Abstract
Chromosome painting is one of the most powerful and spectacular tools of modern molecular cytogenetics, enabling complex analyses of nuclear genome structure and evolution. For many years, this technique was restricted to the study of mammalian chromosomes, as it failed to work in plant genomes due mainly to the presence of large amounts of repetitive DNA common to all the chromosomes of the complement. The availability of ordered, chromosome-specific BAC clones of Arabidopsis thaliana containing relatively little repetitive genomic DNA enabled the first chromosome painting in dicotyledonous plants. Here, we show for the first time chromosome painting in three different cytotypes of a monocotyledonous plant—the model grass, Brachypodium distachyon. Possible directions of further detailed studies are proposed, such as the evolution of grass karyotypes, the behaviour of meiotic chromosomes, and the analysis of chromosome distribution at interphase.
Introduction
Chromosome painting (CP), also known as chromosomal in situ suppression, is one of the most powerful and spectacular tools of molecular cytogenetics. It refers to the method of visualisation of specific chromosome regions or entire chromosomes using fluorescence in situ hybridisation (FISH) with chromosome-specific DNA probes (Pinkel et al. 1988; Ried et al. 1998). For many years, this technique was successfully applied only to studies of animal chromosomes, giving unprecedented insight into the structural and functional compartmentalisation of the nucleus (Cremer et al. 2008; Cremer et al. 1993; Kurz et al. 1996). It has also been widely used for molecular cytotaxonomy, for example of primates (Muller et al. 1999; Wienberg et al. 1990), mutagenicity testing (Marshall and Obe 1998) and radiation biology (Gray et al. 1992). High resolution CP-based analyses have become especially important to pre-, neo- and postnatal clinical diagnostics of various chromosomal aberrations associated with human diseases and malignancies (Gray and Pinkel 1992; Popp et al. 1991; Tkachuk et al. 1991; Weise et al. 2003; Xu et al. 2010).
For more than a decade, efforts to apply CP in plants failed to yield satisfactory results, which was attributed to the presence of vast amounts of ubiquitous repetitive DNA in most plant genomes (Schubert et al. 2001). The first true painting of plant chromosomes was achieved by Lysak et al. (2001) in the dicotyledonous model organism, Arabidopsis thaliana, whose nuclear genome had just been sequenced (Arabidopsis Genome Initiative 2000). The availability of genome sequence and ordered libraries of genomic DNA in BAC (Bacterial Artificial Chromosome) vectors, along with some features of A. thaliana genome, such as its compactness, low amount of repetitive DNA and low number of chromosomes, were key prerequisites for successful painting of its chromosomes. Initially restricted to the chromosomes of A. thaliana (Pecinka et al. 2004), CP in later studies was also successfully applied to other closely related members of the Brassicaceae. This comparative chromosome painting (CCP) allowed unprecedented analyses of the evolution of the genomes of this group of plants at the chromosomal level (Lysak et al. 2006; Lysak et al. 2010; Mandakova and Lysak 2008).
In contrast to the dicots, chromosome painting has not been applied so far to any monocotyledonous plant. Despite the availability of its long-published genomic sequence (Goff et al. 2002), no chromosome painting has been performed in rice. Another species of Poaceae, Brachypodium distachyon, emerged several years ago as a model system for temperate cereals and grasses. Like A. thaliana, B. distachyon possesses numerous ‘model’ attributes, including inter alia a small (∼300 Mb) genome with a low (x = 5) basic chromosome number, small physical stature, self-fertility, a short life cycle and undemanding growth requirements (Draper et al. 2001; Garvin et al. 2008). The genome of B. distachyon has been extensively studied at the chromosomal level using fluorescence in situ hybridisation (FISH) with a wide range of DNA probes, which enabled unambiguous identification of all the chromosomes and their arms of the complement (Hasterok et al. 2004; Hasterok et al. 2006).
The well-established cytogenetic platform, combined with resources such as BAC DNA libraries and bioinformatic data from the recently completed genome sequencing project (Febrer et al. 2010; Gu et al. 2009; International Brachypodium Initiative 2010), opened up the possibility of painting for the first time the chromosomes of a monocot species. In this paper, we present the current status of CP in B. distachyon, which allows reliable recognition and tracking of individual chromosome regions during different stages of meiosis. Additionally, we show that CCP with BAC pools representing individual B. distachyon chromosomes can be used as an effective tool for studying karyotype evolution of its close relatives in the genus Brachypodium. Possible future prospects are also discussed.
Materials and methods
Plant material and chromosome preparations
Three cytotypes of B. distachyon were used in this study. Detailed information on the plant material is provided in Table 1. Seeds were sown at high density in compost. All plants were grown as described in Jenkins and Hasterok (2007). The plants of cytotypes ABR114 and ABR113 did not require vernalisation and usually reached the generative stage of their life cycle within 1 month after sowing.
Table 1.
The original identities, sources, origins and chromosome numbers of the B. distachyon material used in this study
| Species | Accession no. | Origin | Source | 2n |
|---|---|---|---|---|
| B. distachyon | Bd21 | Iraq | a | 10 |
| ‘B. distachyon’ * | ABR114 | Spain | b | 20 |
| ‘B. distachyon’ * | ABR113 | Portugal | b | 30 |
a US Department of Agriculture–National Plant Germplasm System, USA, b Institute of Biological, Environmental and Rural Sciences, Aberystwyth University, UK
*According to our previous cytomolecular studies (Hasterok et al. 2004; Hasterok et al. 2006) ABR114 and ABR113 represent, respectively, a different unnamed diploid and an allotetraploid species within the Brachypodium genus. As their proposed taxonomical status is not yet officially accepted, we refer in this paper to ABR114 and ABR113 as the cytotypes of ‘B. distachyon’. The cytotype with 2n = 10 chromosomes is referred to as B. distachyon without apostrophes
Mitotic chromosome preparations were made using the methodology described by Hasterok et al. (2006). Preparation of anther tissue for meiotic chromosome squashes was adopted from Jenkins and Hasterok (2007) with minor modifications. In brief, immature inflorescences were collected, fixed immediately in fresh 3:1 absolute methanol:glacial acetic acid mixture for 3 × 24 h at room temperature (RT) and stored at −20°C until required. Individual anthers were isolated and washed in 10 mM citric acid–sodium citrate buffer and digested enzymatically for 2 h at 37°C in a mixture comprising 10% (v/v) pectinase (Sigma), 0.65% (w/v) cellulase Onozuka R-10 (Serva), 0.5% (w/v) cellulase (Calbiochem), 0.15% (w/v) cytohelicase and 0.15 % (w/v) pectolyase in 10 mM citrate buffer. Several anthers per slide were squashed in drops of 45% acetic acid, frozen, postfixed in 4% formaldehyde for 10 min, washed briefly in distilled water and air dried.
