Abstract
Sex determination mechanisms differ among animal species, but it is not clear how these differences evolve. New sex determiners may arise in response to sexual conflicts, which occur when traits benefit one sex but hinder the other. Here we identified the genetic basis for the orange-blotch (OB) color pattern, a trait under sexually antagonistic selection in the cichlid fish of Lake Malawi, East Africa. The OB phenotype is due to a cis-regulatory mutation in the Pax7 gene. OB provides benefits of camouflage to females, but disrupts the species-specific male color patterns used for mate recognition. We suggest that the resulting sexual conflict at OB has been resolved by selection for a novel female sex determination locus, promoting its invasion into populations with an ancestral male sex determination system.
When an autosomal mutation modulates a trait to the benefit of one sex and the detriment of the other, the resulting sexually antagonistic selection creates a genetic conflict. This conflict can be resolved if expression of the phenotype is limited to the sex that gains a fitness benefit from the mutation. (1, 2). Sexually dimorphic pigmentation is a hallmark of the radiation of cichlid fishes in Lake Malawi, one of the three major East African Great Lakes (Malawi, Tanganyika, and Victoria). Competition for females has resulted in sexual selection for brightly colored, conspicuous males (Fig. 1B). In contrast, female coloration is generally drab and brown, making them inconspicuous to predators (3) (Fig. 1A).
Figure 1.
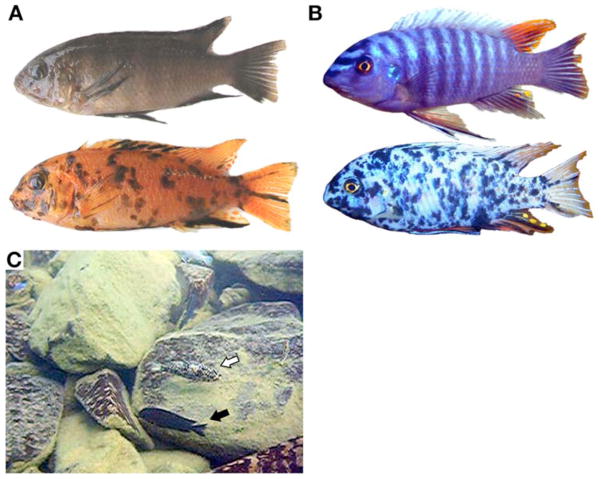
OB pigmentation affects different aspects of fitness in females and males. (A) Typical female BB (top) and OB (bottom) morphs, represented by Tropheops ‘orange chest’. (B) Common BB male nuptial coloration (top) and rare OB male (bottom) of Labeotropheus fuelleborni. (C) OB (white arrow) and BB (black arrow) L. fuelleborni females are differently cryptic over the boulder reef habitat at Nankoma Island (13°53′30″S, 34°36′43″E; frame from Movie S1).
An exception to uniformly brown pigmentation in female rock-dwelling cichlids in Lake Malawi is the orange-blotch (OB) phenotype (4). OB is found almost exclusively in females, and usually co-segregates with the more widespread brown barred (BB) female phenotype in local populations (3). The OB phenotype is characterized by dark melanophore blotches of variable number and size on a background of orange xanthophores (Fig. 1A, and fig. S1). The blotching provides an alternative form of crypsis by background-matching the mottled rock substrate that the cichlids inhabit (Fig. 1C; Movie S1). The variable high-contrast blotching may also provide camouflage by disruptive coloration, independent of background matching (5). OB males are rare and their characteristic nuptial colors are highly disrupted (Fig. 1B) (4). Given the strong reliance on specific male nuptial color cues for mate recognition and attraction (6), the disruption of pattern is predicted to lower the fitness of OB males. The OB phenotype has been observed in more than 20 species in four genera in Lake Malawi. Similar sex-associated blotched phenotypes are found in cichlids from Lake Victoria, but the cichlids of Lake Tanganyika appear to lack comparable phenotypes (7).
The OB locus was previously localized to a 5 cM region of Malawi cichlid linkage group five (LG5) (8). In the absence of a cichlid genome sequence, we took a comparative mapping approach to refine the map interval, using the Tetraodon genome as a guide (Fig. 2A, and fig. S2B). We identified additional markers in the region by aligning cichlid genome survey sequences to the Tetraodon genome (9, 10), and resequencing a panel of cichlid DNA to identify polymorphisms. Genotyping these new markers confirmed the broad-scale synteny between these species.
