genoscope, France).
In parallel, two densely infected E. siliculosus cultures (∼20 mg FW each) were harvested in RNALater®, without prior dissection. The “young” culture (4 weeks after inoculation) contained all Eu. dicksonii development stages, including young intracellular thalli, mature dehiscent sporangia and encysted spores at the surface of algal cells. In contrast, the “old” culture (2 months after inoculation) predominantly contained mature dehiscent sporangia and encysted spores. Total RNA was extracted and subjected to cDNA synthesis as above. For each sample, 50,000 sequence reads were obtained using 454 pyrosequencing.
Bioinformatic analysis
Data assembly
The sequence reads were first assembled separately for each individual dataset using CAP3 (default parameters) [24], and then further assembled, also using CAP3, (default parameters, without resorting to quality file) into the final hybrid assembly discussed throughout the manuscript. Unigenes containing poor quality sequence reads (manual assessment of quality) were removed from the final dataset. A total of 5889 unassembled EST sequences (dissected library) were submitted to the EST db at EMBL and were assigned accession numbers FR839767-FR845655. Pyrosequencing reads (old culture and young culture libraries) were submitted to the EMBL short-read assembly (SRA) database. Sequences from each of the three databases were assembled into a total of 9847 unigenes.
Sequence Analysis
Unigenes were mapped to the E. siliculosus nuclear, chloroplastic and mitochondrial genomes with the use of GenomeThreader [25] to produce the ‘host’ dataset. GenomeThreader was applied with default parameters, except for the minimum alignment length and percent identity, which were set to 90% and 95% resp. Non-mapping unigenes were assigned to Eu. dicksonii after blastn analysis (98% to 100% sequence similarity) to remove potential contaminants, producing the ‘pathogen’ dataset. Open reading frames were predicted with OrfPredictor [26], based on a blastx analysis run against the NCBI Genbank non- redundant (nr) database and the E. siliculosus genome database (http://bioinformatics.psb.ugent.be/genomes/view/Ectocarpus-siliculosus). Sequence similarity searches were carried out using local blast tools [27], the NCBI nr database (28-07-2010 version) and several predicted proteomes of fully sequenced Eukaryotes. These include the proteomes of six oomycete species sequenced to date: Phytophthora infestans, [28]; Phytophthora sojae, [14]; Phytophthora ramorum, [14]; Pythium ultimum, [29]; Hyaloperonospora arabidopsidis [30] and Saprolegnia parasitica (draft genome, publically available at; (www.broadinstitute.org/annotation/genome/Saprolegnia_parasitica/MultiHome.html). Additional proteomes included were: the model brown alga and host species used in the current study, Ectocarpus siliculosus; the marine centric diatom Thalassiosira pseudonana; the marine pennate diatom Phaeodactylum tricornutum; the coccolith-bearing haptophyte Emiliania huxleyi; the Apicomplexan malaria parasite Plasmodium falciparum; the model plant Arabidopsis thaliana; the amoeboflagellate Naegleria gruberi; the amphibian pathogenic chytridiomycete fungus Batrachochytrium dendrobatidis and the plant pathogenic ascomycete fungus Mycosphaerella fijiensis. Predicted protein domains were identified using a standalone InterProScan program [31] (default settings) and the NCBI conserved domain database [32]. E. siliculosus transposable elements were identifed and named according to [5] and [33]. Their sequences were retrieved from the following URL: http://urgi.versailles.inra.fr/Data/Transposable-elements/Ectocarpus.
The Emboss tool infoseq [34] was used to generate statistics for host and pathogen datasets independently.
Signal peptide predictions were carried out using SignalP version 3.0 [35] with a hidden Markov model probability cutoff of 0.6, and 1160 candidate secreted proteins were obtained.
Effector protein motif predictions
Searches for RXLR and CRN motifs were carried out according to the methods of [36]. Briefly, string searches were carried out using the expression RxLR –x (1,40)- [ED] [ED] [KR] using custom build python scripts. This string search was also performed just with the RxLR expression, and was performed on all sequences, as well as those with predicted signal peptides. String searches were also performed with both the CRN motifs from Phytophthora species and Pythium ultimum. HMM profile searches for all these motifs were also carried out using the HMMer 2.2 package as described in [36].
Multiple alignments were conducted using the program CLUSTALW [37] (default parameters) and visualised using Geneious (version 4.8; [38]). Distance trees were constructed using Geneious with neighbour joining algorithms using 1000 bootstrap replications.
Results
Cytological observations of Eu. dicksonii infected E. siliculosus
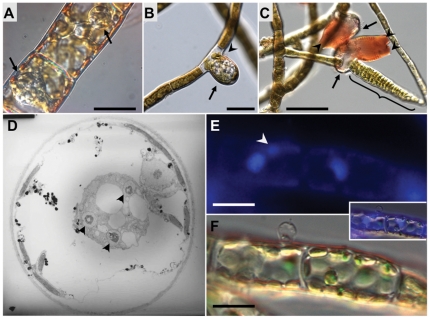
The morphological development and ultrastructural cytology of Eu. dicksonii infecting both E. siliculosus and the related filamentous phaeophyte alga Pylaiella littoralis has been investigated previously [7]. Infection is initated by secondary cysts, which attach to the surface of the host cell. Upon germination, the algal cell wall is breached, and cytoplasm from the pathogenic cell is transfered into the host cytoplasm. The initial phase of parasite development as a non-walled thallus is reminiscent of fungal pathogens of higher plants, such as Plasmodiophora brassicae and Olpidium brassicae (Figure 1A, [7]). The Eu. dicksonii thallus develops as a spherical intracellular syncitium, progressively filling the individual algal cell and ultimately causing its hypertrophy (Figure 1B). The sporangium develops by differentiation of the syncitium that releases new infectious zoospores (Figure 1C). The Eu. dicksonii thallus never propagates to other cells within the algal filament. During infection, the host nucleus and Golgi apparatus are closely associated with the pathogen thallus (Figure 1D–F).
Figure 1. Co-localisation of E. siliculosus nuclei and Eu. dicksonii thalli.
A, B, C: Successive stages of Ectocarpus infection by Eurychasma dicksonii. The Eu. dicksonii thallus develops as a spherical intracellular syncitium (A, arrows), progressively filling any individually infected algal cell, ultimately causing its hypertrophy (B, arrow). The syncitium then differentiates into a sporangium (C, arrows, Congo red staining) that releases new infectious spores into the medium via its apical apertures (C, arrowheads). The original infectious spore at the surface of the infected Ectocarpus cell is visible in B (arrowhead). The structure designated with a brace in C is an algal plurilocular sporangium containing parthenogenetic zoospores. Bars: A, B: 20 mm; C: 50 mm. D. Eu. dicksonii syncyitium (3 nuclei visible, arrowheads) developing next to the nucleus of a highly vacuolated E. siliculosus cell (strain CCAP 1310/299). Picture: courtesy of Dr S. Sekimoto. E & F. DAPI staining (E) and corresponding Nomarski image (F) of the microscopic sexual development stage (gametophyte) of the kelp Macrocystis pyrifera infected with Eurychasma dicksonii. The left hand side algal cell contains a very young Eurychasma thallus (one nucleus visible, white arrowhead) derived from the protoplasm of the empty spore visible at its surface. Insert: merged images. Scale bar: 10 um.
cDNA library quality control and statistics
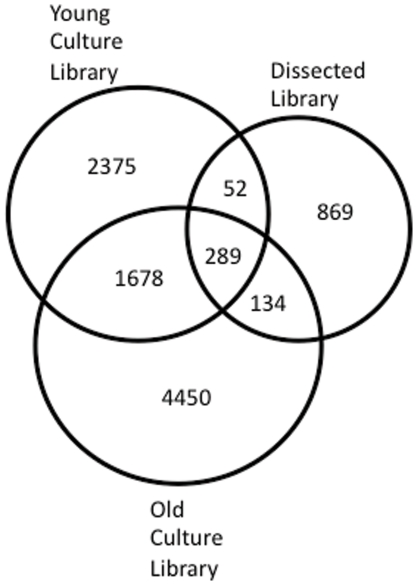
3000 clones from microdissected Eu. dicksonii-infected E. siliculosus cells were Sanger-sequenced (Genoscope, France) and assembled into contigs (CAP3). This resulted in the identification of 1424 unigenes. Pyrosequencing reads, from both the ‘young culture’ and ‘old culture,’ were assembled at the NERC National Molecular Genetics Facility (Liverpool, UK), resulting in two data sets of 4778 unigenes (young culture) and 6840 unigenes (old culture). Unigenes from these three datasets were combined to produce a single dataset, and assembled to produce 9873 unigenes totalling over 3.5 Mb of sequence data (Table 1). 28 sequences (15 from the young culture library, 12 from the old culture library, and 1 singlet from the dissected library) were discarded on the basis of poor sequence (containing regions of more than 20 bases called as Ns). 38 singlets, originating from the dissected library, were also removed on the basis of containing less than 90 nt of readable sequence. The final total hybrid assembly (discussed throughout the text) of 9873 unigenes therefore contained 9658 contigs and 215 singlets. The total 9873 unigene dataset was made up of 295 contigs with representatives from all three libraries, 1680 contigs with representatives in both the young culture and old culture libraries, 52 contigs with representatives in both the young culture and dissected libraries, 136 contigs with representatives in both the dissected and old culture library and 7710 unigenes that were only present in one of the three libraries (Figure 2).
