Abstract
We evaluated a 32 year-old woman whose oculocutaneous albinism, bleeding diathesis, neutropenia, and history of recurrent infections prompted consideration of the diagnosis of Hermansky-Pudlak syndrome type 2 (HPS-2). This was ruled out due to the presence of platelet delta granules and absence of AP3B1 mutations. Since parental consanguinity suggested an autosomal recessive mode of inheritance, we employed homozygosity mapping, followed by whole exome sequencing, to identify two candidate disease-causing genes, SLC45A2 and G6PC3. Conventional di-deoxy sequencing confirmed pathogenic mutations in SLC45A2, associated with oculocutaneous albinism type 4 (OCA-4), and G6PC3, associated with neutropenia. The substantial reduction of SLC45A2 protein in the patient’s melanocytes caused the mis-localization of tyrosinase from melanosomes to the plasma membrane and also led to the incorporation of tyrosinase into exosomes and secretion into the culture medium, explaining the hypopigmentation in OCA-4. Our patient’s G6PC3 mRNA expression level was also reduced, leading to increased apoptosis of her fibroblasts under ER stress. This report describes the first North American patient with OCA-4, the first culture of human OCA-4 melanocytes, and the use of homozygosity mapping followed by whole exome sequencing to identify disease-causing mutations in multiple genes in a single affected individual.
Introduction
Until recently, molecular diagnostics for patients with rare disorders relied upon sequencing of genes identified as candidates by virtue of the clinical phenotypes. Now, massively parallel (“next generation”) sequencing allows for unfocused detection of disease-causing mutations, without the bias introduced by phenotype. This critical advantage can be particularly important for patients with clinical manifestations that point to complex diseases. The process that is generally employed involves single nucleotide polymorphism (SNP) arrays followed by the next generation sequencing technique of whole exome sequencing (Ng et al., 2010; Ng et al., 2009; Otto et al., 2010). This reduces the amount of data generated and is more cost effective than whole genome sequencing (Teer and Mullikin, 2010).
We illustrate the benefit of next generation sequencing in a patient with oculocutaneous albinism (OCA) and congenital neutropenia. OCA is a group of autosomal recessively inherited hypopigmentary disorders that affect melanin pigment synthesis or trafficking of pigmentary enzymes or organelles (King et al., 2002) in the skin, hair and eyes (Gronskov et al., 2007). Four genes are associated with OCA: TYR (OCA-1) (Tomita et al., 1989), OCA2 (OCA-2) (Rinchik et al., 1993), TYRP1 (OCA-3) (Boissy et al., 1996) and SLC45A2 (OCA-4) (Newton et al., 2001). Mutations in these genes rarely cause phenotypic features other than albinism, but certain syndromes have oculocutaneous albinism as an obligate component. Hermansky-Pudlak Syndrome (HPS), for example, is characterized by OCA and a bleeding diathesis. There are 8 known human HPS genes (HPS1–8) (Anikster et al., 2001; Dell'Angelica et al., 1999; Li et al., 2003; Morgan et al., 2006; Oh et al., 1996; Suzuki et al., 2002; Zhang et al., 2003), whose protein products are involved in the biogenesis of lysosome-related organelles (Huizing et al., 2008). HPS commonly has other phenotypic features such as pulmonary fibrosis, which is associated with subtypes 1 and 4 (Oh et al., 1996; Suzuki et al., 2002).
Congenital neutropenia is a heterogeneous group of hereditary disorders characterized by life-threatening bacterial infections early in life due to a paucity of mature neutrophils (Boztug et al., 2008; Welte et al., 2006). Mutations in several genes, including ELA2, HAX1, WAS, GFI1, and recently G6PC3 (Boztug et al., 2009; Devriendt et al., 2001; Horwitz et al., 1999; Klein et al., 2007; Person et al., 2003), cause such conditions. Neutropenia also occurs in patients with HPS-2 disease, due to mutations in AP3B1, which encodes the beta-3A subunit of adaptor complex-3 (AP-3) (Dell'Angelica et al., 1999). Most congenitally neutropenic patients, including those with HPS-2, respond to recombinant human granulocyte colony-stimulating factor (rhG-CSF), which increases neutrophil counts and decreases the number and severity of infections (Bonilla et al., 1989).
In this article we describe a patient of consanguineous parentage who presented with findings that suggested the possible diagnosis of HPS-2, i.e., OCA and neutropenia. When no mutations were found in AP3B1, we performed homozygosity mapping on a SNP array and whole exome sequencing using the patient’s genomic DNA. This allowed us to identify disease-causing mutations in two different disease associated genes, i.e., SLC45A2 causing OCA-4, and G6PC3 causing neutropenia. Neither of these genes would have been pursued according to standard practice. We conclude that SNP array analysis, along with whole exome sequencing, can be useful in identifying disease-causing mutations in a single affected individual, especially if the family is consanguineous and suitable for homozygosity mapping.
Results
Case report
A 32 year old Caucasian female presented to the NIH Clinical Center with a history of inflammatory bowel disease, primary neutropenia and thrombocytopenia, and a surgically repaired atrial septal defect. Her medical history revealed that she was diagnosed with OCA shortly after birth and subsequent eye examinations confirmed the presence of ocular albinism (Figure 1A). The patient was hospitalized for E. coli sepsis in the neonatal period with a white cell count as high as 12 K/µl; her platelet count was 130 K/µl. In her first year of life she suffered from recurrent ear and bladder infections. At three years of age, she was hospitalized for pneumonia and was leukopenic, with a white blood cell count of 2,800 K/µl. Thereafter, she continued to have recurrent lower urinary tract infections and was placed on prophylactic antibiotics.