Selection of BAC clones for chromosome painting
BAC clones used for CP were selected from the five assemblies of FPCs (FingerPrinted Contigs) previously aligned to the B. distachyon karyotype (Febrer et al. 2010; International Brachypodium Initiative 2010). Clones from centromeric and pericentromeric regions were excluded from the painting experiments as they were likely to contain large amounts of highly repetitive and dispersed DNA sequences which might cause unspecific hybridisation signals. For the same reason, the BACs constituting the assemblies were examined for repetitive DNA content. Low repeat BAC clones were identified using the same method as described by Febrer et al. (2010). With a few exceptions, clones chosen for painting experiments contained less than 30% of repeats. The number of BACs selected for each arm of every chromosome in the complement is given in Table 2. The total length of BAC clones spanning a chromosome arm ranged from 9.7% of total arm length for chromosome 5 short (top) arm (Bd5S) to nearly 34% for the long (bottom) arm of B. distachyon chromosome 3 (Bd3L; Table 2).
Table 2.
Characteristics of the BAC pools used for painting B. distachyon chromosome arms
| Chromosome | Bd1 | Bd2 | Bd3 | Bd4 | Bd5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Arm | S | L | S | L | S | L | S | L | S | L |
| Number of clones selected | 70 | 72 | 26 | 29 | 42 | 54 | 20 | 39 | 4 | 19 |
| Average distance between neighbouring clones (kbp) | 347 | 358 | 874 | 707 | 385 | 377 | 594 | 406 | 381 | 543 |
| Percentage of arm length covered by BAC inserts | 28.1 | 28.7 | 13.1 | 12.3 | 24.6 | 33.9 | 12.8 | 19.5 | 9.7 | 13.5 |
Selected BACs were divided into pools of eight to ten clones each. Instead of isolating DNA from each clone separately, liquid cultures of bacteria carrying clones belonging to the same pool were mixed and DNA was isolated. Individual pools were labelled with either digoxigenin- or tetramethyl-rhodamine-dUTP and in order to verify their position on B. distachyon chromosomes, hybridised independently to pachytene or zygotene nuclei (data not shown). In most cases, the hybridisation sites of the selected clones corresponded to their predicted positions on the FPC-derived physical map. The pool assigned to the interstitial region of the Bd4S yielded dispersed signals in the entire chromosome set, probably due to the presence of highly repetitive and ubiquitous DNA in some of the clones. BACs constituting this pool were then examined separately using FISH, and the clones responsible for cross-hybridisation were identified and removed from the pool before subsequent experiments.
Probe labelling and fluorescence in situ hybridisation
BAC DNA was isolated using a standard alkaline method and labelled by nick translation with digoxigenin-11-dUTP (Roche) for short chromosome arms and with tetramethyl-rhodamine-5-dUTP (Roche) for long chromosome arms as described in Hasterok et al. (2002) with the exception of the experiment shown on Figs. 1f and 3. The list of BACs comprising the pools for individual chromosomes may be requested from the authors.
Fig. 1.
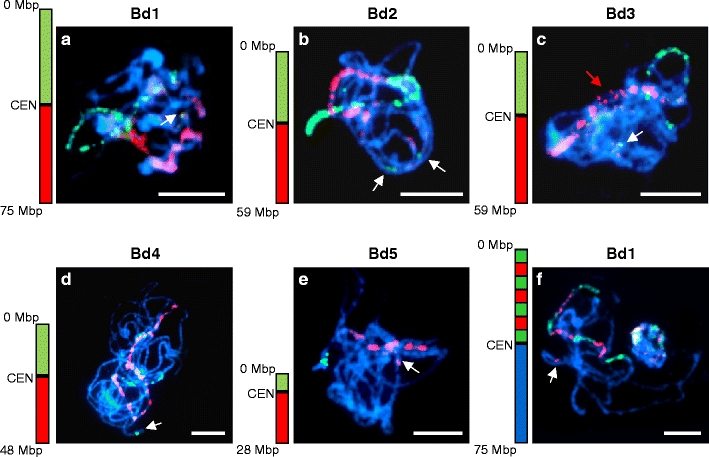
Chromosome painting in PMCs of B. distachyon at late zygotene/pachytene. a–e Painting using BAC pools spanning short (green) and long (red) arms of a given chromosome. The ideograms next to the photographs indicate the approximate physical positions of the pools used. CEN = centromere. a chromosome Bd1; b chromosome Bd2; c chromosome Bd3; d chromosome Bd4; e chromosome Bd5; f painting of the short arm of chromosome Bd1 with 70 BAC clones arranged into seven alternating pools. White arrows indicate additional unexpected signals. The red arrow in c indicates the position of an unpaired chromosome region on Bd3. All chromosomes were counterstained with DAPI. Bars = 5 μm
Fig. 3.