Table 1. Assembly statistics, individual cDNA libraries and final hybrid assembly.
| unigenes (average length; max length) | Ectocarpus: Eurychasma ratio | Non-redundant sequence information | |
| Dissected library | 1432 (722 nts, 2085 nts) | 30%: 70% | 833 kb |
| “Young” culture | 4780 (273 nts, 3645 nts) | 79%: 21% | 1.4 Mb |
| “Old” culture | 6842 (298 nts, 2807 nts) | 74%: 26% | 2.1 Mb |
| Hybrid assembly | 9873 (350 nts, 5341 nts) | 69%: 31% | > 3.5 Mb |
Figure 2. Venn Diagram showing numbers of overlapping sequences from each library within the total dataset.
Unigenes were mapped to the E. siliculosus genome as described in [5]. 6787 unigenes were mapped to the E. siliculosus genome and, therefore, predicted to be of host origin. 3152 unigenes did not map to the E. siliculosus genome. 66 poor quality or short sequences were excluded, leaving 3086 sequences ascribed to Eu. dicksonii. This resulted in the creation of two datasets, ‘host sequences’ containing 6787 unigenes (Figure S1), and ‘pathogen sequences’ (Figure S2) containing 3086 unigenes. Strikingly, unigenes from the host dataset have a mean GC of 51.9% whereas those from the pathogen dataset have a much lower mean GC of 40.5% (Table 2).
Table 2. Characteristics of host and pathogen datasets.
| Total Dataset | Host sequences | Pathogen sequences | |
| Total unigenes | 9873 | 6787 | 3086 |
| Longest contig (nt) | 5341 | 4513 | 5341 |
| Shortest contig (nt) | 90 | 95 | 92 |
| Mean contig length (nt) | 355 | 325 | 409 |
| Mean GC (%) | 48.2 | 51.9 | 40.5* |
| Predicted secreted proteins | 1190 (11.9%) | 839 (12.4%) | 351 (11.6%) |
Highly expressed transcripts
Major up-regulation of Ectocarpus transposable elements
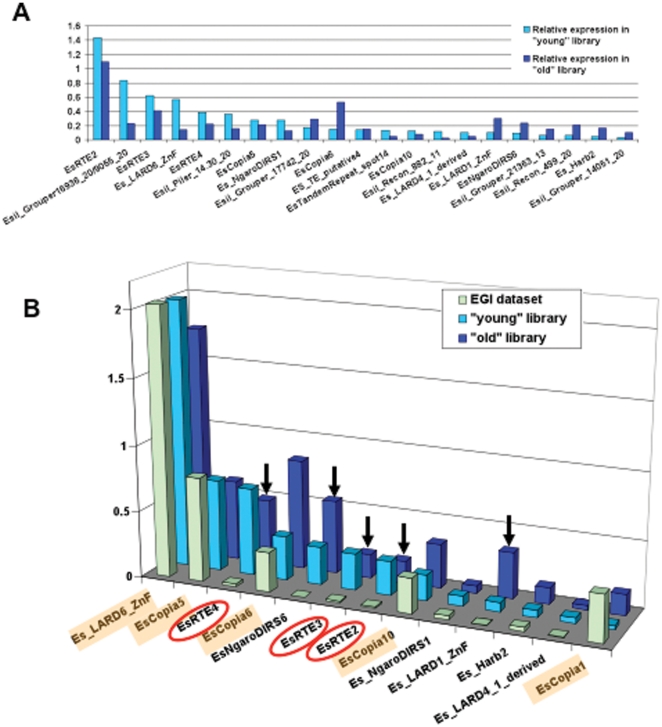
We used the number of sequence reads normalised over contig length as a proxy to evaluate relative gene expression levels in both the “young” and “old” pyrosequenced libraries. Unsurprisingly, the vast majority of the most abundantly expressed transcripts are host housekeeping genes. However, 16 (resp. 25) of the top 150 most expressed unigenes in the young and old libraries are E. siliculosus transposable elements (TEs). E. siliculosus TEs identified among the top most 150 highly expressed contigs in at least one library were extracted, and their expression level is plotted on Figure 3A. We further compared the expression level of TEs annotated by [5] between the two Eu. dicksoniii-infected libraries and the Ectocarpus genome initiative EST collection (Figure 3B). The latter mostly encompasses distinct development stages of unstressed Ectocarpus. Whereas pyrosequencing read counts and EST numbers are not directly comparable between the EGI dataset [5] and ours, differential expression of some TEs is nevertheless clearly discernible. Indeed, the five most expressed TEs identified by Cock and co-authors are also well represented in both Eu. dicksonii-infected libraries. However, the retroelements RTE2, RTE3 and RTE4, the LTR element NgaroDIRS6, and the LARD element EsLARD1_ZnF, virtually silent in unstressed conditions, appear strongly induced in the “young” and “old” pyrosequenced libraries. Interestingly, RTE2, RTE3 and RTE4 also belong to the top 10 most abundant repeats in the E. siliculosus genome, suggesting stress-induced transposing activity in the E. siliculosus genome [33].
Figure 3. Over-representation of Ectocarpus transposable elements (TEs) in Eurychasma-infected libraries.
A. Relative expression of Ectocarpus TEs in the pyrosequenced “young” and “old” Eurychasma-infected libraries. Total read counts were normalised over contig length in order to identify the most expressed unigenes in each library. Expression values are given for all TEs belonging to the top 150 most highly expressed unigenes in at least one library. If several contigs matched the same TE in a given library, their relative expression levels were summed. TE nomenclature is as per Maumus (2009). B. Differential expression pattern of Ectocarpus TEs between the pyrosequenced “young” and “old” Eurychasma-infected libraries vs. the Ectocarpus Genome Initiative EST collection (“EGI dataset”, Cock et al, 2010). For each library, sequence read counts were normalised over the genome coverage of each TE in order to control for the leaky transcription hypothesis. The 5 most highly expressed TEs in the EGI dataset are highlighted in orange; those circled in red belong to the 10 most abundant repeated elements identified in the Ectocarpus genome. Arrows point to TEs highly induced in the “young” and “old” infected cultures.
Highly expressed Eurychasma transcripts
The 10 most abundant sequences within the dissected library are of pathogen origin, with the exception of a conserved unknown E. siliculosus protein, demonstrating the efficacy of microdissection for enriching the sample in pathogen RNAs (Table 3). The majority of the most abundant sequences are housekeeping and ribosomal genes. However, three unigenes of Eu. dicksonii origin are highly abundant in this library and are proteins with unknown functions (Table 3). Contig500203 encodes a methionine and lysine rich unknown protein. Contig500580 encodes a predicted protein with similarity to a group of unknown oomycete proteins. These proteins are a group of at least 8 proteins present in the Saprolegnia parasitica genome, but absent in the genomes of other sequenced oomycetes. These appear to be uncharacterised cytoplasmic proteins, which may be ancestral proteins lost from higher oomycetes. Contig50031 appears to encode a lysine and alanine rich protein or protein fragment, with no similarity to known proteins.