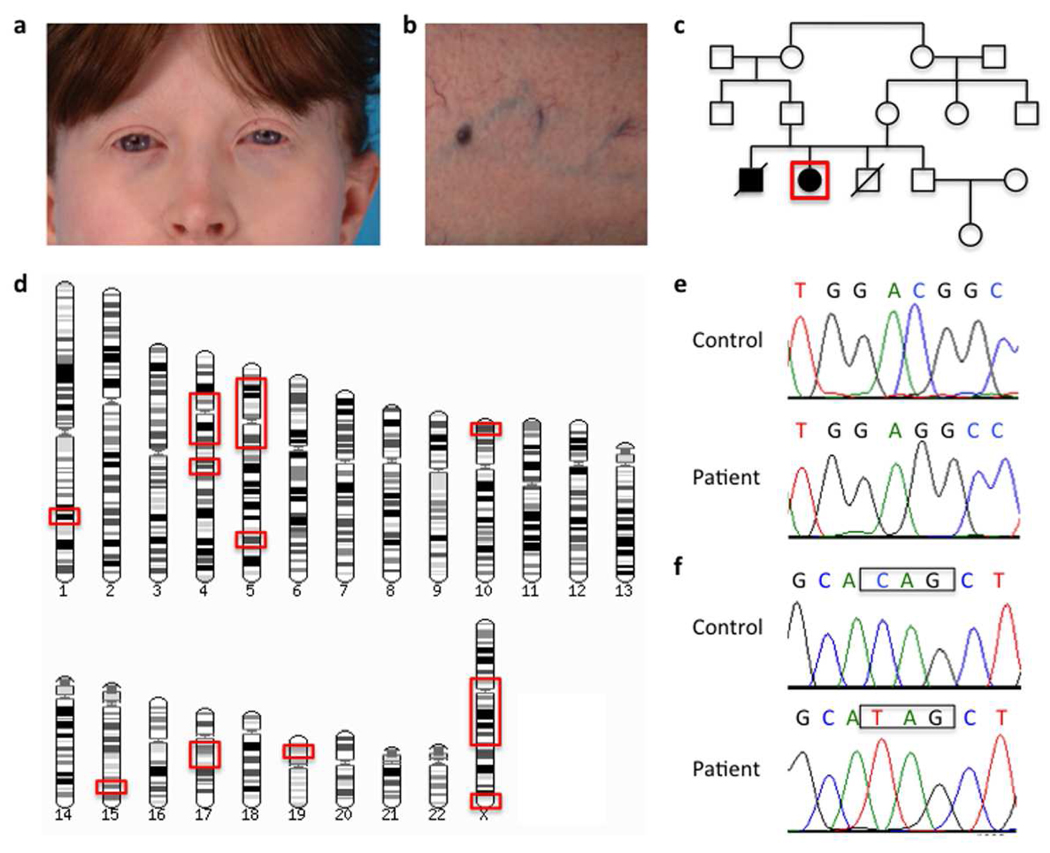
Figure 1. Clinical findings and DNA analysis.
a) Fair skin and classic red hair in our patient, typical for OCA-4 patients.
b) Prominent superficial venous pattern seen in all previously described G6PC3 mutation-positive neutropenia patients.
c) Pedigree showing affected and unaffected individuals; our patient is depicted by the red box. Note the consanguineous background of the family.
d) Regions of homozygosity in the genome of the patient (shown within red boxes).
e) Chromatogram of the mutation in SLC45A2 (c.986delC, p.T329RfsX68).
f) Chromatogram of the mutation in G6PC3 (c.829C>T, p.Q277X). Control sequences are shown for comparison.
Throughout childhood and adolescence, she had symptoms of colitis, chronic abdominal pain, bloody diarrhea, recurrent skin infections, easy bruising, and heavy menstrual periods. She was eventually diagnosed with Crohn’s disease at age 16 and failed medical therapy, resulting in a hemi-colectomy at age 17. Despite this surgery she continued to have symptoms of inflammatory bowel disease and has received antibiotics and immunosuppressive therapy intermittently.
Because of her persistent cytopenias and history of infections, the patient underwent a bone marrow aspiration at age 16 that showed arrested neutrophil development; granulocytic precursors were markedly increased, but matured only to the myelocyte and metamyelocyte stage. The patient began receiving rhG-CSF (Neupogen®), resulting in improved neutrophil counts (Supplementary Table 1) and fewer infections, especially skin abscesses. The absolute neutrophil count at age 29 was recorded as 0; neutrophil release and chemotaxis tests were abnormal. The first report of significant thrombocytopenia occurred at age 29, with a platelet count of 94. Thereafter, platelet counts ranged from 21 to 57 K/µl and averaged 36.6 K/µl. Platelet aggregation studies showed an abnormal secondary aggregation response, with no response to mini-dose ADP or adrenaline. However, whole-mount electron microscopy of her platelets showed the presence of dense bodies. Despite her laboratory abnormalities, the patient did not experience serious bleeding, nor did she receive platelets or DDAVP, even with major surgeries such as an atrial septal defect repair at age 31.
Upon admission to the NIH at age 32, the proband underwent a screening examination that revealed prognathism and abnormal skin findings. She had ecchymoses and petecchiae on her extremities, an eczematous rash on her upper extremities, and fine telangiectasias on her arms and chest. A prominent superficial venous pattern was present on her legs (Figure 1B). There was no evidence of skin infections. Her eye exam was consistent with ocular albinism, and her visual acuities were 20/160 in the right eye and 20/800 in the left eye. Her pulmonary, liver, and renal function tests were normal.
A hematological evaluation was performed to investigate the cytopenias. Her total leukocyte and neutrophil counts were low, but the remainder of her differential was normal, and her red blood cell and reticulocyte indices were normal (Supplementary Table 2). A peripheral smear showed neutropenia; the granulocytes contained primary granules, though some were hypogranular. Rare Dohle bodies were also noted, and the platelets were enlarged.
While the patient was receiving rhG-CSF, the marrow aspirate showed hypercellularity, myeloid hyperplasia with a left shift, and increased megakarocytes. The differential was 75% myeloid precursors, 19% erythroid precursors, 5% lymphocytes, and 1% blasts. Primary granules persisted and some mature granulocytes had cytoplasmic vacuoles. Rare pelgeroid bodies were noted; there was no stainable iron.