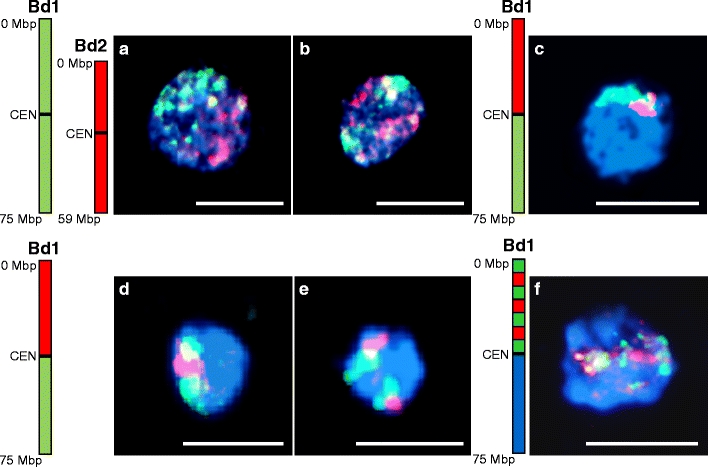
Identification of chromosome territories in interphase nuclei of B. distachyon anther tissue using chromosome painting. a–b Simultaneous painting of chromosomes Bd1 (green) and Bd2 (red). a Interphase nucleus showing association of homologous chromosomes while territories occupied by heterologues remain separated; b interphase nucleus showing separation of all four chromosome territories; c–e painting of the short (red) and long (green) arm of chromosome Bd1; c association of territories of both arms; d association of territories of the short arm; e separation of territories occupied by chromosome Bd1; f chromosome painting of interphase nuclei with seven alternating BAC pools specific to different regions of Bd1 short arm: the result is visible as a mixture of red and green signals and the territories occupied by particular pools are not discernible. The drawings next to the photographs indicate approximate positions of the BAC pools used for painting. CEN = centromere. All chromosomes were counterstained with DAPI. Bars = 5 μm
The FISH procedure was carried out according to the protocol of Jenkins and Hasterok (2007) with minor modifications. The BAC DNAs were pooled, precipitated and dissolved in hybridisation mixture consisting of 50% deionised formamide and 10% dextran sulphate in 2× saline sodium citrate (SSC). Chromosome preparations and pre-denatured (10 min at 75°C) hybridisation mixtures were denatured together for 4.5 min at 73°C and allowed to hybridise in a humid chamber for about 20 h at 37°C. Post-hybridisation washes were performed in 10% formamide in 0.1× SSC at 42°C, which is equivalent to 79% stringency. The hybridisation signals were detected by anti-digoxigenin-fluorescein conjugated antibodies (Roche) or—in the case of tetramethyl-rhodamine-5-dUTP—directly visualised. For comparative chromosome painting, the hybridisation time was occasionally extended up to 48–55 h. The chromosomes were mounted and counterstained in Vectashield (Vector Laboratories) containing 2.5 mg/ml of 4′,6-diamidino-2-phenylindole (DAPI; Serva).
All microphotographs were acquired using a monochromatic CCD camera attached to a wide-field epifluorescence microscope and digitally processed and superimposed using Wasabi (Hamamatsu Photonics), Photoshop (Adobe) and Picture Publisher (Micrografx/Corel) software.
Results
Painting of B. distachyon chromosomes
To paint a chromosome along its entire length, pools of BAC DNA were combined into arm-specific probes. Discrimination between the arms of a chromosome was enabled by differential labelling of the probes specific for its short and long arm. Labelled DNA was mapped to pachytene or late zygotene spreads which provide the best resolution for physical mapping by fluorescence in situ hybridisation. Every painting probe gave a strong and distinct hybridisation signal allowing unambiguous identification and tracking of homologous chromosome pairs (Fig. 1). In some cases, unspecific signals on other chromosomes were also observed. Most of such signals were faint and presumably could be attributed to the presence of repetitive DNA in the painting probe. However, signals of high fluorescence intensity also appeared in chromosomal positions in addition to those expected (Fig. 1a–f; white arrows). This suggests misalignment of several clones in the FPC contig map. Due to exclusion from the experiments of high repeat clones from the pools, the BACs used for CP rarely overlapped, but usually were separated from each other by tens or hundreds of kilobases. These breaks in the tiling path of clones are visualised on chromosomes as blue, DAPI-stained gaps without signals. The exclusion of centromeric and pericentromeric BACs, as well as clones containing 45S rDNA repeats, was particularly noticeable as large unlabelled chromosomal segments (Fig. 1c, e). The discrimination between different chromosome regions was achieved by differential labelling of BAC pools. Simultaneous hybridisation of 70 BAC clones arranged into seven alternating pools resulted in a banding pattern of red and green segments along the short arm of chromosome Bd1 (Fig. 1f). Similar results were obtained after differential labelling of BAC pools assigned to other chromosomes of the karyotype (data not shown). The size of a particular block was always consistent with the length of the BAC assembly covered by the respective pool, and the sequential order of the pools along the chromosome was easily discernible.
A pool of 40 clones corresponding to the distal part of the short arm of chromosome Bd1 was hybridised to chromosomes at different stages of meiosis (Fig. 2). The low condensation and frequent entanglement of the chromatin at zygotene (Fig. 2a) often made it difficult to track the painted region among other chromosomes. At pachytene (Fig. 2b), the arrangement of the labelled part of chromosome Bd1 was much easier to follow. Gaps resulting from excluded BACs were visible at both zygotene and pachytene nuclei. During later stages of meiosis (Fig. 2c–e), the gaps were no longer detectable due to increasing condensation of the chromosomes. Similar observations were made when the same painting probe was applied to mitotic metaphase chromosomes (Fig. 2f), which are 30–40 times as condensed as pachytene bivalents.
Fig. 2.
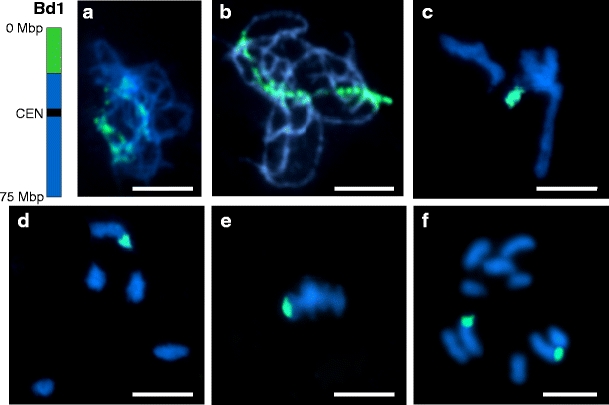
Tracking a distal region of Bd1 short arm (green) at different stages of meiosis and mitotic metaphase using chromosome painting with a BAC pool containing 40 clones spanning 19.5 Mbp. The ideogram next to the photographs indicates the approximate physical position of the pool on the chromosome. CEN = centromere. a zygotene; b pachytene; c diplotene; d diakinesis; e meiotic metaphase I; f mitotic metaphase. All chromosomes were counterstained with DAPI. Bars = 5 μm
In interphase nuclei of anther tissues, chromosome painting identified the territories occupied by different chromosomes. Simultaneous hybridisation of differentially labelled chromosomes Bd1 and Bd2 revealed frequent association of homologues (Fig. 3a), while heterologous chromosomes were usually separated (Fig. 3a–b). Analyses of the respective positions of the short and long arms of Bd1 indicated that association of both (Fig. 3c) or only one chromosome arm (Fig. 3d) is common in interphase nuclei, although two distinct nuclear domains corresponding to each of the Bd1 homologues were also occasionally observed (Fig. 3e). When seven alternating BAC pools specific for different regions of the Bd1 short arm were used as a hybridisation probe, the result was visible as a mixture of green and red signals that could not be attributed to particular pools (Fig. 3f).