Table 3. Top 10 most highly expressed unigenes in the Eu. dicksonii-enriched microdissected library.
| Contig | Length (bp) | Members | Origin | Hybrid assembly | Top blastn/x hit custom database* or Genbank nr |
| 500130 | 93 | 80 | Pathogen | In contig2041 | PITG_00179 40S ribosomal protein S12 P. infestans 6e-44 |
| 500773 | 1473 | 65 | Pathogen | In contig2006 | XP_002908783.1 beta 1 tubulin P. infestans e = 0 |
| 500203 | 831 | 49 | Pathogen | In contig1846 | No hit |
| 5001047 | 1701 | 47 | Pathogen | 5001047 | 18S rRNA E. dicksonii |
| 500580 | 1176 | 43 | Pathogen | In contig1954 | SPRG_15343T0 predicted protein S. parasitica 4e-10 |
| 500569 | 756 | 40 | Pathogen | In contig1953 | XP_001809251 similar to I-connectin T. castaneum 7e-12 |
| 500393 | 1442 | 33 | Pathogen | 500393 | 28S rRNA E. dicksonii |
| 500904 | 937 | 28 | Pathogen | In contig2041 | PITG_00179 40S ribosomal protein S12 P. infestans 6e-44 |
| 500588 | 1258 | 26 | Host | 500588 | Esi0110_0040 conserved hypothetical protein |
| 50031 | 713 | 24 | Pathogen | In contig1807 | No hit |
*custom database made up of proteomes described in Figure 5.
Functional Annotation
The 9873 unigene set was annotated by comparison to the NCBI non-redundant (nr) protein database (28-07-2010 version) and the E. siliculosus genome database (http://bioinformatics.psb.ugent.be/webtools/bogas/overview/Ectsi; [5] using BLAST analyses [25]. Overall, approximately 20% of the sequences in the total dataset showed similarity to previously described genes in the NCBI nr protein database, using an e value cutoff of <1e-05, indicating that this is largely a unique dataset.
Functional annotation of host genes
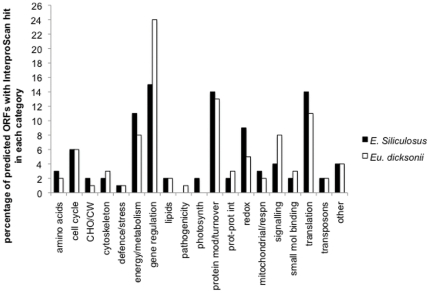
The E. siliculosus unigene set (host sequences) was initially annotated by direct comparison to the E siliculosus genome sequence [5]. Blastx analysis (expectation value <1e -05) of this dataset to the E. siliculosus predicted proteome resulted in 1299 (19.2%) of sequences matching predicted E siliculosus proteins. 46% (3163) have a significant blastn hit to genomic mRNA sequences, which include the 3′ UTRs, with the remainder of the sequences hitting mitochondrial, chloroplastic or intergenic sequences. Protein sequences were predicted using ORFPredictor [26] using the E. siliculosus blastx results and checked for agreement with the E. siliculosus genome database. Predicted protein sequences were analysed for the presence of an N-terminal signal sequence, using both Hidden Markov Models and Neural Network algorithms in SignalP [35]. 839 (12.4%) of the host unigenes were predicted to contain a signal peptide. Predicted proteins were also analysed for functional domains using InterProScan [31]. 822 unigenes (12%) of the dataset had one or more hits to functional domains using InterProScan. All the 822 predicted ORFs with InterproScan hits were within the 1299 sequences predicted to match protein-coding regions of the E. siliculosus genome. 63% of the unigenes that are derived from E. siliculosus protein coding regions, therefore, contain functional domains as identified by InterproScan. Functional annotation of host genes was checked with the HECTAR predictions for full-length sequences in the E. siliculosus genome database [5]. Predicted ORFs in this dataset were assigned to functional categories based on GO annotations of the InterProScan/HECTAR predictions (Figure 4). The largest category was gene regulatory proteins (15% of the total), including predicted transcription factors, translation factors, DNA and RNA binding proteins and proteins containing one or more coiled coil domains. 14% of predicted ORFs were classified as involved in translation, with approximately two thirds of these being ribosomal proteins. 14% of predicted proteins were also classified as being involved in protein modification, targeting or turnover including chaperones, and proteins involved in ubiquitination and proteolysis.
Figure 4. Classification of unigenes based on identification of functional domains using InterproScan.
Functional categories assigned to the 822 E. siliculosus (host) and 1397 Eu. dicksonii (pathogen) predicted ORFs (12% and 43% of total unigenes, respectively) with an Interpro hit. Numbers assigned to each category are percentages of the 822 and 1397 predicted ORFs. Amino acids includes amino acid synthesis and metabolism; cell cycle genes also include mitosis, DNA and RNA synthetic genes; CW/CHO includes cell wall biosynthesis and carbohydrate synthesis and metabolism; cytoskeleton includes transcripts involved in cytoskeletal rearrangements; defence and stress includes transcripts involved in general and specific defence or stress responses; energy/metabolism includes transcripts involved in energy production and cellular metabolism but does not include those involved specifically in respiration; gene regulation includes transcripts involved in DNA and RNA binding, transcription factors and transcripts with coiled-coil domains, lipids includes both lipid biosynthesis and metabolism; pathogenicity includes transcripts with a predicted function in pathogenicity or virulence, photosynth includes photosynthetic machinery; protein mod/turnover includes transcripts involved in protein folding, targeting, modification and degradation; prot-prot int includes transcripts predicted to be involved in protein-protein interactions; redox includes transcripts involved in homeostasis, detoxification and those classified as redox antioxidants; mitochondrial/respn transcripts are predicted to be mitochondrial and/or involved in respiration; signalling includes transcripts with a predicted role in signalling or signal transduction; small mol binding includes those transcripts which bind small molecules; translation includes those transcripts with a role in translation, including ribosomal proteins; transposons includes sequences both of transposons and those of putative retroviral origin; transcripts which could not be assigned to any one of the above categories are classified in the section termed other.
Functional annotation of pathogen genes
The Eu. dicksonii unigene dataset (containing sequences that did not map to the host genome) was analysed and ORFs were predicted as described above for E. siliculosus.
InterProScan analysis was performed on Eu. dicksonii predicted ORFs, in the same manner as on the host dataset. 1397 of the 3086 (44%) pathogen unigenes were predicted to contain one or more functional domain using InterProScan. As seen with the host dataset, the largest class of proteins were those predicted to contain gene regulatory domains, including those with coiled-coil domains. This class represented almost a quarter (24%) of all pathogen sequences with functional domains (Figure 4). 8% of sequences were classified as being involved in signalling in the pathogen dataset, compared to just 4% of host sequences. Interestingly, proteins predicted to be involved in pathogenicity and suppression of immunity were also identified in the pathogen dataset, but were absent from host sequences. A higher percentage of proteins classified as having a role in redox reactions, including those involved in detoxification and homeostasis, were identified within the host dataset (9%) in comparison to the pathogen dataset (5%). It is anticipated that several of these proteins function in cellular defence mechanisms.
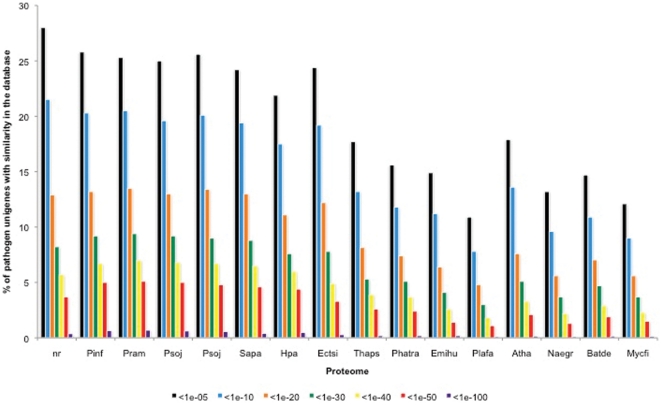
In addition to blastx similarity searches with the NCBI nr database, fifteen eukaryotic proteomes (listed in the methods section) were added to the analysis (Figure 5). Up to 25% of the pathogen sequences showed significant similarity (< 1e-05) to proteins from sequenced oomycetes. Interestingly about the same number (28% at <1e -05) showed significant similarity to proteins in the NCBI nr dataset and to the algal host E. siliculosus predicted proteome (24.4% at 1e-05), indicating that this dataset is largely made up of unique, uncharacterised sequences. As expected, the Eu. dicksonii dataset showed the most similarity to the oomycete proteomes, and to E. siliculosus. A lesser degree of similarity was seen with the diatom, haptophyta and apicomplexa proteomes. Surprisingly, a higher level of similarity (up to 18%, at 1 e-05) was observed between the pathogen unigenes and the proteome of the model plant Arabidopsis thaliana, than with some of the chromalveolata, such as Plasmodium falciparum with which there was only 11% similarity (at 1e-05). Likewise, up to 15% sequence similarity was seen between Eu. dicksonii unigenes and distantly related organisms such as the amoeboflagellate Naegleria gruberi and true fungi (Figure 5).