Figure 2.
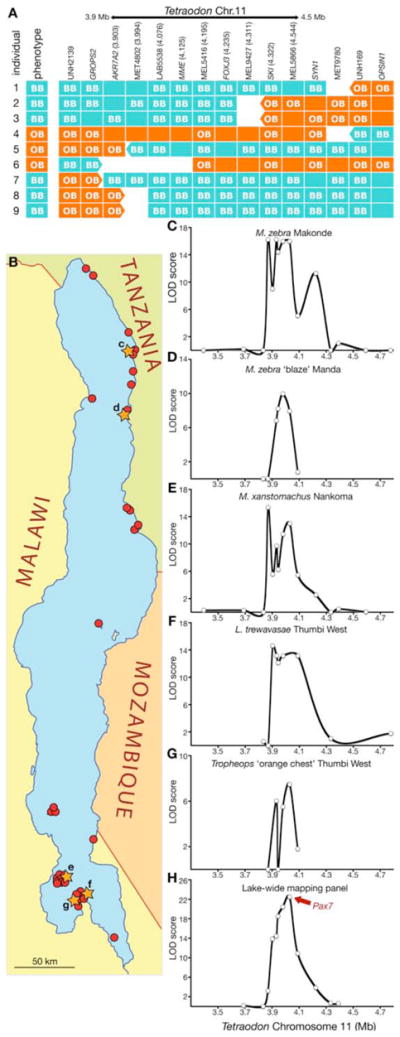
Genetic mapping in families and natural populations. (A) Individual recombinant chromosomes (rows) at the OB locus from pedigreed families, with “OB” and “BB” indicating genotypes of inherited maternal alleles, and empty blocks indicating non-informative markers. Marker names and Mb position on Tetraodon Chr.11 are indicated at top of marker columns. Breakpoint analysis across multiple recombinants defines an OB interval between markers AKR7A2 and SKI; one recombinant refines the interval to AKR7A2 and MET4802. Italics indicate gene-associated markers. (B) Collecting sites in Lake Malawi. The populations analyzed in panels C-G are marked with orange stars (c-g). Localities combined for the lake-wide mapping panel (H) are marked with red dots. (C-G) Association of markers with OB and OB-like phenotypes in populations, graphed as LOD of association versus position on Tetraodon Chr.11. (C) M. zebra Makonde (24 BB, 24 WhB & Wh); (D) M. zebra ‘blaze’ Manda (24 BB, 16 OB & O); (E) M. xanstomachus Nankoma (26 BB, 22 OB); (F) L. trewavasae Thumbi West (24 BB, 24 OB & O); (G) Tropheops ‘orange chest’ Thumbi West (32 BB, 18 OB). (H) Multi-species lake-wide mapping panel (34 BB, 44 OB class; Table S2).
Pedigreed families from several species of the genera Labeotropheus and Metriaclima were genotyped for newly identified microsatellite markers on LG5 (46 families, 678 individuals, 349 OB individuals; table S1). Breakpoint analysis localized OB to an interval of less than 1 cM, corresponding to a region from 3.9–4.0 Mb on Tetraodon chromosome 11 (Fig. 2A). We then used association mapping in natural populations to pinpoint the causative locus. Single nucleotide polymorphism (SNP) markers were developed spanning the region, including an intronic SNP marker for each annotated gene within the OB interval. We measured linkage disequilibrium (LD) between OB and SNPs within natural populations of Labeotropheus, Metriaclima, and Tropheops from the northern and southern parts of Lake Malawi (Fig. 2B). Within each population, a peak of LD was found in a region overlapping the interval defined by breakpoint analysis (Fig. 2C-G). Given the shared pattern of association across study populations, we created a lake-wide mapping panel to increase the effective size of the mapping population. This lake-wide panel contains BB and OB individuals from 36 distinct populations of 12 species segregating OB (Table S2). Analysis of marker data across these populations increased resolution and statistical significance of LD beyond that available from any single population (Fig. 2H).