Comparative chromosome painting
The feasibility of using the BAC pools for comparative physical mapping of other Brachypodium species was tested by cross-hybridising pools that represented each chromosome of Bd21 to pachytene bivalents of two ‘B. distachyon’ cytotypes: diploid ABR114 (2n = 20) and allotetraploid ABR113 (2n = 30). The probe comprising BAC clones specific for chromosome Bd1 identified two bivalents in ABR114 PMCs at pachytene (Fig. 4a1). Both bivalents were painted green and red, corresponding to the short and long arms of Bd1, respectively. In both bivalents, terminal regions of the bivalents proximal to the green segments were unlabelled. Hybridisation of the same BAC pools to ABR113 highlighted three bivalents (Fig. 4b1). The painted bivalents differ in size, one (designated I) was at least twice as long as the other two (II and III). The hybridisation pattern on the three homoeologues was similar, with the green signal corresponding to the short arm of Bd1 abutting the red signal representing the long arm of Bd1. However, whilst the bivalents numbered I and II were painted along their entire length, bivalent number III was only partially decorated by the probe for Bd1. The border between labelled and unlabelled regions of bivalent II is clearly visible (Fig. 4b1, red arrow), but the length of the non-painted chromosome segment was difficult to discern due to the entanglement of the pachytene chromosomes.
Fig. 4.
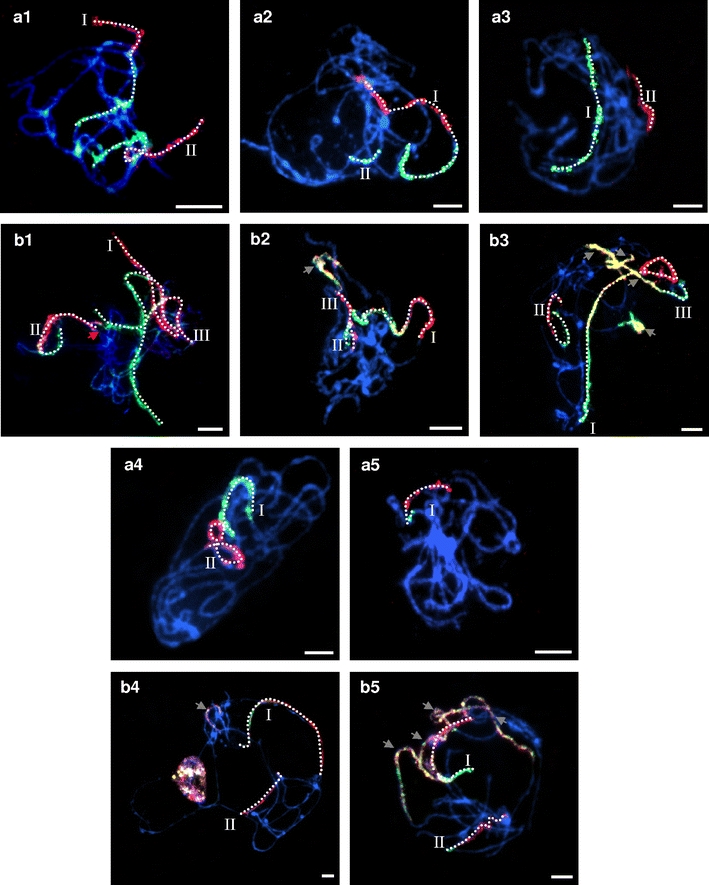
Comparative chromosome painting of ‘B. distachyon’ cytotypes ABR114 and ABR113 using BAC pools spanning the short (green) and long (red) arms of the five B. distachyon chromosomes. a1–a5 chromosome painting of ABR114 pachytene chromosomes; b1–b5 chromosome painting of ABR113 pachytene. Roman numerals indicate homoeologues of a given chromosome in the cytotypes. The red arrow in b1 indicates the border between labelled and unlabelled regions of the homoeologue III in ABR113. The labelled bivalent axes have been highlighted with dots to emphasise continuity. Grey arrows indicate non-specific hybridisation. All chromosomes were counterstained with DAPI. Bars = 5 μm
Painting with probes specific for the long and short arms of Bd2 revealed two bivalents in ABR114 (Fig. 4a2). One of them (designated I) has long tracts of green and red signal, representing the short and long arms of Bd2, respectively. A short fragment of another bivalent (II) was painted green only. Analysis of the Bd2 hybridisation pattern to ABR113 (Fig. 4b2) showed contiguous painting of the entire arms of one bivalent (I). Additional short green (II) and short red (III) segments were found to belong to the same bivalent. An additional pair of bivalents is consistently labelled as a mosaic of green, red and colocalising yellow signals.
The painting probes comprising BAC clones specific for short and long arm of chromosome Bd3 identified two bivalents in ABR114 PMCs at pachytene (Fig. 4a3). One of them (I) hybridised only with the pool representing the short arm of Bd3, whilst the other (II) only with the pool for the long arm. Bd3-specific probes painted three pachytene bivalents of ABR113 (Fig. 4b3). Bivalents I and II differed significantly in terms of signal length, but both had green and red segments corresponding to the short and long arms of Bd3. The former also has an interstitial yellow tract, presumably delimiting sequences in common between the two pools. Bivalent III has a short green segment adjoining a longer yellow tract, presumably representing non-specific repeat sequences common to the two arms. A short green and yellow tract was also observed.
Chromosome painting of Bd4 probes to ABR114 chromosomes highlighted two bivalents (Fig. 4a4), one (I) hybridising with the Bd3 short arm probe, and the other (II) with the long arm probe. The two tracts do not belong to the same bivalent. The probe comprising BAC clones specific for Bd4 identified only two bivalents in ABR113 (Fig. 4b4). One bivalent (I), presumably derived from a B. distachyon-like genome, hybridised with both the Bd4-short arm and Bd4-long arm probe, whilst the other (II) showed only red fluorescence.