Figure 5. Comparison of Eu. dicksonii sequences to the non-redundant protein database at NCBI and proteomes of fully sequenced organisms.
Unigenes were blasted using the BLASTX algorithm to the nr database and to the proteomes of Phytophthora infestans (Pinf), Phytophthora ramorum (Pram), Phytophthora sojae (Psoj), Pythium ultimum (Pult), Saprolegnia parasitica (Sapa), Hyaloperonospora arabidopsidis (Hpa), Ectocarpus siliculosus (Ectsi), Thalassisosira pseudonana (Thaps), Phaeodactylum triconutum (Phatr), Emiliana huxleyi (Emilhu), Plasmodium falciparum (Plafa), Arabidopsis thaliana (Atha), Naegleria gruberi (Naegr), Batrachochytrium dendrobatidis (Batde) and Mycosphaerella fijiensis (Mycfi). For each E-value class the percentage of unigenes showing similarity is indicated.
351 (11.6%) of the pathogen sequences were predicted to contain a signal peptide. 179 of these also had one or more hits to InterProScan domains, including non-specific ‘seg’ hits. At least 30 of those containing known domains were in fact membrane proteins, or fragments of membrane proteins that were likely picked up with the SignalP algorithms because they are not full length. In the majority of cases, it is not possible to conclude whether our contigs represent full-length sequences with real signal peptides, or simply fragments of proteins with hydrophobic or transmembrane spanning regions. However, 10 sequences from the Eu. dicksonii dataset are predicted to encode full-length, unknown, secreted proteins that do not contain transmembrane domains (Table 4). These predicted proteins either have no sequence similarity to known proteins or domains, or have an Interpro hit to the non-specific ‘seg’ database. There is no significant sequence similarity between the 10 predicted proteins. However, three of them have similarity to uncharacterised secreted proteins from other oomycetes. These proteins may possibly be ancient oomycete effectors, however further work is required to investigate the precise function of these proteins. Other potentially secreted pathogenicity factors, which contain conserved domains are listed in Table S1.
Table 4. Predicted full-length secreted Eu. dicksonii proteins of unknown function.
| Contig | Length (aa) | Signal Peptide (aa) | Top blastx hit custom database* or Genbank nr |
| 1471 | 154 | 22 | no hits |
| 1573 | 91 | 41 | conserved hypothetical (134aa) protein Anaerococcus tetradius ZP_03930892 2e-12. |
| 908 | 132 | 34 | no hits |
| 1962 | 140 | 18 | conserved hypothetical (102aa) protein Phytophthora infestans PITG_08303 1e-10. |
| 1984 | 231 | 15 | no hits |
| 402981 | 113 | 21 | conserved hypothetical (115aa) protein Phytophthora infestans PITG_08745 0.01e. |
| 500818 | 138 | 15 | no hits |
| 500933 | 130 | 29 | no hits |
| 405461 | 157 | 24 | hypothetical protein Psta_475a (266aa) Pirellula staleyi YP_003373260 5e-08. |
| 4YO14FM1 | 141 | 34 | no hits |
*custom database made up of proteomes described in Figure 5.
Insights into oomycete infection of brown algae
Pathogen cell surface proteins
Mucins are typically high molecular weight cysteine, threonine, or serine non-glycosylated proteins that act as lubricants, protectants, signal tranducers or adhesins [39] and have been identified in the cell walls of higher oomycetes such as P. ramorum [40] and P. infestans [41]. We identified three gene fragments (contig1838, contig403969 and contig500361) with similarity to mucins or mucin-like sequences in the pathogen dataset (Table S1). We also identified a unigene sequence, contig500361, from the dissected library with a fibronectin type-3 domain (Table S1). Contig500361 is missing an initial start codon, but the entire protein fragment is predicted to be on the outside of the cell, anchored to the cell membrane, using both the new Eukaryotic subcellular predictor, Euk-mPLoc 2.0, [42] and TMHMM [43]. Contig500361 is not predicted to contain a signal sequence for secretion, according to SignalP [33] however it is likely that the N-terminal section of the protein is missing from Contig500361. Fibronectin is a high molecular weight extracellular matrix protein with diverse functions including roles in adhesion, growth, differentiation and wound healing [44]. Contig500361 is, therefore, likely to be part of a large surface protein in Eu. dicksonii.
Host and pathogen cell wall interactions
Higher oomycetes secrete cell wall degrading enzymes, to aid in penetration of host cells [45]. Since Eu. dicksonii penetrates the host algal cell to initiate infection, we looked for cell wall degrading enzymes within the Eu. dicksonii unigenes, as well as for algal genes that may be involved in strengthening or remodelling the host cell wall to prevent penetration.
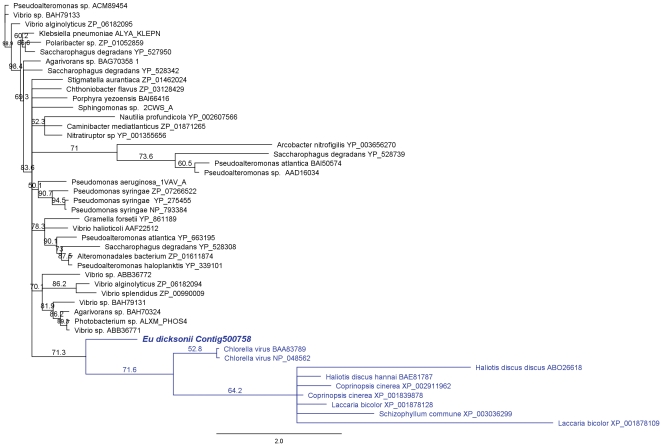
An Eu. dicksonii unigene had similarity to genes involved in the degradation of alginates, which are a key component of the brown algal cell wall matrix [46]. Contig500758 was predicted by InterproScan to contain a full alginate lyase 2 domain and has weak similarity to a 222 aa alginate lyase from Pseudomonas syringae pv. phaseolicola (Table S1). Phylogenetic analysis showed that it also had similarity to alginate lyase sequences from Chlorella virus and algal gastropods (Figure 6). However, it is likely that this gene is not full length, and therefore the prediction of signal peptides, specific substrates or catalytic residues is speculative without first obtaining the full length gene sequence. Interestingly, the Hyaloperonospora arabidopsidis, Saprolegnia parasitica, Phytophthora infestans, Phytophthora ramorum and Phytophthora sojae genomes do not contain homologues of Contig500758, indicating that this may be uniquely produced for the degradation of the algal host wall by Eu. dicksonii, and thus directly relate to its host spectrum. Contig500758 has similarity to alginate lyases from algal grazers (Haliotis species) and fungi and groups within a clade of alginate lyases from these organisms (Figure 6).
Figure 6. A Eu. dicksonii candidate pathogenicity effector has similarity with alginate lyases of brown algal grazers and fungi.
The conserved amino acid sequence region between eukaryotic, bacterial and viral alginate lyases present in the NCBI non-redundant database and the predicted protein sequence of the Eu. dicksonii gene fragment Contig500758 were aligned using MUSCLE. A dendrogram was produced using Geneious 4.8 with a neighbour-joining algorithm and 1000 bootstrap iterations. Branches with less than 50% bootstrap support were collapsed. Genbank accession numbers of the sequences used in the alignment are indicated on the tree. The Eu. dicksonii sequence groups with a clade of eukaryotic alginate lyases from fungi and abalone (Haliotis sp.), highlighted in blue. The latter also contains sequences from a green algal virus, thought to have been acquired by horizontal gene transfer.