Other immunological, autoimmune, and infectious disease studies were normal. She had a minimal decrease in her total IgA level, but other tests of immune competency were normal, and an assay for HIV was negative. Her soluble IL-2R level was also normal. The antinuclear antigen level, extractable nuclear antigen panel, and assays for antibodies to platelets and neutrophils were all negative.
The patient’s parents were first cousins (Fig. 1C). Her brother had albinism, colitis, and a history of recurrent infections. Though his medical history was similar to the proband’s, he was never compliant with treatment and became progressively more ill in his late 20s and early 30s, resulting in a colostomy at age 32. Afterward, he developed recurrent infections and was noncompliant with rhG-CSF therapy. At age 37 he was hospitalized for sepsis and endocarditis, and died shortly afterward from multi-system organ failure due to septic shock.
Molecular diagnostics
Due to the presentation of HPS-2 related symptoms of oculocutaneous albinism, bleeding and neutropenia, all coding exons and flanking intronic sequences of AP3B1 were sequenced using conventional di-deoxy sequencing. No disease causing mutations were detected, and further analysis of mRNA and protein in dermal fibroblasts and melanocytes revealed no change in expression compared to control cells (data not shown). Since our current definition of HPS requires the absence of platelet dense bodies, and this patient had a normal contingent of δ-granules, we considered the possibility that this could represent a new disease.
SNP array analysis
The patient is a result of a first cousin union (Figure 1C), so autosomal recessive inheritance was suspected. Therefore, a SNP-chip microarray was carried out on the patient’s genomic DNA to identify regions of her genome that were homozygous and could harbor the disease-causing mutation. Indeed, approximately 10% of her genome was homozygous, including the centromeric areas of chromosomes 4 and 5 (Figure 1D and Supplementary Figure 1). These data, however, were not very informative, since the regions of homozygosity were so large.
Whole exome sequencing
We next subjected the patient’s DNA to whole exome sequencing. This technique covers the 1.22% of the human genome corresponding to the Consensus Conserved Domain Sequences database (CCDS) and greater than 1000 non-coding RNAs (Gnirke et al., 2009). After alignment of the patient’s DNA sequence fragments to the UCSC reference genome (build 18) there were 62,235 variations and after filtering out known SNPs using dbSNP (http://www.ncbi.nlm.nih.gov/snp/), there were 13,950 variants. This number was reduced to 1502 variations after filtering for variants likely to affect the protein sequence. After further filtering for regions of homozygosity from the SNP-chip data, the number of homozygous variations was reduced to 30 (Supplementary Table 3). From there a candidate gene approach was used to identify potentially pathogenic mutations using the potential detriment (CD_Pred) score as a guide (Johnston et al., 2010).
The most likely mutations identified were a known mutation in G6PC3 (c.829C>T, p.Q277X), which had previously been seen in a French patient (Boztug et al., 2009), and a novel mutation in SLC45A2 (c.987delA, p.A330PfsX68). Both of these mutations were in the expected homozygous state. Conventional di-deoxy sequencing of G6PC3 confirmed the mutations and explained the patient’s neutropenia (Figure 1F), but the mutation identified by whole-exome sequencing for SLC45A2 was incorrect. The actual mutation in SLC45A2 was c.986delC, p.T329RfsX68 (Figure 1E), a previously described mutation in five German OCA-4 patients (Rundshagen et al., 2004). An explanation for the discrepancy would be the mis-alignment of the whole exome reads due to presence of a known SNP (rs2287949) immediately after the deletion.
SLC45A2 and oculocutaneous albinism
To ascertain if the mutation in SLC45A2 was likely to be pathogenic, expression analysis of mRNA and protein was carried out. Quantitative real-time PCR (qRT-PCR) also showed significantly reduced mRNA levels (14.1% ± 1.24% compared to control, p<0.001; Figure 2a). SLC45A2 protein was undetectable in the patient’s melanocytes compared to control cells by western blotting (Figure 2b). Immunofluorescence microscopy of normal human melanocytes revealed punctate cytoplasmic staining for SLC45A2 that co-localized to PMEL17 positive stage II melanosomes (Figure 2c). Since the patient had no detectable SLC45A2 protein expression, no signal was seen in this channel, confirming the specificity of the antibody. Nevertheless, the PMEL17 localization appeared comparable to that of wild-type cells, suggesting that biogenesis and trafficking of PMEL17 positive organelles is not defective.