Bd5-specific paints hybridised with only one bivalent of ABR114 (Fig. 4a5; I), whereas they hybridised to two bivalents in ABR113 (Fig. 4b5; I and II). In addition, some of the other bivalents comprise patchworks of green, red and yellow signals revealing the sites of sequences common to several of the chromosomes of the complement.
A summary of the pattern of hybridisation of the five chromosome-specific paints to the two ‘B. distachyon’ cytotypes is shown in Fig. 5. Since the continuity of unlabelled bivalents cannot be tracked with certainty in most cases, the diagrammatic representations of such bivalents have been left open ended. The relative lengths of bivalents in the three cytotypes is subject to stage-specific variation.
Fig. 5.
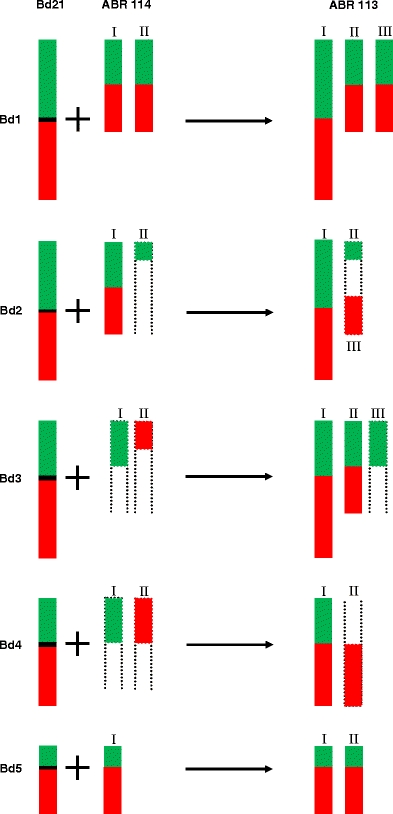
Diagrammatic summary of the pattern of chromosome painting in pachytene bivalents of Bd21, ABR114 and ABR113. Hybridisation with short-arm probes is shaded green, and long arm probes red. Centromeres are marked with horizontal black bars. Bivalents that cannot be traced from end to end have dotted open-ended lines. Relative lengths are only approximate. Roman numerals correspond to those in Fig. 4
Discussion
CP in plants has not been demonstrated since the first painting of A. thaliana chromosome 4 by Lysak et al. (2001) and subsequent painting of the rest of the complement as well as the complements of related species within the Brassicaceae (Lysak and Lexer 2006). The main requirement for successful chromosome painting is the availability of a contiguous tiling path of sequenced large-insert clones integrated with the karyotype of a particular species. Moreover, the effectiveness and specificity of the technique depend upon eliminating highly repetitive DNA sequences from the painting probe. CP is potentially useful, therefore, only in species with small genomes and with repetitive DNA localised in pericentromeric regions. B. distachyon, a recently established model grass species, has one of the smallest genomes in the Poaceae and very low repetitive DNA content. Recently developed resources for investigating its genome, including genetic, physical and cytogenetic maps fully integrated with genome sequence assemblies (Febrer et al. 2010; International Brachypodium Initiative 2010), have made this species an ideal candidate for CP, thus enabling the application of this method to study monocots.
A crucial step in making CP of B. distachyon chromosomes feasible was the selection of the chromosome-specific, single-locus BAC clones. One of the available approaches to identify clones that contain low amount of interspersed DNA repeats and thus presumably map to single locus in the genome is dot-blot or Southern-blot hybridisation of labelled genomic DNA to all analysed BACs and subsequent selection of clones that have less intensive hybridisation signals (Jenkins and Hasterok 2007; Kim et al. 2005; Pedrosa-Harand et al. 2009; Walling et al. 2006). However, it was demonstrated that the strategy based on in silico sequence scanning can be equally efficient in the evaluation of repeated sequence content in large insert clones (Kim et al. 2005). In the case of A. thaliana, the suitability of the clones for CP was double-tested using bioinformatic analyses of BAC sequences as well as dot-blot hybridisation (Lysak et al. 2003). In the presented work, BACs from FPC contigs were assayed for interspersed repeat content by comparison of their sequence data with the Pooideae-specific Repeat Element Database (http://mips.helmholtz-muenchen.de/plant/recat/) from MIPS. Only clones containing less than 30% of DNA repeats within their inserts were included in the painting probes. As it turned out, this approach was in most cases sufficient to guarantee the chromosomal specificity of selected BACs. As a result of setting the threshold of permitted repeated DNA amount at 30%, the number of clones selected for painting was much lower than the number of clones used for CP of A. thaliana chromosomes (Pecinka 2005), despite the genome size of B. distachyon being over twice that of A. thaliana (Arabidopsis Genome Initiative 2000; International Brachypodium Initiative 2010). These clones rarely constituted a contiguous tiling path, but rather were distributed along chromosomes at intervals of ∼347 to ∼874 kbp on average, depending on the chromosome arm (Table 2). This lack of continuity of probe sequence was reflected by the discontinuity of the hybridisation signal but did not hamper the identification and tracking of any painted chromosome. It proves that CP is feasible even where the total length of BAC inserts used for painting is less than one third the length of the painted chromosome if the clones used are spread uniformly along its arms (Table 2).
The painting probes enabled not only discrimination of individual chromosomes of B. distachyon but also delimited different chromosome regions. As shown in Fig. 2, any chromosomal segment can be unambiguously identified and observed in PMCs at different developmental stages. In late zygotene cells, CP highlighted paired and unpaired chromosome regions (Figs. 1c and 4b1, red arrows). This means that CP can be used to investigate in detail the pairing behaviour of homologous chromosomes during early prophase I. Coupled with immunolabelling of recombination proteins, there is the potential to correlate the assembly of meiotic bivalents with recombination events, as has been recently demonstrated for barley (Phillips et al. 2010). The development of a similar assay for B. distachyon based on immunolocalisation of key meiotic proteins and chromosome painting could allow physical mapping of recombination events to specific chromosomes and their segments with unprecedented resolution and precision.