We also identified a mannuronan C5-epimerase, the enzyme that catalyses the final step in alginate biosynthesis in the E. siliculosus sequences (Contig290, E. siliculosus Esi0495_0002). In kelps, C5 epimerases belong to a small family, with some isoforms induced in protoplast and elicitor-treated cultures [47]–[48]. The representation of the otherwise lowly expressed Esi0495_0002 gene [5] in our dataset suggests that alginate biosynthesis may be important to strengthen the host cell wall as a response to Eu. dicksonii infection. However, none of the other Ectocarpus enzymes involved in alginate biosynthesis [49] were represented in our dataset.
Although not full length, two additional Eu. dicksonii contigs encode glucanases that may be involved in breakdown of host cell wall glucans or cellulose. The predicted protein sequence of Contig500983 contains a partial exo-beta-1,3-glucanase domain (Table S1). It has similarity to glucan 1,3 beta-glycosidase and endo-1,3-beta glucanase from P. infestans, both of which are secreted proteins. Contig403087 is similar to the secreted putative exo-1,3-beta-glucanase from P. infestans (Table S1).
There are 9 putative cellulose synthase genes in E. siliculosus [5]. We identified three of these in our host dataset, suggesting a role for host cellulose synthesis during infection by Eu. dicksonii. Contig1852 mapped to host gene Esi0004_0105, Contig404144 mapped to host gene Esi0185_0053, whilst Contig301614 and Contig1623 mapped to the 3′ untranslated region of Esi0231_0017 (Table S1).
Eu. dicksonii thalli are first formed in a non-walled state, and then mature to walled thalli as infection progresses [7]. Therefore we expect cell wall biosynthesis to be an important part of the infection process of this oomycete. Oomycetes have traditionally been described as cellulosic organisms, however, chitin and chitosaccharides have been identified in the cell walls of several oomycetes, as have chitin synthases [50], [51]. We were not able to identify genes with similarity to cellulose synthases in Eu. dicksonii. However, we did identify a putative chitin synthase, namely Contig500448, which displays significant similarity to the chitin synthase 1 gene from the oomycete Saprolegnia monoica [52], [53] (Table S1).
Brown algal cell walls also contain sulfated fucans (fucoidans). Possible enzymes in the biochemical pathway for the synthesis and sulfation of fucans have recently been identified in the E. siliculosus genome [49], however, we were unable to identify any of these enzymes in our E. siliculosus dataset. At least two kinds of glycosidases act on fucans, flucan sulfate hydrolases and fucoidonase [54] however we were unable to detect pathogen transcripts with similarity to these enzymes in our dataset.
Searching for known oomycete effector protein families
All oomycete avirulence genes cloned to date contain a signal peptide for secretion and the RXLR amino acid motif at the N terminus of the protein. Hundreds of effector molecules containing this motif have been predicted in the genomes of sequenced oomycetes [28]. We therefore mined our dataset for RXLR-like sequences. We were unable to unambiguously identify transcripts that fulfill the criteria for putative RXLR effectors, as identified by [55] and [28].
The Crinkler (CRN) protein family elicits host responses in P. infestans host interactions, and contain an LFLAK motif. These effector proteins have been identified in all Phytophthora species sequenced to date [14], [28] and in the legume pathogen Aphanomyces euteiches [36]. A group of similar proteins has also been identified in the Pythium ultimum genome, with a related, but divergent, motif [29]. It is therefore possible that CRN proteins are ancestral effector proteins present throughout the oomycete lineage. However, we were unable to identify CRN proteins in our dataset. Thus, complete transcriptome or full genome sequencing will be needed to prove or disprove the absence of RXLR and CRN effectors in Eu. dicksonii.
Genes encoding other potential pathogenicity factors in Eu. Dicksonii
A vast array of pathogenicity factors and potential effector molecules, which may be involved during infection, or which trigger host defences, have now been identified in higher oomycete pathogens [16], [56]. Pathogenicity determinants are, either, presented at the cell surface, or secreted and/or actively transported into the host cell, to manipulate the host. We therefore, mined our dataset for potentially secreted proteins with similarity to known effectors or pathogenicity factors.
Contig400638 encodes a protein with a predicted protein tyrosine phosphatase-like (PTPLA) domain and a signal peptide for secretion. It has significant similarity to a conserved hypothetic protein from P. infestans and to a protein tyrosine phosphatase-like protein from Rattus norvegicus (Table S1). PTPLA proteins are key regulatory proteins, involved in regulating signal transduction by removal of phosphate from tyrosine residues in proteins such as MAP kinases. It is, therefore, possible that Eu. dicksonii secretes a PTPLA protein to interfere with host defence signal transduction, perhaps blocking transduction of host signaling that would lead to an algal defence response.
Contig500988 encodes a protein with a signal peptide and a histone deacetylase domain. The predicted protein has significant similarity to histone deacetylases from protozoan parasites (Table S1). Histone deacetylases may be involved in transcriptional regulation by removal of acetyl groups from the lysine residues of histones.
Sequences derived from Eu. dicksonii with similarity to cyclophilins/immunophilins, which may also be involved in pathogenicity were also identified (Table S1).
Eu. dicksonii genes involved in growth regulation and programmed cell death
Several Eu. dicksonii sequences, which are not predicted to be secreted, or which are not full-length, are similar to genes involved in growth inhibition or programmed cell death in other models. Singlet 8YN09FM1 encodes a protein fragment which contains a PHD domain (Table S1). PHD folds into an interleaved type of Zn-finger chelating Zn ions in a similar manner to RING-finger domains. Several PHD-finger proteins have now been identified that bind modules of methylated histone H3 and thereby inhibit transcription [57], [58], [59]. 8YN09FM1 also contains an inhibitor of growth domain (Table S1), which binds chromatin and acts as a transcription regulator. 8YN09FM1 does not encode a full-length protein, and does not contain the predicted start of the protein. It is therefore not possible to determine if this protein is secreted to interfere with host growth during infection, or is targeted to regulate Eu. dicksonii growth, temporarily silencing genes possibly as a stealth mechanism, upon onset of infection. Alternately, this could be the product of a generalized stress response in Eu. dicksonii.
Contig400862 and contig402793 also encode fragments of Eu. dicksonii translation factor(s) possibly involved in programmed cell death (Table S1). Neither contig is full-length and both are missing the predicted start of the gene(s), so it is not possible to predict the presence of signal peptides for secretion. The similar P. infestans protein (XP_002899252; PITG_14133) does not contain a signal peptide. Therefore Contig_400862 and contig_402793 may also represent oomycete gene(s) targeted internally rather than to the host.
Insights into pathogen physiology, stress responses and metabolism
We identified a unigene encoding a protein fragment predicted to be an hypoxia-induced protein. Contig_404084 from Eu. dicksonii was similar to HIGi hypoxia-inducible domain family member 2A from Xenopus laevis and the predicted protein fragment contained two transmembrane domains and an HIG_1_N response to hypoxia domain (Table S1). We also identified three sequences with thioredoxin-like domains within our pathogen dataset (Table S1). Several gene fragments with functional domains classified as transporters were also identified in Eu. dicksonii (Table S1). None of these sequences were full length, or predicted to contain signal peptides, however they may play an important role in the uptake of nutrients from the host.
One pathogen sequence, Contig_500199, predicted to encode a secreted lipase was identified in the dissected library, indicating host lipids may be an important nutrient source for Eu. dicksonii (Table S1). We identified 13 sequences, which we were unable to determine as secreted (1.3% of those with functional domains; 0.4% of total dataset) of Eu. dicksonii origin, that are involved in lipid metabolism, and 29 (2.3% of those with a functional domain and 1% of total dataset) involved in amino acid biosynthesis and metabolism (Figure 4), highlighting the importance of these processes in oomycete metabolism as previously reported in higher oomycetes [60], [61].
Host responses to Eu. Dicksonii
Contig1359 encodes the full-length putative protein inhibitor Esi0079_0059 from E. siliculosus. Esi0079_0059 contains a proteinase inhibitor I13 domain (Table S1) and is predicted to exhibit serine-type endopeptidase inhibitor activity. This protein also contains a signal peptide for secretion from host cells. Esi0079_0059, is therefore, likely to be secreted as a defence response to protect host proteins from Eu. dicksonii secreted peptidases.
Contig303625 encodes the fragment of an E. siliculosus nuclear movement domain protein (Esi0000_0481). This protein contains a p23_NUDC_like domain (Table S1), required for movement of the nucleus of other eukaryote species. Interestingly, the expression of this unigene correlates with a close association between the host nucleus and the infecting Eu. dicksonii thallus. The latter is observed at the earliest detectable stage of infection, suggesting an early migration of the host nucleus towards the infection site (Figure 1).