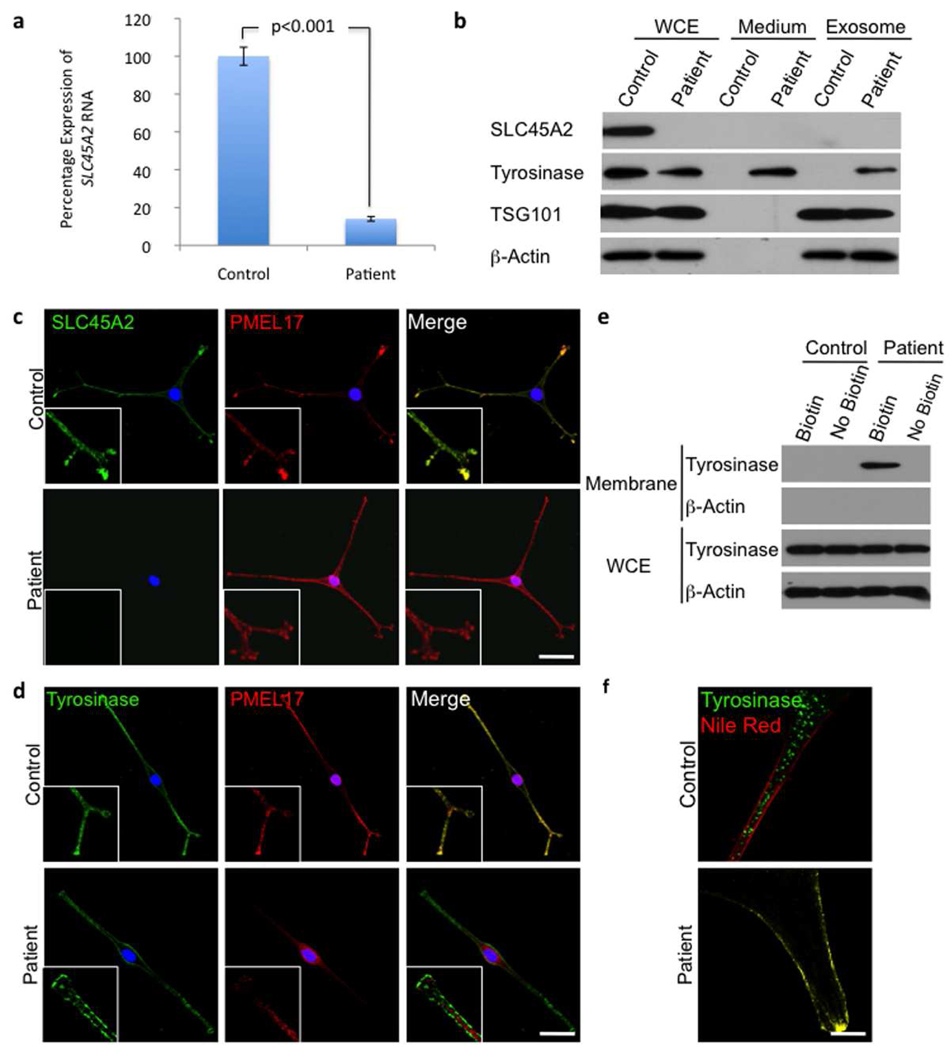
Figure 2. Functional analysis of the OCA-4 phenotype.
a) Quantitative real-time PCR results for SLC45A2 mRNA expression in patient compared to control melanocytes. Values shown are percentage expression of SLC45A2 in patient cells compared to control cells, normalized by ACTB (Error bars = ± 1 SEM, n=3, p<0.001).
b) Western blots of whole cell extracts (WCE), conditioned medium and exosome pellets from control and patient melanocytes. There is no detectable SLC45A2 protein expression and tyrosinase expression is reduced in patient cells compared with control cells. Tyrosinase is present in the medium from patient cells, suggesting that this protein is abnormally secreted from these cells; it is also found in the excreted exosome pellet. The media were collected from an equal number of cells and equal volumes of concentrated proteins were loaded onto the gel. Loading was controlled by assessing β-actin expression in the whole cell lysate of the corresponding cells. The western blot with the exosome marker TSG101 demonstrates that exosomes were isolated from the conditioned medium.
c) Control and patient cells stained for SLC45A2 and the melanosome marker PMEL17, showing co-localization (inserts) of the 2 proteins in control cells. Consistent with the western blot, no staining for SLC45A2 is seen in patient cells but PMEL17 looks comparable to control cells.
d) Control and patient cells stained for tyrosinase and PMEL17, show the co-localization (inserts) of the 2 proteins in control cells. No co-localization is seen in patient cells, and tyrosinase appears predominantly in the plasma membrane. Nuclei are stained with DAPI; scale bar represents 20 µm.
e) Plasma membrane protein biotinylation assay. Only the patient’s cells showed tyrosinase on the plasma membrane. No β-Actin was detected in the membrane fraction (demonstrating purity). Whole cell lysates showed tyrosinase expression in control and patient’s cells (loading controlled by β-actin).
f) High magnification images of control and patient cells stained for tyrosinase and the plasma membrane marker Nile Red. Tyrosinase membrane localization in the patient is evidenced by the co-localization of tyrosinase and Nile Red. Scale bar represents 5 µm.
In normal melanocytes, tyrosinase predominantly localizes to PMEL17 positive cytoplasmic structures, whereas in patient cells there was no co-localization (Figure 2d). No endoplasmic reticulum or trans-Golgi network (TGN) retention of tyrosinase was observed in the patient’s melanocytes, as seen in OCA-1 and OCA-3 cells (Halaban et al., 2000; Toyofuku et al., 2001). In patient cells, tyrosinase was primarily localized to the plasma membrane as observed by immunofluorescence microscopy (Figure 2d) and specifically co-localized with the plasma membrane marker Nile Red (Figure 2f). Furthermore, after separation of plasma membrane proteins by biotinylation before cell lysis, tyrosinase was only found in the membrane fraction of patient cells (Figure 2e). Expression of tyrosinase in the patient’s melanocytes was slightly reduced in whole cell extracts compared to control cells; tyrosinase was detectable only in the growth medium of patient’s cells (Figure 2b). Furthermore, isolating exosomes from the conditioned medium revealed that the secreted tyrosinase was present in exosomes (Figure 2b).
It is possible that tyrosinase and other proteins are being secreted into the medium as a result of the reduced G6PC3 expression. However, Coomassie or silver staining of the gels containing whole cell lysates and conditioned medium revealed that there were no differences in total amounts of secreted proteins between patient and control (Supplementary Figure 2a and b). This suggests that the aberrant protein secretion is tyrosinase-specific, and therefore likely due to the SLC45A2 defect.