The analysis of chromosome territories in somatic and premeiotic interphase nuclei is another possible application of CP. Although the spatial distribution of particular chromosome domains has been extensively studied in human and animal cells (for review, see Cremer and Cremer 2010), this topic has not been explored to the same extent in plants. Painting of A. thaliana chromosomes significantly broadened our knowledge of nuclear architecture in plants, showing that save for NOR-bearing chromosomes there is no association of homologues at interphase (Pecinka et al. 2004) and that the three-dimensional arrangement of nuclear chromosome domains is similar in meristematic and differentiated tissues (Berr and Schubert 2007). With the advent of painting probes for B. distachyon, it would be interesting and profitable to compare territorial organisation of chromosomes in the nuclei of a dicot and a monocot plant.
After successful painting of B. distachyon chromosomes, we focused on determining whether the same probes had utility for comparative mapping in the genus Brachypodium as in A. thaliana and its relatives. The effectiveness of cross-mapping single BAC clones from B. distachyon genomic libraries to the wide range of Brachypodium species has been demonstrated before (Hasterok et al. 2006; Wolny et al. 2011). In order to test whether large BAC pools can be cross-mapped with the same efficiency as single clones, we hybridised the painting probes (Bd1-5) to the chromosomes of two ‘B. distachyon’ cytotypes, known as ABR114 (2n = 20) and ABR113 (2n = 30). These were considered until recently to be polyploid derivatives of diploid B. distachyon (2n = 10); ABR114 being autotetraploid, and ABR113 being autohexaploid cytotypes (Robertson 1981). Recent cytomolecular analyses suggest that ABR114 is rather a distinct diploid species, while ABR113 is in fact an allotetraploid formed by natural hybridisation between two diploid progenitors, one of which is similar to B. distachyon with 2n = 10 chromosomes and the other resembling ABR114 (Hasterok et al. 2004; Hasterok et al. 2006; Wolny et al. 2011). As the taxonomical status of the two cytotypes has not yet been officially established, comparative chromosome painting should shed more light on their true phylogeny.
CCP with the Bd1 painting probe showed that B. distachyon chromosome 1 is represented in the ABR114 genome by two different chromosomes bearing close structural similarity to one another as well as to their homoeologue in the B. distachyon karyotype. Results of the experiment with the Bd4-specific painting probes are in concordance with the data obtained earlier (Hasterok et al. 2006) for single B. distachyon BAC clones, as we observed signals specific for the short and for the long arm of Bd4 on two different bivalents in ABR114. A similar pattern was observed for Bd3-specific probes in ABR114. However, Bd5 probes specific for the short and long arms were represented in ABR114 on a single bivalent homoeologous to a B. distachyon-like genome. Based on earlier experiments of landing single B. distachyon BAC clones (Hasterok et al. 2006), it would be expected that the painting pattern of the Bd2-specific probes in ABR114 should resemble that of Bd3 and Bd4. Surprisingly, in ABR114 signals for the short and for the long arm of Bd2 are present on the same bivalent. Additionally, there is a small fragment corresponding to a part of the short arm on another bivalent. One possible explanation is that the short green fragment on the bivalent designated II resulted from a local translocation event and the BAC clone ABR1-41-E10 used in the previous experiment landed in the translocated region. The results of CCP seem to support our previous conclusion that multiple fusion/fission events involving centromeric regions of the chromosomes were one of the mechanisms shaping the present structure of the karyotypes of ABR114 and B. distachyon (Hasterok et al. 2006). The painting patterns of Bd1-, Bd2- and Bd5-specific probes in ABR114 do not contradict the fusion/fission hypothesis because it cannot be ruled out that different chromosomes of the B. distachyon karyotype have evolved in different ways and that various chromosomal rearrangements, e.g., translocations or duplications also played an important role in the evolution of the karyotypes of B. distachyon and ABR114.
The most convincing evidence for a hybrid origin of ABR113 came from genomic in situ hybridisation (GISH) with total nuclear DNA from both putative progenitors. Hybridisation of genomic DNA from the diploid (2n = 10) B. distachyon highlighted the ten largest chromosomes of the complement, while the genomic DNA from ABR114 distinguished the other 20 smaller chromosomes (Hasterok et al. 2004). Comparative mapping of single locus BAC clones from B. distachyon libraries revealed that for the vast majority of clones, the number of loci observed in the putative parental genomes of the allotetraploid is additive (Hasterok et al. 2006). CCP of ABR113 has corroborated the evidence for its parental origin from ABR114 and B. distachyon. Similar to the data obtained with mapping single BACs, the number of bivalents in ABR113 painted with the Bd1 probe was equal to the sum of the painted chromosomes in the putative ancestors. Also, the size and structure of homoeologues closely resembled their counterparts in the respective parental genomes. The longest bivalent is presumably derived from the B. distachyon genome. The two shorter ones might originate from ABR114. The absence of the unlabelled segment at the terminal position on one of the shorter bivalents in comparison to ABR114 chromosomes might be the result of chromosome restructuring which is a frequent phenomenon in polyploid genomes. It was also the case for CCP with Bd5 probe, where the number of painted chromosomes was equal to the sum of ABR114 and B. distachyon genomes. The analysis of orthologous gene pairs in diverse grass genomes, such as rice, wheat and sorghum, demonstrated that chromosome 5 is the most conserved (International Brachypodium Initiative 2010). The CCP analysis of Bd2, Bd3 and Bd4 in ABR113 revealed significant chromosome rearrangements. Probably, the longest chromosomes in all cases were from a B. distachyon-like ancestral genome, while different fragments of the short and long arm originated from translocations of ABR114 chromosomes. Multiple chromosome rearrangements during the evolution of genomes has been demonstrated in allopolyploids of Brassica napus (Udall et al. 2005) as well as in some species in the Tragopogon (Lim et al. 2008) and Lilium (Xie et al. 2010) genera. Painting with probes that allow only discrimination between chromosome arms obviously cannot reveal all chromosomal rearrangements or the exact locations of breakpoints. CP with denser probes targeting particular chromosome regions may uncover karyotypical changes with higher resolution and facilitate studies of chromosomal evolutionary processes.