Contig2091 encodes an E. siliculosus protein (Esi0035_0036) currently un-annotated in the genome database. This protein contains a GRIM_19 domain, which promotes cell death via apoptosis in animals.
Expression of an E. siliculosus viral sequence
Intriguingly, Contig_403835 encodes a fragment of Esi0052_0171 a gene that belongs to the lysogenic virus inserted into the Ectocarpus genome. However, a comprehensive transcriptomic analysis reported by [5] revealed that the inserted viral genome is transcriptionally silent throughout the life cycle of Ectocarpus, even following the application of several stress treatments (hyperosmotic, hypoosmotic and oxidative stress). Hence, this result suggests that although lysogeny has yet to be observed in the E siliculosus strain used in the present study, the expression of inserted viral genes may still be triggered by infection from another pathogen. Unfortunately, the gene encoded by Esi0052_0171 (EsV-1-191) does not contain any conserved domains or have similarity to any functionally characterised protein, giving little clue as to what its function could be.
Discussion
In this study we describe an extensive characterisation of an EST collection of the brown alga E. siliculosus infected with the marine oomycete Eurychasma dicksonii. Given its wide host range and worldwide distribution, Eu. dicksonii is likely to be important to the ecology of coastal habitats where brown algae are predominant primary producers. Furthermore, Eu. dicksonii belongs to the most early branching clade within the oomycete lineage, and is therefore of considerable interest to decipher the origin and evolution of pathogenicty in this group. This study represents the first large scale sequencing study undertaken outside the best studied oomycete orders (namely Pythiales, Peronosporales and Saprolegniales). The latter, however, only represent a fraction of the lineage's diversity. Thus, and perhaps not so surprisingly, only 1314 (42% of a total 3074) pathogen unigenes shared sequence similarity with known oomycete genes. 1760 of the pathogen unigenes had no similarity to any known sequence, and no defining protein domains with which to predict function. In a similar study of the Aphanomyces euteiches transcriptome, Gaulin et al [36] found that approximately 70% of EST sequences showed similarity to previously described genes in the NCBI non-redundant protein database and around 80% showed similarity to Phytophthora proteins. Within the sequences we identified of Eu. dicksonii origin, only a maximum of 28% showed similarity to previously described proteins. Therefore, our Eu dicksonii gene dataset is largely unique and indicates how far away we are from identifying the full repertoire of oomycete genes, despite the availability of several genomes from plant and fish pathogenic species. Additionally, it is clear from this study that Eu. dicksonii shares some genetic similarity with higher order oomycetes, but is in fact very unique genetically, in comparison to these other sequenced oomycetes.
Making use of the recently completed E. siliculosus genome [5], we were able to unambiguously map 6787 of our unigenes to the algal host. Of these sequences, only approximately 20% mapped to protein coding regions. E. siliculosus contains large 3′UTR regions and mapping of EST datasets generated in previous projects has resulted in large proportions mapping to these 3′ UTRS [5]. Therefore the result that up to 80% of our sequences map outside of the protein coding regions of E. siliculosus genes is consistent with previously reported data and may go part-way to explaining the low number of host sequences with significant similarity in the NCBI nr protein database.
The unusually high level of expression of a broad range of TEs in E. siliculosus is also fully consistent the observations reported by the Ectocarpus Genome Initiative [5]. Importantly however, the most highly expressed TEs in our libraries only partially overlap those identified in this earlier work. Our results suggest an environmental, most probably stress-dependent, transcriptional regulation of TEs in E.siliculosus We conclude that, whilst methylation has not been detected in E. siliculosus, TE expression is probably tightly regulated in the genome by an as yet uncharacterised mechanism. Finally, the fact that the retroelements RTE2, RTE3 and RTE4 are the most highly induced in infected libraries and among the most abundant repeats in the E. siliculosus genome points to a stress-induced transposing activity, which remains to be investigated.
We found a GC content in host sequences of 51.9% consistent with the E. siliculosus genome GC content of 53.6%. However, analysis of our Eu. dicksonii dataset revealed a much lower GC content, at 40.5%. This figure appears to be representative of real protein encoding Eu. dicksonii genes sequenced so far, and is not a result of poor quality or non-protein coding sequencing. This result is in stark contrast with the GC content of other sequenced oomycete genomes, which is approximately 58% GC [14], [28]–[30]. A breadth of possible explanations, including pathogenic lifestyle, have been formulated to account for low genome GC content [62]. Hence, the biological significance of the GC-impoverished Eu. dicksonii transcriptome remains unclear . Interestingly, it is known that unlike other oomycetes, Eu. dicksonii uses a previously reported stop codon to encode tryptophan in mitochondrial genes [7]. This bias towards the use of UGA, rather than UGG in mitochondria is consistent with the widely held view that AT-rich genomes are prone to Trp-UGA codon reassignment.
Several hundred candidate effector molecules carrying a RXLR or CRN motif have now been identified in the genomes of oomycetes pathogenic on plants [16], [28] and one putative RXLR-effector was found in Saprolegnia parasitica, which is a pathogen of fish [63]. It is widely assumed that the RXLR motif mediates the translocation of pathogenicity effectors into host cells [55], [64]. It is also functionally interchangeable with the PEXEL translocation signal of apicomplexa pathogens [65], [66]. Despite Eu. dicksonii being an obligate intracellular biotrophic pathogen, we were unable to unambiguously identify oomycete effector molecules in our dataset. Thus, it is possible that Eu. dicksonii does not contain RXLR proteins, and uses an alternate signal for translocation of effectors into host cells. Alternately, since the current study does not represent a complete Eu. dicksonii transcriptome, and since many of our sequences are not full length, RXLR effectors may be present, but unidentifiable using the current dataset. Furthermore, if such effector sequences are expressed at low levels we may not pick them up in EST libraries derived from infected host material. Full transcriptome or genome sequencing of Eu. dicksonii is therefore required to comprehensively search for the ancient origins of RXLR, CRN or other effectors within the oomycete lineage.
Whilst RXLR or CRN-type effectors were not identified in this study, we were able to identify sequences as potential pathogenicity factors, including both genes not previously described as oomycete pathogenicity determinants, and genes with similarity to pathogenicity factors from oomycetes and other organisms.
A novel putative pathogenicity factor is the protein encoded by Contig500758, a predicted alginate lyase. This sequence does not have homologues in sequenced oomycete genomes and appears to be most similar to alginate lyases from fungi and brown algal grazers such as abalone (Haliotis ssp., Figure 6). Since alginates are the major structural component of the brown algal cell wall, we hypothesize that this Eu. dicksonii protein may be a major pathogenicity determinant. Several other sequences with similarity to cell wall degrading enzymes were also identified in the pathogen dataset. It has been reported that Eu. dicksonii has an extremely wide host range, infecting all brown algae tested so far [6]. It is possible that this infection is achieved primarily through the pathogen's ability to degrade host alginate, and other cell wall structural components, and thereby enter the host cell. Several other proteins involved in production or degradation of both host and pathogen cell walls were also identified in this study, indicating the importance of the cell wall in oomycete-host interactions. Enzymes involved in alginate and cellulose biosynthesis were identified in the host sequence set, whilst a putative chitin synthase was identified in Eu. dicksonii. This suggests that chitin or chitosaccharides may be ancient and important components of the oomycete cell wall that may have either been lost in higher oomycetes, or which fulfil subtle functions in structuring the cell wall.
We identified three E. siliculosus putative cellulose synthases in our host dataset. Evidence for secreted cellulose degrading enzymes in Eu. dicksonii was not as conclusive. It is possible that E. siliculosus strengthens the cell wall using cellulose in response to pathogen attack and degradation of alginates. Esi0185_0053 has a CesA_CelA-like domain, and Esi0231_0017 has a cellulose synthase domain, along with a glycosyl transferase GTA family domain. However, Esi0004_0105 only contains a glycosyl transferase GTA family domain. Since the annotation of these genes is purely based on bioinformatic analysis, their exact enzymatic function is not known. Therefore, it is possible that at least one of these genes could function in the production of the related defence molecule callose, rather than cellulose.