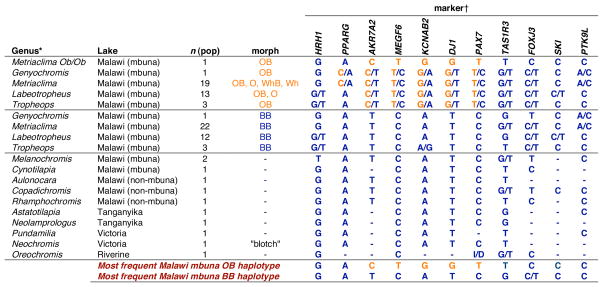
The maintenance of strong LD in the lake-wide mapping panel suggests the presence of a single, shared haplotype of SNP genotypes across the flock. Indeed, we identified a common haplotype signature found largely intact in OB individuals of all species, indicating a single origin of the causative OB mutation in Lake Malawi (Table 1). The OB haplotype block extends from Akr7A2 to Pax7 in most individuals, and based on comparative genomic information includes at least five genes (Fig. S2). Comparison to outgroup species suggests that BB chromosomes represent the ancestral haplotypes across this interval (Table S3). The single blotched cichlid examined from Lake Victoria also carries an ancestral haplotype, suggesting an independent evolutionary origin for the Victorian OB-like phenotype (Table 1, Neochromis ‘blotch’ morph). Our finding of a single origin of OB in Lake Malawi cichlids contrasts with a previous phylogenetic analysis that suggested multiple origins (11).
Table 1.
The most prevalent SNP genotypes at the OB locus in the four genera segregating OB and in outgroup genera. The most frequent OB and BB haplotypes are indicated separately at the bottom. SNP genotypes indicating the OB-specific haplotype signature are labeled in orange.
|
Details of the species and localities sampled for this table are found in Tables S2 and S3.
Intronic SNP markers labeled by gene are a subset of those used in LD analysis (Fig. 2, C-H).
The OB haplotype is a genetic marker allowing us to make additional conclusions about the relationship between genotype and phenotype. Individuals in some populations are heavily pigmented with melanophore blotches, whereas individuals in other populations lack blotches almost entirely (the orange or “O” morph). Sex-linked white-blotch and white morphs (“WhB” and “Wh”), similar to OB and O morphs but lacking orange background coloration, also exist in the Malawi flock (Fig. S1). Individuals of these morphs also carry the OB haplotype, and can be definitively attributed to the OB locus (Table 1). Discrete morphs with different degrees of blotching sometimes segregate within a population, covering a similar phenotypic range as that found across species. OB individuals from the three study populations segregating multiple blotch phenotypes are all heterozygous for the same SNP haplotype block (Fig. 2C, 2D, and 2F), suggesting that differences between morphs are not due to gene dosage. Discrete morphs may result from allelic variants within the OB haplotype, or genetic modifiers of OB segregating in some populations.
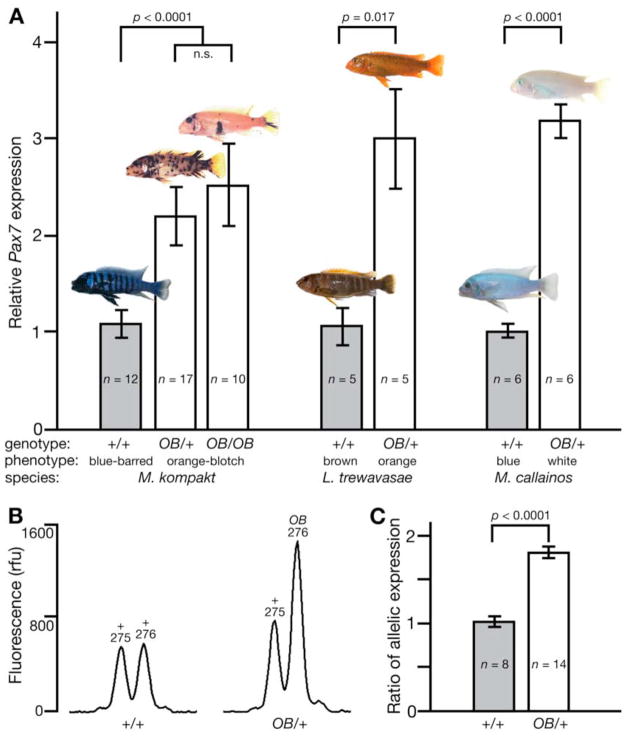
The peak of LD in the lake-wide mapping panel corresponds to a non-coding SNP within a Pax3/7 subfamily gene. These transcription factors play key roles in specifying pigment cell lineages from neural crest precursors (12, 13). Gene phylogeny and conserved synteny identify the OB-associated gene as cichlid Pax7 (Fig. S2). OB individuals have significantly higher expression of Pax7 in tailfin, regardless of the degree of melanophore blotching or relative numbers of other pigment cell populations (Fig. 3A). Allele-specific expression analysis demonstrates that the OB allele of Pax7 is specifically upregulated (Fig. 3B, C). Since no OB-associated differences were found in Pax7 coding sequence from the species used for expression analysis, we suggest that cis-regulatory differences in Pax7 account for the OB pigmentation phenotype (14).