Our data indicate that comparative chromosome painting with BAC pools from B. distachyon is a useful tool to investigate phylogenetic relationships between different Brachypodium species and the mechanisms which shape their karyotypes. The genus Brachypodium comprises relatively few species of different genome sizes and complexities. It includes diploid species with basic chromosome numbers of 5, 7, 8, 9 and 10 (Robertson 1981) as well as allopolyploids (Hasterok et al. 2004; Wolny and Hasterok 2009). Because of this diversity in chromosome number and ploidy, Brachypodium could become an interesting model system for studying karyotype evolution and divergence in grasses. It has yet to be determined whether or not B. distachyon-specific BAC pools could be used for comparative analysis of grasses outside the genus. Presumably, CCP can be applied successfully to the genomes of species such as Lolium, Bromus or Festuca that are phylogenetically relatively closely related to Brachypodium. It will be much more problematic to paint the large and complex genomes of crop cereals, such as barley and wheat or more distantly related species such as sorghum or rice. Cross hybridisation of B. distachyon single-locus BAC clones to the chromosomes of Triticale and rice did not yield hybridisation signals. As these clones were identified as syntenic to rice chromosome 6, it is likely that the homologous rice sequences are interspersed with blocks of species-specific repeats which preclude the effective visualisation of the probe signal (Hasterok et al. 2006). On the other hand, it was demonstrated that BAC clones from sorghum genomic libraries can effectively hybridise to maize chromosomes (Koumbaris and Bass 2003) despite the fact that the genome of maize is three times larger than the genome of sorghum (Paterson et al. 2009). Moreover, successful mapping of two B. distachyon BAC clones to barley chromosomes has been recently reported by Ma et al. (2010). These results are promising in the context of comparative chromosome painting in Triticeae species. Undoubtedly, the development of interspecific chromosome painting with B. distachyon-specific BAC pools significantly broadens the repertoire of cytogenetic techniques available for the analysis of grass genomes.
Acknowledgements
The authors are grateful to Dr. Martin Lysak and Ms. Terezie Mandakova from Masaryk University, Brno, Czech Republic, for their invaluable advice on the chromosome painting procedure as well as to two anonymous reviewers for their helpful comments on the manuscript. This work was supported by the Polish Ministry of Science and Higher Education Grant N N303 570738 to DI, EW, KL and RH as well as by the Biological and Biotechnological Sciences Research Council Grant BB/E004725/1 to MWB and the John Innes Centre Core Strategic Grant. DI and AB acknowledge scholarships from the UPGOW project co-financed by the European Social Fund.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Dominika Idziak and Alexander Betekhtin contributed equally to this work.
References
- Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Berr A, Schubert I. Interphase chromosome arrangement in Arabidopsis thaliana is similar in differentiated and meristematic tissues and shows a transient mirror symmetry after nuclear division. Genetics. 2007;176:853–863. doi: 10.1534/genetics.107.073270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harbor Press Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, et al. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harb Symp Quant Biol. 1993;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- Cremer M, Grasser F, Lanctot C, Muller S, Neusser M, et al. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol Biol. 2008;463:205–239. doi: 10.1007/978-1-59745-406-3_15. [DOI] [PubMed] [Google Scholar]
- Draper J, Mur LA, Jenkins G, Ghosh-Biswas GC, Bablak P, et al. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 2001;127:1539–1555. doi: 10.1104/pp.010196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febrer M, Goicoechea JL, Wright J, McKenzie N, Song X, et al. An integrated physical, genetic and cytogenetic map of Brachypodium distachyon, a model system for grass research. PLoS One. 2010;5:e13461. doi: 10.1371/journal.pone.0013461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin DF, Gu YQ, Hasterok R, Hazen SP, Jenkins G, et al. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Sci Plant Genome. 2008;48:S69–S84. [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Gray JW, Pinkel D. Molecular cytogenetics in human cancer diagnosis. Cancer. 1992;69:1536–1542. doi: 10.1002/1097-0142(19920315)69:6+<1536::AID-CNCR2820691306>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Gray JW, Lucas JN, Pinkel D, Awa A. Structural chromosome analysis by whole chromosome painting for assessment of radiation-induced genetic damage. J Radiat Res (Tokyo) 1992;33(Suppl):80–86. doi: 10.1269/jrr.33.SUPPLEMENT_80. [DOI] [PubMed] [Google Scholar]
- Gu YQ, Ma Y, Huo N, Vogel JP, You FM, et al. A BAC-based physical map of Brachypodium distachyon and its comparative analysis with rice and wheat. BMC genomics. 2009;10:496. doi: 10.1186/1471-2164-10-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasterok R, Langdon T, Taylor S, Jenkins G. Combinatorial labelling of DNA probes enables multicolour fluorescence in situ hybridisation in plants. Folia Histochem Cytobiol. 2002;40:319–323. [PubMed] [Google Scholar]
- Hasterok R, Draper J, Jenkins G. Laying the cytotaxonomic foundations of a new model grass, Brachypodium distachyon (L.) Beauv. Chromosome Res. 2004;12:397–403. doi: 10.1023/B:CHRO.0000034130.35983.99. [DOI] [PubMed] [Google Scholar]
- Hasterok R, Marasek A, Donnison IS, Armstead I, Thomas A, et al. Alignment of the genomes of Brachypodium distachyon and temperate cereals and grasses using bacterial artificial chromosome landing with fluorescence in situ hybridization. Genetics. 2006;173:349–362. doi: 10.1534/genetics.105.049726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Jenkins G, Hasterok R. BAC ‘landing’ on chromosomes of Brachypodium distachyon for comparative genome alignment. Nat Protoc. 2007;2:88–98. doi: 10.1038/nprot.2006.490. [DOI] [PubMed] [Google Scholar]