G6PC3 and neutropenia
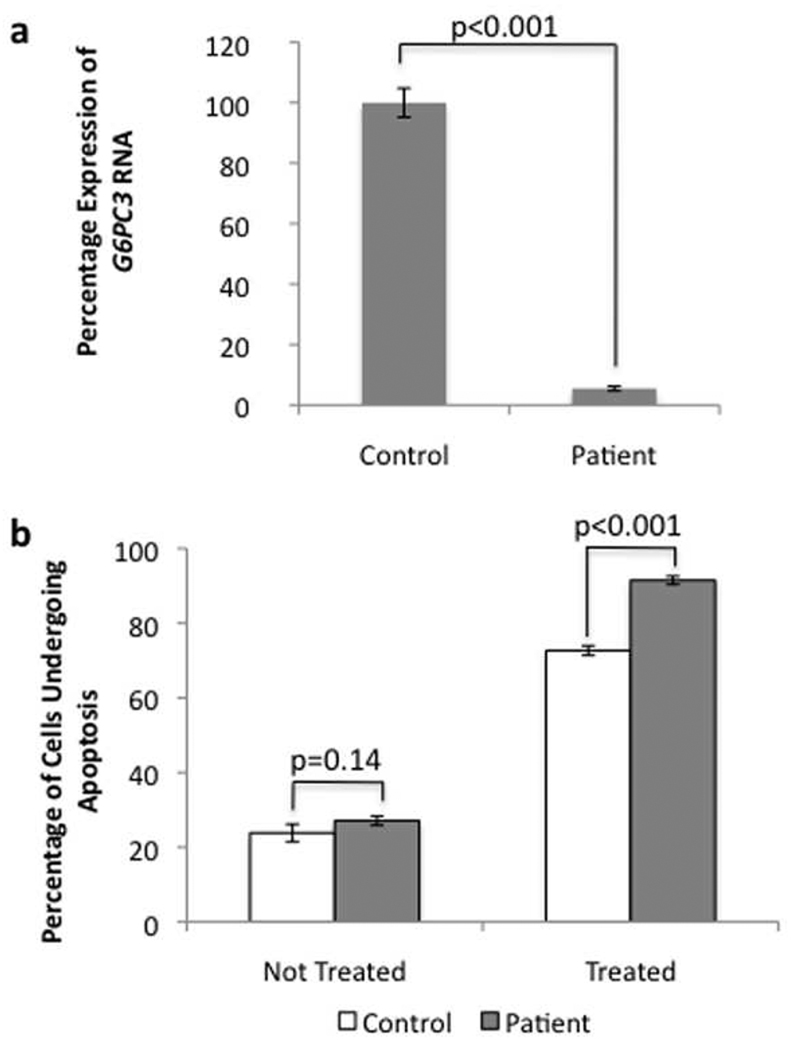
The G6PC3 mutation was further investigated. Quantitative RT-PCR revealed a significant reduction in the expression of the transcript (5.47% ± 0.75% compared to control, p<0.001; Figure 3a), probably due to nonsense-mediated decay. Because G6PC3 deficiency is related to increased spontaneous apoptosis due to ER stress (Boztug et al., 2009; Cheung et al., 2007), we studied the rate of spontaneous apoptosis in our patient’s fibroblasts. The percentage of patient cells that were undergoing apoptosis was higher than in control cells (23.8% ± 2.3 for control, 27.1% ± 1.2 for patient; p=0.14), but this difference was not statistically significant (Figure 3B). However, when the cells were subjected to ER stress using 5mM dithiothreitol, the percentage of the patient’s cells undergoing apoptosis was significantly greater than for control cells (91.5% ± 1.1 vs 72.6% ± 1.3; p<0.001).
Figure 3. Functional analysis of the G6PC3 phenotype.
a) Quantitative real-time PCR results for G6PC3 mRNA expression in patients compared to control melanocytes. Values shown are percentage expression of G6PC3 in patient cells compared to control cells, normalized by ACTB (Error bars = ± 1 SEM, n=3, p<0.001).
b) Flow cytometry results using the Annexin V apoptosis assay for control and patient cells with and without ER stress induction (Error bars = ± 1 SEM, n=3). Without 5mM dithiothreitol treatment no signicant difference can be seen; with treatment, there is a clear increase in apoptosis in patient cells.
Discussion
The application of SNP array analysis, homozygosity mapping, and whole exome sequencing to a patient with OCA and neutropenia allowed us to diagnose two different disorders in a single, unique patient. This was possible in part because the patient was the product of a consanguineous mating. Furthermore, the identified mutations were present in genes previously associated with similar phenotypes (Boztug et al., 2009; Rundshagen et al., 2004), allowing for greater confidence that these mutations are causing the phenotypes seen in our patient.
One of the homozygous variants identified in our patient involved SLC45A2, associated with OCA-4. Reduced mRNA expression, consistent with nonsense-mediated decay, combined with absence of SLC45A2 protein detected by western blotting and immunofluorescence in cultured melanocytes, strongly suggested that the SLC45A2 mutation, c.986delC, is pathogenic. This case represents the first North American patient shown to have OCA-4, and to our knowledge, her melanocytes are the only such cultures obtained from an individual with OCA-4.
Relatively little is known about the function of SLC45A2 in human melanocytes, although mutations in the gene were identified as the cause of OCA-4 nearly a decade ago (Newton et al., 2001). We showed that human SLC45A2-deficient melanocytes exhibited a normal distribution of PMEL17-tagged melanosomes, similar to what was observed in the Underwhite (uw) mouse model of OCA-4 (Costin et al., 2003). We also demonstrated that, in our patient’s cells, tyrosinase was mislocalized to the plasma membrane, suggesting that SLC45A2 functions in the correct trafficking of melanocyte-specific proteins to melanosomes, probably at the level of TGN sorting. Moreover, the presence of tyrosinase, in the growth medium of the patient’s cells, and specifically in exosomes (Figure 2b), suggests that tyrosinase is also prematurely secreted out of the cell without passing through melanosomes. These findings explain the hypopigmentation phenotype of OCA-4, and are consistent with observations in melanocytes from the uw OCA-4 mouse model (Costin et al., 2003). However, our results should be interpreted with caution, since the patient’s cells have a mutation in another disease-causing gene, with unknown effects on melanocytes.
The second mutation in our patient involves G6PC3. Three genes are known to encode enzymes that have glucose-6-phosphatase activity, i.e., G6PC1, G6PC2 and G6PC3. All three proteins have the same enzymatic activity, i.e., removal of the phosphate group from glucose-6-phosphate to produce glucose. G6PC1 is expressed in the liver, kidney and small intestine and catalyzes the essential hydrolysis of glucose-6-phosphate in the gluconeogenic and glycogenolytic pathways (Lei et al., 1993). G6PC2 is expressed only in pancreatic islet cells where it is thought to function in glucose-dependent insulin secretion by controlling free glucose levels (Martin et al., 2001; Petrolonis et al., 2004). In contrast, G6PC3 is ubiquitously expressed, as evidenced by a wide range of affected tissues in patients with G6PC3 mutations. For example, such patients have atrial septal defects and low platelet counts (Boztug et al., 2009), features that were also observed in our patient.