Figure 3.
Pax7 expression is increased in OB individuals independent of species and morph. (A) Pax7 expression relative to BB expression level within each species, measured by quantitative real-time PCR of juvenile tailfin RNA and normalized to β-actin expression. OB genotypes and expressed phenotypes are indicated along the x-axis. (B, C) Allele-specific expression using RT-PCR of a size polymorphism in the 5′UTR of Pax7. (B) Representative size separation traces of products from +/+ and OB/+ individuals, with allele type and size indicated above peaks. (C) The ratio of OB- to non-OB-sized allele expression. All p values from pairwise t-tests.
In addition to overt pigmentation differences between BB and OB individuals, close examination reveals cell-level differences in tailfin melanophores. Melanophore number is reduced in three-month-old M. ‘kompakt’ OB/+ individuals relative to BB broodmates (BB (n = 7), 414±173; OB/+ (n = 10), 118±77; Welch’s t-test: p = 0.0032). In contrast, contracted melanophore size is larger in OB/+ individuals (BB (n = 7), 26.5±8.6 pixels2; OB/+ (n = 10), 50.6±14.3 pixels2; t-test: p = 0.0013). The finding that upregulation of Pax7 in cichlids is correlated with a fewer-but-larger melanophore phenotype is particularly interesting given that downregulation of Pax3 in zebrafish produces a more-but-smaller melanophore phenotype (13). In both cichlids and zebrafish, higher Pax3/7 dosage correlates with development of fewer, larger melanophores.
In our analysis of families and natural populations, we have been unable to genetically separate OB from the female sex determiner (W) known to be in the same region of LG5. Only one species in our laboratory stocks, M. ‘kompakt’, occasionally produces OB/+ males. However, these OB males inherit the maternal OB haplotype intact, and when bred, their OB-carrying chromosome determines female sex in offspring (Table S4) (14). We conclude that these OB males are genetically female at the W locus and sex-reversed by another mechanism. Mating of M. ‘kompakt’ OB/+ males to OB/+ females produces viable OB/OB homozygotes in expected Mendelian ratios (21 of 89 individuals from five broods; χ2 = 1.36, p = 0.5067). Thus OB homozygotes are fully viable, at least on some genetic backgrounds. However, we have not observed OB homozygous individuals in natural populations (0 of 199 wild OB individuals), which suggests that OB males have low mating success.
The maintenance of the OB haplotype over evolutionary timescales might be explained by selection for tight linkage between OB (Pax7) and the W sex determiner (15). Linkage and suppression of recombination between OB and W might be further increased by a W-specific inversion encompassing both genes, a rearrangement of the kind hypothesized to be an early stage of sex chromosome evolution (16). However, no obvious candidate gene for sex determination is within the interval (14), and we found no evidence of structural rearrangements.
We suggest that OB originated once as a novel allele upregulating Pax7 expression, and was incorporated into multiple species across four genera of the Malawi cichlid flock by sorting of an ancestral polymorphism and/or hybridization events after the radiation of species. The OB phenotype positively affects the fitness of females by providing an alternative form of crypsis against the mottled background of their habitat. Conversely, OB reduces male fitness by disrupting their nuptial pigmentation patterns. The genetic conflict created by sexually antagonistic selection on the OB phenotype may be resolved by sexually dimorphic expression of the trait. In the case of the OB locus, the sexual conflict has been resolved by tight genetic linkage with a dominant female sex determiner, rather than by evolution of modifier genes to sex-limit expression of the phenotype during development. Thus, the OB-W haplotype has invaded populations in a manner consistent with a model of sex chromosome evolution by sexual conflict (17).