- Kim JS, Klein PE, Klein RR, Price HJ, Mullet JE, et al. Molecular cytogenetic maps of sorghum linkage groups 2 and 8. Genetics. 2005;169:955–965. doi: 10.1534/genetics.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumbaris GL, Bass HW. A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J. 2003;35:647–659. doi: 10.1046/j.1365-313X.2003.01829.x. [DOI] [PubMed] [Google Scholar]
- Kurz A, Lampel S, Nickolenko JE, Bradl J, Benner A, et al. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol. 1996;135:1195–1205. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KY, Soltis DE, Soltis PS, Tate J, Matyasek R, et al. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae) PloS one. 2008;3:e3353. doi: 10.1371/journal.pone.0003353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Lexer C. Towards the era of comparative evolutionary genomics in Brassicaceae. Pl Syst Evol. 2006;259:175–198. doi: 10.1007/s00606-006-0418-9. [DOI] [Google Scholar]
- Lysak MA, Fransz PF, Ali HB, Schubert I. Chromosome painting in Arabidopsis thaliana. Plant J. 2001;28:689–697. doi: 10.1046/j.1365-313x.2001.01194.x. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Pecinka A, Schubert I. Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Res. 2003;11:195–204. doi: 10.1023/A:1022879608152. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, et al. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci U S A. 2006;103:5224–5229. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Mandakova T, Lacombe E. Reciprocal and multi-species chromosome BAC painting in crucifers (Brassicaceae) Cytogenet Genome Res. 2010;129:184–189. doi: 10.1159/000312951. [DOI] [PubMed] [Google Scholar]
- Ma L, Vu GT, Schubert V, Watanabe K, Stein N, et al. Synteny between Brachypodium distachyon and Hordeum vulgare as revealed by FISH. Chromosome Res. 2010;18:841–850. doi: 10.1007/s10577-010-9166-3. [DOI] [PubMed] [Google Scholar]
- Mandakova T, Lysak MA. Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae) Plant Cell. 2008;20:2559–2570. doi: 10.1105/tpc.108.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R, Obe G. Application of chromosome painting to clastogenicity testing in vitro. Environ Mol Mutagen. 1998;32:212–222. doi: 10.1002/(SICI)1098-2280(1998)32:3<212::AID-EM3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Muller S, Stanyon R, O’Brien PC, Ferguson-Smith MA, Plesker R, et al. Defining the ancestral karyotype of all primates by multidirectional chromosome painting between tree shrews, lemurs and humans. Chromosoma. 1999;108:393–400. doi: 10.1007/s004120050391. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Pecinka A (2005) Chromosome painting and arrangement of interphase chromosome territories in Arabidopsis thaliana [PhD thesis]. Halle (Saale), Germany: Martin-Luther-Universität Halle-Wittenberg, pp 92
- Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, et al. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma. 2004;113:258–269. doi: 10.1007/s00412-004-0316-2. [DOI] [PubMed] [Google Scholar]
- Pedrosa-Harand A, Kami J, Gepts P, Geffroy V, Schweizer D. Cytogenetic mapping of common bean chromosomes reveals a less compartmentalized small-genome plant species. Chromosome Res. 2009;17:405–417. doi: 10.1007/s10577-009-9031-4. [DOI] [PubMed] [Google Scholar]
- Phillips D, Nibau C, Ramsay L, Waugh R, Jenkins G. Development of a molecular cytogenetic recombination assay for barley. Cytogenet Genome Res. 2010;129:154–161. doi: 10.1159/000314335. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, et al. Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci U S A. 1988;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp S, Jauch A, Qiu JY, Smialek B, Cremer T, et al. Translocation (8;21) in acute nonlymphocytic leukemia delineated by chromosomal in situ suppression hybridization. Cancer Genet Cytogenet. 1991;57:103–107. doi: 10.1016/0165-4608(91)90195-Z. [DOI] [PubMed] [Google Scholar]
- Ried T, Schrock E, Ning Y, Wienberg J. Chromosome painting: a useful art. Hum Mol Genet. 1998;7:1619–1626. doi: 10.1093/hmg/7.10.1619. [DOI] [PubMed] [Google Scholar]
- Robertson IH. Chromosome numbers in Brachypodium Beauv. (Gramineae) Genetica. 1981;56:55–60. doi: 10.1007/BF00126930. [DOI] [Google Scholar]
- Schubert I, Fransz PF, Fuchs J, de Jong JH. Chromosome painting in plants. Methods Cell Sci. 2001;23:57–69. doi: 10.1023/A:1013137415093. [DOI] [PubMed] [Google Scholar]
- Tkachuk DC, Pinkel D, Kuo WL, Weier HU, Gray JW. Clinical applications of fluorescence in situ hybridization. Genet Anal Tech Appl. 1991;8:67–74. doi: 10.1016/1050-3862(91)90051-r. [DOI] [PubMed] [Google Scholar]
- Udall JA, Quijada PA, Osborn TC. Detection of chromosomal rearrangements derived from homologous recombination in four mapping populations of Brassica napus L. Genetics. 2005;169:967–979. doi: 10.1534/genetics.104.033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling JG, Shoemaker R, Young N, Mudge J, Jackson S. Chromosome-level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics. 2006;172:1893–1900. doi: 10.1534/genetics.105.051466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise A, Heller A, Starke H, Mrasek K, Kuechler A, et al. Multitude multicolor chromosome banding (mMCB)—a comprehensive one-step multicolor FISH banding method. Cytogenet Genome Res. 2003;103:34–39. doi: 10.1159/000076286. [DOI] [PubMed] [Google Scholar]
- Wienberg J, Jauch A, Stanyon R, Cremer T. Molecular cytotaxonomy of primates by chromosomal in situ suppression hybridization. Genomics. 1990;8:347–350. doi: 10.1016/0888-7543(90)90292-3. [DOI] [PubMed] [Google Scholar]
- Wolny E, Hasterok R. Comparative cytogenetic analysis of the genomes of the model grass Brachypodium distachyon and its close relatives. Ann Bot. 2009;104:873–881. doi: 10.1093/aob/mcp179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolny E, Lesniewska K, Hasterok R, Langdon T. Compact genomes and complex evolution in the genus Brachypodium. Chromosoma. 2011;120:199–212. doi: 10.1007/s00412-010-0303-8. [DOI] [PubMed] [Google Scholar]
- Xie S, Khan N, Ramanna MS, Niu L, Marasek-Ciolakowska A, et al. An assessment of chromosomal rearrangements in neopolyploids of Lilium hybrids. Genome. 2010;53:439–446. doi: 10.1139/G10-018. [DOI] [PubMed] [Google Scholar]
- Xu W, Li JY, Liu Q, Zhu Y, Pan JL, et al. Multiplex fluorescence in situ hybridization in identifying chromosome involvement of complex karyotypes in de novo myelodysplastic syndromes and acute myeloid leukemia. Int J Lab Hematol. 2010;32:e86–e95. doi: 10.1111/j.1751-553X.2008.01101.x. [DOI] [PubMed] [Google Scholar]