The G6PC3 mutation identified in our patient, c.829C>T, was previously described in a French patient in the heterozygous state (Boztug et al., 2009). However, since it is a null allele, this mutation was not functionally investigated. Furthermore, expression at the protein level could not be assessed because there is no commercially available antibody against the G6PC3. We did demonstrate a reduction in G6PC3 mRNA expression in our patient’s cells (Figure 3a), consistent with nonsense-mediated decay. This finding strongly implies decreased protein expression as well. Functional G6PC3 is required to maintain neutrophil viability; absence of the enzyme results in reduced neutrophil number and increased spontaneous apoptosis due to ER stress (Boztug et al., 2009; Cheung et al., 2007). We demonstrated a higher rate of spontaneous apoptosis in our patient’s fibroblasts compared to control cells. If the same phenomenon were to occur in neutrophils, the decreased G6PC3 protein expression could explain the neutropenia. It could also account for the relative paucity of mature neutrophils in the blood and bone marrow, since the older cells may be more likely to undergo apoptosis than survive.
Our patient’s inflammatory bowel disease could not readily be attributed to either SLC45A2 or G6PC3 mutations, but may be related to another mutated gene due to consanguinity.
Syndromic, multisystemic pigmentary disorders are excellent examples of diseases whose identification can be difficult, especially when consanguinity introduces the possible involvement of multiple genes. One approach in this situation is to perform homozygosity mapping based upon SNP array results, followed by whole exome sequencing.
Materials and Methods
Patient
The patient was enrolled in protocol 76-HG-0238 (www.clinicaltrials.gov, NCT00369421), “Diagnosis and Treatment of Inborn Errors of Metabolism”, approved by the NHGRI Institutional Review Board. Written, informed consent was obtained in adherence to the Helsinki Guidelines.
SNP arrays and dideoxy sequencing
For SNP genotyping, genomic DNA was run on a Human 1M-Duo DNA Analysis BeadChip and the data analyzed using the GenomeStudio software (both Illumina, San Diego, CA). For dideoxy sequencing, primers were designed to cover all coding exons and flanking intronic regions of AP3B1, G6PC3 and SLC45A2 (primer sequences available upon request). Direct sequencing was carried out using the di-deoxy termination method (ABI BigDye Terminator v3.1) on an ABI 3130×l DNA sequencer (Applied Biosystems, Austin, TX). Results were analyzed using Sequencher v4.9 software (Gene Codes Corporation, Ann Arbor, MI). All mutations were verified bi-directionally.
Whole exome sequencing and variant analysis
Solution hybridization exome capture was carried out using the SureSelect Human All Exon System (Agilent Technologies, Santa Clara, CA), which employs biotinylated RNA baits to hybridize to sequences that correspond to exons. The manufacturer’s protocol version 1.0 compatible with Illumina paired end sequencing was used, with the exception that DNA fragment size and quality was measured using a 2% agarose gel stained with SYBR Gold instead of using an Agilent Bioanalyzer. Flowcell preparation and 101-bp paired end read sequencing were carried out as per protocol for the GAIIx sequencer (Illumina Inc, San Diego CA) (Bentley et al., 2008). A single 101-bp paired-end lane on a GAIIx flowcell was used per exome sample to generate sufficient reads to align the sequence. Image analysis and base calling on all lanes of data were performed using Illumina Genome Analyzer Pipeline software (GAPipeline versions 1.4.0 or greater) with default parameters. Reads were aligned to a human reference sequence (UCSC assembly hg18, NCBI build 36) using the package called Efficient Large-scale Alignment of Nucleotide Databases (ELAND). Genotypes were called at all positions where there were high-quality sequence bases using a Bayesian algorithm called the Most Probable Genotype MPG (Teer et al., 2010). A graphical java tool, VarSifter, was developed by the NIH Intramural Sequencing Center (NISC) to view, sort, and filter the variants.
Tissue culture
Primary patient and control fibroblasts and melanocytes were cultured from a forearm skin biopsy. Fibroblasts were grown in high-glucose (4.5 g/l) DMEM medium supplemented with 10% FBS, 2 mM L-glutamine, MEM nonessential amino acid solution and penicillin-streptomycin. Melanocytes were cultured in Ham’s F10 (Invitrogen, Carlsbad, CA), supplemented with 5% fetal calf serum (Gemini Bio-Products, West Sacramento, CA), 5 µg/l basic fibroblast growth factor (Sigma, St. Louis, MO), 10 µg/l Endothelin (Sigma), 7.5 mg/l 3-Isobutyl-1-methylxanthine (Sigma), 30 µg/l Choleratoxin (Sigma), 3.3 µg/l Phorbol 12-myristate 13-acetate (Sigma), 10 ml Pen/Strep/ Glutamine (Invitrogen) and 1 ml Fungizone (Invitrogen).
Protein extraction and immunoblotting
Cells were grown to confluency in 25-cm2 flasks, washed twice with ice-cold PBS and scraped into 250 µl of cell lysis buffer containing 50 mM Tris-HCl (at pH7.5), 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1 mM EDTA, 1 mM EGTA, 0.27 M sucrose, 1% Triton X-100 and Complete, Mini Protease Inhibitor Cocktail (Roche Diagnostics). Cell lysates were centrifuged (15,000 × g, for 15 min at 4°C); supernatants were removed for immunoblotting or plasma-membrane protein biotinylation experiments. Twenty micrograms of total protein, as determined by the Dc Protein assay (BioRad, Hercules, CA), were loaded onto 4–20 % Tris-Glycine gels. After blotting, the PVDF membranes (Invitrogen) were probed with a rabbit polyclonal antibody against SLC45A2 (ProteinTech, Chicago, IL) or mouse monoclonal antibodies against tyrosinase (Clone T311; Santa Cruz Biotechnology, Santa Cruz, CA) and TSG101 (Clone 4A12; Abcam, Cambridge, MA). A mouse monoclonal antibody against β-actin was used as a loading control (Clone AC-15; Sigma). HRP-labeled secondary anti-mouse or anti-rabbit antibodies were used (Amersham Biosciences, Piscataway, NJ). The antigen–antibody complexes were detected with an Enhanced Chemiluminescence (ECL) kit (Amersham Biosciences, Piscataway, NJ).