The OB-associated ZW sex determining system is epistatically dominant to a putatively ancestral XY locus on LG7, responsible for sex determination in several other Malawi cichlids (Tables S5 and S6) (14). We propose that sexually antagonistic selection at OB (Pax7) provided conditions facilitating the invasion of a tightly linked female sex determiner on LG5 to resolve the genetic conflict. Alternatively, the OB polymorphism may have arisen near a nascent female sex determiner, providing conditions for the OB-W allele to rise in frequency and persist in multiple lineages along with the ancestral XY sex determination system. In either case, the evolutionary success of the allele is the result of the interaction of the pigmentation and sex determination traits.
The OB-W locus is a nascent sex chromosome, and demonstrates the interactive evolution of sexual dimorphism and sex determination resulting from resolution of sexual conflicts. Sexually antagonistic selection is usually thought to arise from processes of sexual selection (18). Our results show that a novel trait increasing fitness under natural selection can readily create strong sexual conflicts, especially if the trait negatively impacts sexually selected characters in the opposite sex. Phenotypes rarely fall into a simple dichotomy of sexually and naturally selected traits, nor do they impact overall male and female fitness equally. Thus, multiple aspects of a phenotype can trigger sexual conflicts, and ultimately the evolution of new sex determiners. We suspect that similar genetic conflicts are important in the evolution of many other traits, and have likely contributed to the extraordinary rates of diversification and speciation in African cichlid fishes.
Supplementary Material
Acknowledgments
We thank O. Seehausen for providing cichlid tissues from Lake Victoria, and S. Salzberg for assistance in aligning cichlid genome sequences to the Tetraodon genome. This work was supported by an NIH fellowship (#F32HD051383) to RR and grants from NSF (#DEB- 0445212) and NIH (#R01HD058635) to TDK. Sequences generated in this study are deposited in Genbank under accession numbers GQ403984-GQ403986.
References
- 1.Fisher RA. Biol Rev. 1931;6:345–368. [Google Scholar]
- 2.Rice WR. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 3.Konings A. Malawi Cichlids in their Natural Habitat. 4. Cichlid Press; El Paso, TX: 2007. pp. 19–20. [Google Scholar]
- 4.Holzberg S. J Zool Syst Evol Res. 1978;16:171–186. [Google Scholar]
- 5.Schaefer HM, Stobbe N. Proc R Soc B. 2006;273:2427–2432. doi: 10.1098/rspb.2006.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Oppen MJH, et al. Mol Ecol. 1998;7:991–1001. [Google Scholar]
- 7.Lande R, Seehausen O, van Alphen JJ. Genetica. 2001;112–113:435–443. [PubMed] [Google Scholar]
- 8.Streelman JT, Albertson RC, Kocher TD. Mol Ecol. 2003;12:2465–2471. doi: 10.1046/j.1365-294x.2003.01920.x. [DOI] [PubMed] [Google Scholar]
- 9.Loh YH, et al. Genome Biol. 2008;9:R113. doi: 10.1186/gb-2008-9-7-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurtz S, et al. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allender CJ, Seehausen O, Knight ME, Turner GF, Maclean N. PNAS. 2003;100:14074–14079. doi: 10.1073/pnas.2332665100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacosta AM, Muniesa P, Ruberte J, Sarasa M, Domínguez L. Pigment Cell Res. 2005;18:243–251. doi: 10.1111/j.1600-0749.2005.00238.x. [DOI] [PubMed] [Google Scholar]
- 13.Minchin JEN, Hughes SM. Dev Biol. 2008;317:508–22. doi: 10.1016/j.ydbio.2008.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Detailed information is supplied as supporting material on Science Online.
- 15.Rice WR. Evolution. 1987;41:911–914. doi: 10.1111/j.1558-5646.1987.tb05864.x. [DOI] [PubMed] [Google Scholar]
- 16.Charlesworth D, Charlesworth B, Marais G. Heredity. 2005;95:118–28. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- 17.van Doorn GS, Kirkpatrick M. Nature. 2007;449:909–912. doi: 10.1038/nature06178. [DOI] [PubMed] [Google Scholar]
- 18.Arnqvist G, Rowe L. Sexual Conflict. Princeton Univ. Press; Princeton, NJ: 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.