Rearrangement of actin microfilaments to focus on the infection site, and movement of the host nucleus to the site of attack is a well-documented feature of plant-oomycete and plant-fungal interactions (reviewed in [67]). However, little is known of the brown algal response to infection. In the present study, we identified an E. siliculosus candidate nuclear movement protein within our dataset. Furthermore, the Eu. dicksonii thallus is always observed in close proximity to the host nucleus and Gogli, ([7]; Figure 1 this study). This association between Eu. dicksonii and its host nucleus is already established at the earliest detectable stages of infection, and is conserved across the brown algal genera Ectocarpus, Macrocystis and Pylaiella. These observations suggest that migration of the host nucleus towards the parasitic thallus is a physiologically-relevant feature underpinning the infection process.
Further host responses to infection by Eu. dicksonii might be mediated by the secretion of a proteinase inhibitor, presumably targeted towards proteinases produced by the pathogen. These findings highlight the power of the production of EST libraries from infected host material to build up a picture of the dynamics of the interactions between E. siliculosus and Eu. dicksonii during infection. However, since many of the oomycete sequences identified in this study do not show similarity to previously described genes, future challenges include functional characterisation of such genes and identification of further pathogenicity determinants in this organism.
Supporting Information
Ectocarpus assembled contigs.
(TXT)
Eurychasma assembled contigs.
(TXT)
Sequences providing insights into Eu. dicksonii infection of E. siliculosus.
(XLS)
Acknowledgments
Leighton Pritchard (Scottish Crop Research Institute, Dundee, UK) is gratefully acknowledged for help with the development of custom python scripts used to sort and analyse sequence data. Dieter G. Müller (University of Constance, Germany) and Florian Maumus (INRA Versailles, France) are gratefully acknowledged for helpful suggestions and for the provision of biological material. Satoshi Sekimoto (University of British Columbia, Canada) kindly provided original electron microscopy pictures of Eu. dicksonii.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is supported by the University of Aberdeen (PvW), the Biotechnology and Biological Sciences Research Council (BBSRC) (LGB, PvW), the Royal Society (PvW), the Total Foundation (FCK, PvW) and the Natural Environment Research Council (NERC) via the SOFI initiative (award NE/F012705/1) (LGB, CMMG, FCK and PvW), through Oceans 2025 WP4.5 (FCK), a New Investigator Grant (NE/D521522/1) (FCK) and a sequence allocation from the NERC Molecular Genetics Facility (Pilot Project MGF 211) (CMMG, PvW and FCK). The authors are also supported by the European Commission via a Marie Curie Intra-European Fellowship (MIEF-CT-2006-022837) (CMMG), a European Reintegration Grant (PERG03-GA-2008-230865) (CMMG), and an EU ECOSUMMER PhD fellowship (MEST-CT-2005-20501) (MS, FCK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gachon CMM, Sime-Ngando T, Strittmatter M, Chambouvet A, Kim GH. Algal diseases: spotlight on a black box. Trends Plant Sci. 2010;15:633–639. doi: 10.1016/j.tplants.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Lafferty KD, Allesina S, Arim M, Briggs CJ, De Leo G, et al. Parasites in food webs: the ultimate missing links. Ecology Letters. 2009;11:533–546. doi: 10.1111/j.1461-0248.2008.01174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frada M, Probert I, Allen MJ, Wilson W, de Vargas C. The “Cheshire Cat” escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc Natl Acad Sci U S A. 2008;105:15944. doi: 10.1073/pnas.0807707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinberger F. Pathogen-induced defense and innate immunity in macroalgae. Biological Bulletin. 2007;213:290–302. doi: 10.2307/25066646. [DOI] [PubMed] [Google Scholar]
- 5.Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, et al. The Ectocarpus genome and the independent evolution of multicellularity in the brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- 6.Müller DG, Küpper FC, Küpper H. Infection experiments reveal broad host ranges of Eurychasma dicksonii (Oomycota) and Chytridium polysiphoniae (Chytridiomycota), two eukaryotic parasites in marine brown algae (Phaeophyceae). Phycological Research. 1999;47:217–23. [Google Scholar]
- 7.Sekimoto S, Beakes GW, Gachon CMM, Müller DG, Küpper FC, et al. The development, ultrastructural cytology and molecular phylogeny of the basal oomycete Eurychasma dicksonii infecting the filamentous phaeophyte algae Ectocarpus siliculosus and Pylaiella littoralis. Protist. 2008;159:299–318. doi: 10.1016/j.protis.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Sparrow FK. The University of Michigan Press; 1960. Aquatic phycomycetes, 2nd ed. [Google Scholar]
- 9.Gachon CMM, Strittmatter M, Müller DG, Kleinteich J, Küpper FC. Differential host susceptibility to the marine oomycete pathogen Eurychasma dicksonii detected by real-time PCR: not all algae are equal. Applied and Environmental Microbiology. 2009;75:322–328. doi: 10.1128/AEM.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Küpper FC, Müller DG. Massive occurrence of the heterokont and fungal parasites Anisolpidium, Eurychasma and Chytridium in Pylaiella littoralis (Ectocarpales, Phaeophyceae). Nova Hedwigia. 1999;69:381–9. [Google Scholar]
- 11.Strittmatter M, Gachon CMM, Küpper FC. Ecology of lower oomycetes. In: Lamour K, Kamoun S, editors. Oomycete genetics and genomics: diversity, interactions and research tools. Hoboken: John Wiley and Sons Inc. 2009;25-46 [Google Scholar]
- 12.Küpper FC, Maier I, Müller DG, Loiseau-de Goër S, Guillou L. Phylogenetic affinities of two eukaryotic pathogens of marine macroalgae, Eurychasma dicksonii (Wright) Magnus and Chytridium polysiphoniae Cohn. Cryptogamie Algologie. 2006;27:165–184. [Google Scholar]
- 13.Baldauf S. The deep roots of Eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 14.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RHY, et al. Phytophthora Genome Sequences Uncover Evolutionary Origins and Mechanisms of Pathogenesis Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 15.Grenville-Briggs LJ, van West P. The Biotrophic Stages of Oomycete-Plant Interactions. Advances in Applied Microbiology. 2005;57:217–243. doi: 10.1016/S0065-2164(05)57007-2. [DOI] [PubMed] [Google Scholar]
- 16.Kamoun S. Molecular genetics of pathogenic oomycetes. Eukaryotic Cell. 2003;2:191–199. doi: 10.1128/EC.2.2.191-199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van West P, Appiah AA, Gow NAR. Advances in research on root pathogenic oomycetes. Physiological & Molecular Plant Pathology. 2003;62:99–113. [Google Scholar]
- 18.Phillips AJ, Anderson VL, Robertson EJ, Secombes CJ, van West P. New insights into animal pathogenic oomycetes. Trends in Microbiology. 2008;16:13–19. doi: 10.1016/j.tim.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 19.van West P. Saprolegnia parasitica, an oomycete pathogen with a fishy appetite:new challenges for an old problem. Mycologist. 2006;20:99–104. [Google Scholar]
- 20.Haverkort AJ, Boonekamp PM, Hutten R, Jacobsen E, Lotz LAP, et al. Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Res. 2008;51:47–57. [Google Scholar]
- 21.Müller DG, Gachon CMM, Küpper FC. Axenic clonal cultures of filamentous brown algae. Initiation and maintenance. Cahiers de Biologie Marine. 2008;49:59–65. [Google Scholar]
- 22.West JA, McBride DL. Long term and diurnal carpospore discharge patterns in the Ceramiaceae, Rhodomelaceae and Delesseriaceae (Rhodophyta). Hydrobiologia. 1999;398399:101–114. [Google Scholar]
- 23.Katsaros C, Galatis B. Immunofluorescence and electron microscopic studies of microtubule organisation during the cell cycle of Dictyota dichotoma (Phaeophyta, Dictyotales) Protoplasma. 1992;169:75–84. [Google Scholar]
- 24.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Research. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gremme G, Brendel V, Sparks ME, Kurtz S. Engineering a software tool for gene structure prediction in higher organisms. Information and Software Technology. 2005;47:965–978. [Google Scholar]