Conditioned Medium and Exosome Isolation
To prepare serum-free conditioned medium, 3 × 106 cells per flask were washed three times and incubated for 24 h at 37°C with 15 ml serum-free culture medium. The medium was harvested and centrifuged at 1250 × g for 10 min at 4°C The supernatant was ultra-centrifuged at 200,000 × g for 60 minutes to isolate exosomes. The exosome pellets were re-suspended in 2× Lamelli loading buffer. The supernatant was 30× concentrated at 4°C in Centriprep YM-10 tubes (Millipore, Billerica, MA). The resulting protein concentrates, exosome pellets and whole cell extracts from the corresponding cells were subjected to western blotting as above.
Plasma-membrane Protein Biotinylation
For membrane protein biotinylation, control and patient melanocytes were incubated with 500 µl of 0.25 mg/ml EZ-Link Sulfo-NHS-SS-Biotin (Thermo Scientific, Waltham, MA) for 30 min at 4°C. This reaction was quenched by incubating with 1 M NH4Cl for 5 min. Protein was then extracted using the protocol above. The biotin-labeled proteins were separated from the total lysates using streptavidin Dynabeads (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The protein samples were loaded directly onto SDS-PAGE gels for immunoblotting analysis.
qRT-PCR
Total RNA was isolated using RNA-Easy Mini-Kit (Qiagen) according to the manufacturer’s protocol. RNA was treated with a DNase kit (DNA-free) to remove all remaining DNA according to the manufacturer’s protocol (Applied Biosystems, Austin, TX). RNA concentration and purity were measured on the Nanodrop ND-1000 apparatus (Nanodrop Technologies, Wilmington, DE). First strand cDNA was synthesized using a high capacity RNA-to-cDNA kit (Applied Biosystems) according to the manufacturer’s guidelines. Taqman gene expression master mix reagent and Assays-On-Demand (Applied Biosystems) were obtained for G6PC3 (Assay ID Hs00292720_m1), SLC45A2 (Assay ID Hs01125486_m1) and a control gene, ACTB (Assay ID Hs99999903_m1). Q-RT-PCR was performed using 100 ng cDNA, on an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems) using the comparative CT method (ΔΔCT); this method measures relative gene expression (Livak and Schmittgen, 2001). The cycling conditions were as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles at 95° C for 15 s and 60° C for 60 s.
Immunofluorescence microscopy
Cells were grown in 4-well chamber slides and fixed using 4% paraformaldehyde and permeabilized using 0.1% Triton-X-100. Antibodies against SLC45A2 and tyrosinase were purchased for immunoblotting. A mouse monoclonal antibody (clone HMB45) and a rabbit polyclonal against PMEL17 were purchased from LifeSpan BioSciences (Seattle, WA) and Neomarkers (Fremont, CA), respectively. Alexafluor 488 and 555 secondary antibody conjugates and Nile Red were purchased from Invitrogen, and nuclei were counterstained with DAPI. Cells were imaged with a Zeiss 510 META confocal laser-scanning microscope with the pinhole set to 1 Airy unit. A series of optical sections were collected from xy plane and merged into maximum projection images.
Apoptosis assay
Spontaneous apoptosis was measured using the Annexin V and propidium iodide apoptosis assay (Pigault et al., 1994). The Annexin V-FITC apoptosis detection kit was purchased from Sigma and the assay carried out according to the manufacturer’s instructions with 5 × 105 fibroblast cells per treatment group. ER stress was induced with 5 mM dithiothreitol (DTT, Sigma) for 2 h at 37°C. The samples were sorted on a Becton Dickenson FACSCalibur flow cytometer and with green and or red fluorescence were counted and analyzed using FlowJo software.
Supplementary Material
Acknowledgments
We thank S. Anderson, R. Hess, R. Fischer and H. Dorward for their help with the study. This study was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Anikster Y, Huizing M, White J, et al. Mutation of a new gene causes a unique form of Hermansky-Pudlak syndrome in a genetic isolate of central Puerto Rico. Nat Genet. 2001;28:376–380. doi: 10.1038/ng576. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissy RE, Zhao H, Oetting WS, et al. Mutation in and lack of expression of tyrosinase-related protein-1 (TRP-1) in melanocytes from an individual with brown oculocutaneous albinism: a new subtype of albinism classified as "OCA3". Am J Hum Genet. 1996;58:1145–1156. [PMC free article] [PubMed] [Google Scholar]
- Bonilla MA, Gillio AP, Ruggeiro M, et al. Effects of recombinant human granulocyte colony-stimulating factor on neutropenia in patients with congenital agranulocytosis. N Engl J Med. 1989;320:1574–1580. doi: 10.1056/NEJM198906153202402. [DOI] [PubMed] [Google Scholar]
- Boztug K, Appaswamy G, Ashikov A, et al. A syndrome with congenital neutropenia and mutations in G6PC3. N Engl J Med. 2009;360:32–43. doi: 10.1056/NEJMoa0805051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boztug K, Welte K, Zeidler C, et al. Congenital neutropenia syndromes. Immunol Allergy Clin North Am. 2008;28:259–275. doi: 10.1016/j.iac.2008.01.007. vii–viii. [DOI] [PubMed] [Google Scholar]
- Cheung YY, Kim SY, Yiu WH, et al. Impaired neutrophil activity and increased susceptibility to bacterial infection in mice lacking glucose-6-phosphatase-beta. J Clin Invest. 2007;117:784–793. doi: 10.1172/JCI30443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costin GE, Valencia JC, Vieira WD, et al. Tyrosinase processing and intracellular trafficking is disrupted in mouse primary melanocytes carrying the underwhite (uw) mutation. A model for oculocutaneous albinism (OCA) type 4. J Cell Sci. 2003;116:3203–3212. doi: 10.1242/jcs.00598. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Shotelersuk V, Aguilar RC, et al. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- Devriendt K, Kim AS, Mathijs G, et al. Constitutively activating mutation in WASP causes X-linked severe congenital neutropenia. Nat Genet. 2001;27:313–317. doi: 10.1038/85886. [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronskov K, Ek J, Brondum-Nielsen K. Oculocutaneous albinism. Orphanet J Rare Dis. 2007;2:43. doi: 10.1186/1750-1172-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaban R, Svedine S, Cheng E, et al. Endoplasmic reticulum retention is a common defect associated with tyrosinase-negative albinism. Proc Natl Acad Sci U S A. 2000;97:5889–5894. doi: 10.1073/pnas.97.11.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz M, Benson KF, Person RE, et al. Mutations in ELA2, encoding neutrophil elastase, define a 21-day biological clock in cyclic haematopoiesis. Nat Genet. 1999;23:433–436. doi: 10.1038/70544. [DOI] [PubMed] [Google Scholar]
- Huizing M, Helip-Wooley A, Westbroek W, et al. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu Rev Genomics Hum Genet. 2008;9:359–386. doi: 10.1146/annurev.genom.9.081307.164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Teer JK, Cherukuri PF, et al. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am J Hum Genet. 2010;86:743–748. doi: 10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RA, Oetting WS, Creel D, et al. Abnormalities of pigmentation. In: Rimoin DL, Connor JM, Pyeritz RE, editors. Emery and Rimoin's Principles and Practice of Medical Genetics. New York: Churchill Livingstone; 2002. pp. 3731–3785. [Google Scholar]
- Klein C, Grudzien M, Appaswamy G, et al. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease) Nat Genet. 2007;39:86–92. doi: 10.1038/ng1940. [DOI] [PubMed] [Google Scholar]
- Lei KJ, Shelly LL, Pan CJ, et al. Mutations in the glucose-6-phosphatase gene that cause glycogen storage disease type 1a. Science. 1993;262:580–583. doi: 10.1126/science.8211187. [DOI] [PubMed] [Google Scholar]
- Li W, Zhang Q, Oiso N, et al. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nat Genet. 2003;35:84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martin CC, Bischof LJ, Bergman B, et al. Cloning and characterization of the human and rat islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP) genes. J Biol Chem. 2001;276:25197–25207. doi: 10.1074/jbc.M101549200. [DOI] [PubMed] [Google Scholar]
- Morgan NV, Pasha S, Johnson CA, et al. A germline mutation in BLOC1S3/reduced pigmentation causes a novel variant of Hermansky-Pudlak syndrome (HPS8) Am J Hum Genet. 2006;78:160–166. doi: 10.1086/499338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton JM, Cohen-Barak O, Hagiwara N, et al. Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet. 2001;69:981–988. doi: 10.1086/324340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Bailin T, Fukai K, et al. Positional cloning of a gene for Hermansky-Pudlak syndrome, a disorder of cytoplasmic organelles. Nat Genet. 1996;14:300–306. doi: 10.1038/ng1196-300. [DOI] [PubMed] [Google Scholar]
- Otto EA, Hurd TW, Airik R, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Person RE, Li FQ, Duan Z, et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrolonis AJ, Yang Q, Tummino PJ, et al. Enzymatic characterization of the pancreatic islet-specific glucose-6-phosphatase-related protein (IGRP) J Biol Chem. 2004;279:13976–13983. doi: 10.1074/jbc.M307756200. [DOI] [PubMed] [Google Scholar]
- Pigault C, Follenius-Wund A, Schmutz M, et al. Formation of two-dimensional arrays of annexin V on phosphatidylserine-containing liposomes. J Mol Biol. 1994;236:199–208. doi: 10.1006/jmbi.1994.1129. [DOI] [PubMed] [Google Scholar]
- Rinchik EM, Bultman SJ, Horsthemke B, et al. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature. 1993;361:72–76. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- Rundshagen U, Zuhlke C, Opitz S, et al. Mutations in the MATP gene in five German patients affected by oculocutaneous albinism type 4. Hum Mutat. 2004;23:106–110. doi: 10.1002/humu.10311. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Li W, Zhang Q, et al. Hermansky-Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light-ear gene. Nat Genet. 2002;30:321–324. doi: 10.1038/ng835. [DOI] [PubMed] [Google Scholar]
- Teer JK, Bonnycastle LL, Chines PS, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–1431. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, Mullikin JC. Exome sequencing: the sweet spot before whole genomes. Hum Mol Genet. 2010;19:R145–R151. doi: 10.1093/hmg/ddq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y, Takeda A, Okinaga S, et al. Human oculocutaneous albinism caused by single base insertion in the tyrosinase gene. Biochem Biophys Res Commun. 1989;164:990–996. doi: 10.1016/0006-291x(89)91767-1. [DOI] [PubMed] [Google Scholar]
- Toyofuku K, Wada I, Valencia JC, et al. Oculocutaneous albinism types 1 and 3 are ER retention diseases: mutation of tyrosinase or Tyrp1 can affect the processing of both mutant and wild-type proteins. FASEB J. 2001;15:2149–2161. doi: 10.1096/fj.01-0216com. [DOI] [PubMed] [Google Scholar]
- Welte K, Zeidler C, Dale DC. Severe congenital neutropenia. Semin Hematol. 2006;43:189–195. doi: 10.1053/j.seminhematol.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhao B, Li W, et al. Ru2 and Ru encode mouse orthologs of the genes mutated in human Hermansky-Pudlak syndrome types 5 and 6. Nat Genet. 2003;33:145–153. doi: 10.1038/ng1087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.