- 26.Min XJ, Butler G, Storms R, Tsang A. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res Web Server Issue. 2005:W677–W680. doi: 10.1093/nar/gki394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Haas BJ, Kamoun S, Zody MC, Jiang RHY, et al. Genome sequence analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 29.Lévesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, et al. Genome sequence of the necrotrophic plant pathogen, Pythium ultimum, reveals original pathogenicity mechanisms and effector repertoire. Genome Biology. 2010;11:R73. doi: 10.1186/gb-2010-11-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, et al. Signatures of Adaptation to Obligate Biotrophy in the Hyaloperonospora arabidopsidis Genome Science. 2010;330:1549–1551. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, et al. InterPro: the integrative protein signature database. Nucleic Acids Research. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(suppl1):D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maumus F. Transcriptional and epigenetic regulation in the marine diatom Phaeodactylum triconutum. A thesis presented for the degree of Doctor of Philosophy, Université Paris-Sud. 2009;11 [Google Scholar]
- 34.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 35.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nature Protocols. 2007;2:953–971 (2007). doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 36.Gaulin E, Madoui MA, Bottin A, Jacquet C, Mathé C, et al. Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways. PLoS ONE. 1723;3(3):e1723. doi: 10.1371/journal.pone.0001723. doi: 10.1371/journal.pone.0001723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 38.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, et al. 2011. Geneious v4.8, Available: http://www.geneious.com/
- 39.Perez-Vilar J, Hill RL. The structure and assembly of secreted mucins. J Biol Chem. 1999;274:31751–31754. doi: 10.1074/jbc.274.45.31751. [DOI] [PubMed] [Google Scholar]
- 40.Meijer HJG, van de Vondervoort PJI, Yuan Yin Q, de Koster CG, Klis FM, et al. Identification of cell wall-associated proteins from Phytophthora ramorum. . Mol Plant Microbe Interact. 2006;19:1348–1358. doi: 10.1094/MPMI-19-1348. [DOI] [PubMed] [Google Scholar]
- 41.Grenville-Briggs LJ, Avrova AO, Hay RJ, Bruce CR, Whisson SC, et al. Identification of appressorial and mycelial cell wall proteins and a survey of the membrane proteome of Phytophthora infestans. . Fungal Biology. 2010;114:702–723. doi: 10.1016/j.funbio.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Chou KC, Shen HB. A New Method for Predicting the Subcellular Localization of Eukaryotic Proteins with Both Single and Multiple Sites: Euk-mPLoc 2.0. PLoS ONE. 2010;5:e9931. doi: 10.1371/journal.pone.0009931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krogh A, Larsson B, von Heijne, Sonnhammer GELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 44.White E, Baralle F, Muro A. New insights into form and function of fibronectin splice variants. J Pathol. 2008;216:1–14. doi: 10.1002/path.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mcleod A, Smart CD, Fry WE. Characterisation of 1,3-beta-glucanase and 1,3;1,4-beta-glucanase genes from Phytophthora infestans. Fungal Genet Biol. 2003;38:250–263. doi: 10.1016/s1087-1845(02)00523-6. [DOI] [PubMed] [Google Scholar]
- 46.Kloareg B, Quatrano RS. Structure of the cell walls of marine algae and ecophysiological functions of the matrix polysaccharides. Oceanography and Marine Biology: An Annual Review. 1988;26:259–315. [Google Scholar]
- 47.Roeder V, Collén J, Rousvoal S, Corre E, Leblanc C, et al. Identification of stress genes from Laminaria digitata (Phaeophyceae) protoplast cultures by expressed sequence tag analysis. J Phycol. 2005;41:1227–1235. [Google Scholar]
- 48.Tonon T, Rousvoal S, Roeder V, Boyen C. Expression profiling of the mannuronan C5-epimerase multigenic family in the brown alga Laminaria digitata (Phaeophyceae) under biotic stress conditions. Journal of Phycology. 2008;44:1250–1256. doi: 10.1111/j.1529-8817.2008.00580.x. [DOI] [PubMed] [Google Scholar]
- 49.Michel G, Tonon T, Scornet D, Cock JM Kloareg B. The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytologist. 2010;188:82–97. doi: 10.1111/j.1469-8137.2010.03374.x. [DOI] [PubMed] [Google Scholar]
- 50.Bulone V, Chanzy H, Gay L, Girard V, Fevre M. Characterisation of chitin and chitin synthase from the cellulosic cell wall fungus Saprolegnia monoica. Exp Mycol. 1992;16:8–21. [Google Scholar]
- 51.Badreddine I, Lafitte C, Heux L, Skandalis M, Spanu Z, et al. Cell wall chitosaccharides are essential components and exposed patterns of the phytopathogenic oomycete Aphanomyces euteiches Eukaryotic Cell. 2008;7:1980–199. doi: 10.1128/EC.00091-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mort-Bontemps M, Gay L, Fevre M. CHS2 a chitin synthase gene from the oomycete Saprolegnia monoica. Microbiology. 1997;143:2009–2020. doi: 10.1099/00221287-143-6-2009. [DOI] [PubMed] [Google Scholar]
- 53.Guerriero G, Avino M, Zhou Q, Fugelstad J, Clergeot P-H, et al. Chitin synthases from Saprolegnia are involved in tip growth and represent a potential target for anti-oomycete drugs. PLOS Pathogens. 2010;6:1–12. doi: 10.1371/journal.ppat.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berteau O, Mulloy B. Sulfated fucans, fresh perspectives: structures, functions, and biological properties of sulfated fucans and an overview of enzymes active toward this class of polysaccharide. Glycobiology. 2003;13:29R–40R. doi: 10.1093/glycob/cwg058. [DOI] [PubMed] [Google Scholar]
- 55.Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 56.Oliva R, Win J, Raffaele S, Boutemy L, Bozkurt TO, et al. Recent developments in effector biology of filamentous plant pathogens. Cellular Microbiology. 2010;12:705–715. doi: 10.1111/j.1462-5822.2010.01471.x. [DOI] [PubMed] [Google Scholar]
- 57.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, et al. Molecular basis for site-specific readout of histone H3K4Me3 by the BPTF PHD finger of NURF. Nature. 2006;442:100–103. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, et al. Molecular mechanism of histone H3K4me3 regulation by plant homeodomain of ING2. Nature. 2006;442:91–95. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, et al. YNGI PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription of a subset of targetted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bartnicki-Garcia S. Cell wall chemistry, morphogenesis and taxonomy of fungi. Annu Rev Microbiol. 1968;22:87–108. doi: 10.1146/annurev.mi.22.100168.000511. [DOI] [PubMed] [Google Scholar]
- 61.Grenville-Briggs LJ, Avrova AO, Bruce CR, Williams A, Birch PRJ, et al. Elevated Amino Acid Biosynthesis in Phytophthora infestans During Appressorium Formation and Potato Infection. Fungal Genet Biol. 2005;42:244–256. doi: 10.1016/j.fgb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 62.Rocha EPC, Feil EJ. Mutational Patterns Cannot Explain Genome Composition: Are There Any Neutral Sites in the Genomes of Bacteria? PLoS Genet. 2010;6(9):e1001104. doi: 10.1371/journal.pgen.1001104. doi: 10.1371/journal.pgen.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van West P, de Bruijn I, Minor KL, Phillips AJ, Robertson EJ, et al. The putative RxLR effector protein SpHtp1 from the fish pathogenic oomycete Saprolegnia parasitica is translocated into fish cells. FEMS Microbiology. 2010;310:127–137. doi: 10.1111/j.1574-6968.2010.02055.x. [DOI] [PubMed] [Google Scholar]
- 64.Grouffaud S, Whisson SW, Birch PJR, van West P. Towards an understanding of how RxLR effector proteins are translocated from oomycetes into host cells. Fungal Biology Reviews. 2010;24:27–36. [Google Scholar]
- 65.Bhattacharjee S, Hiller NL, Liolios K, Win J, Kanneganti TD, et al. The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathogens. 2006;2:e50. doi: 10.1371/journal.ppat.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grouffaud S, van West P, Avrova AO, Birch PRJ, Whisson SC. Plasmodium falciparum and Hyaloperonospora parasitica effector translocation motifs are functional in Phytophthora infestans. Microbiology. 2008;154:3743–3751. doi: 10.1099/mic.0.2008/021964-0. [DOI] [PubMed] [Google Scholar]
- 67.Takemoto D, Hardham AR. Update: The cytoskeleton as a regulator and target of biotic interactions in plants. Plant Physiol. 2004;136:3864–3876. doi: 10.1104/pp.104.052159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ectocarpus assembled contigs.
(TXT)
Eurychasma assembled contigs.
(TXT)
Sequences providing insights into Eu. dicksonii infection of E. siliculosus.
(XLS)