Abstract
The steroid 20-hydroxy-ecdysone (20-HE) and the sesquiterpenoid Juvenile Hormone (JH) coordinate insect life stage transitions. 20-HE exerts these effects by the sequential induction of response genes. In the nematode Caenorhabditis elegans hormones also play a role in such transitions, but notably, microRNA such as let-7 and lin-4 have likewise been found to help order developmental steps. Little is known about the corresponding function of homologous microRNA in Drosophila melanogaster, and the way microRNA might be regulated by 20-HE in the fly is ambiguous. Here we used Drosophila S2 cells to analyze the effects of 20-HE on D. melanogaster microRNA let-7 and miR-125, the homolog of lin-4. The induction by 20-HE of let-7 and miR-125 in S2 cells is inhibited by RNai knockdown of the ecdysone receptor and, as previously shown, by knockdown of its cofactor broad-complex C. To help resolve the currently ambiguous role of 20-HE in the control of microRNa, we show that nanomolar concentrations of 20-HE primes cells to subsequently express microRNa when exposed to micromolar levels of 20-HE. We then explore the role microRNa plays in the established relationship between 20-HE and the induction of innate immunity. We show that the 3′UTR of the antimicrobial peptide diptericin has a let-7 binding site and that let-7 represses translation from this site. We conclude that 20-HE facilitates the initial expression of innate immunity while it simultaneously induces negative regulation via microRNa control of antimicrobial peptide translation.
Key words: microRNA, ecdysone, innate immunity, let-7, diptericin, inflammation
Introduction
microRNA were first discovered through their effects on the developmental staging of the nematode C. elegans.1,2 These non-coding RNA of about 22 nucleotides bind with incomplete complementarity to the 3′UTR of transcripts to down-regulate protein expression.3,4 microRNA have since been described in many animals, sometimes with a high degree of sequence conservation.5,6 It is also now clear that the potential functions of microRNA go well beyond developmental timing, and includes processes such as cell proliferation and cell death, metabolism, life span, stress resistance and adaptive immunity.7–10 A step toward understanding such diverse consequences of conserved microRNA is to explore their function in additional genetic model systems. Drosophila melanogaster has 148 predicted microRNA,11 including the widely conserved let-7 and mir-125, the homolog of C. elegans lin-4. Early studies of Drosophila let-7 and mir-125 were focused on the expression dynamics of these microRNA through the life cycle and their potential regulation by the steroid hormone 20 hydroxy- ecdysone (20-HE).12–14 A role for these microRNA that is consistent with 20-HE control of metamorphosis was recently revealed in a let-7/mir-125 mutant.15 The gene abrupt, encoding a BTB-POZ Zn-finger nuclear protein, was identified to be a target of let-7. Thus, there are clear developmental patterns and consequences for the expression of let-7 and mir-125, but we still know little about the factors regulating these microRNA and whether they have functions aside from the timing of stage-specific processes.
The D. melanogaster microRNAs let-7 and mir-125 are highly expressed in late larvae and pupae, which is the time when 20-HE rapidly increases to initiate metamorphosis.12–14 These microRNAs are also induced within cultured Drosophila cells when they are experimentally exposed to 20-HE.12–14 In addition, juvenile hormone (JH) was found to repress induction of let-7 in cells treated with 20-HE,14 as might be expected because JH is a classic antagonist of 20-HE. During development JH represses metamorphosis at larval stage transitions,21 and in cells JH inhibits the ability of 20-HE to sensitize the expression of antimicrobial peptides.17
Interpreting the role of ecdysone control of microRNA has been difficult because conflicting conclusions arise from genetic analyses. Consistent with the notion that 20-HE regulates microRNA, levels of let-7 RNA were reduced in mutants of ecdysone synthesis (ecd1) and of the early ecdysone response gene broad-complex C (BR-C), encoded by npr6.13 These mutants also contained less mir-125 and mir-100 RNA.14 In contrast to such evidence for ecdysone control of microRNA expression, different outcomes were reported from analysis of conditional knockdown of the ecdysone receptor.12 Transient expression of EcR-RNAi reduced E74A and E74B, indicating there was effective repression of established ecdysone receptor targets, but this did not affect the levels of let-7 or mir-125. Uncertainty about the role of 20-HE also arises from differences in the temporal expression of let-7 in experimental cell culture relative to its expression in flies.12–14 Both let-7 and mir-125 increased at early stages of pupation, within hours of the physiological increase in 20-HE. But in cultured cells the induction of these microRNA in cell culture was not seen until 25 hours after cells were treated with 20-HE.
It therefore remains an open problem as to whether and how 20-HE modulates Drosophila microRNA, and there has been little exploration of the physiological consequences for any such control. Here we begin to address these issues with an analysis of 20-HE upon microRNA in cultured Drosophila S2 cells. We show that the ecdysone receptor is required in S2 cells for 20-HE to induce expression of let-7 and mir-125, and we find that priming cells with nanomolar concentrations of 20-HE is required for subsequently higher levels of 20-HE to regulate these microRNA. We then put the regulation of let-7 by 20-HE into a functional context by describing how let-7 and 20-HE jointly regulate innate immunity. We show that translation can be repressed by let-7 through a binding site in the 3′UTR of the antimicrobial peptide gene diptericin. Importantly, previous work showed that 20-HE facilitates the expression of diptericin mRNA when cells are exposed to bacterial peptidoglycans.16,17 With our current observations we suggest that 20-HE also induces a limiter for the innate immune response in the form of microRNA. This dual control by 20-HE may serve to activate the immune response while simultaneously modulating its level or duration.
Results
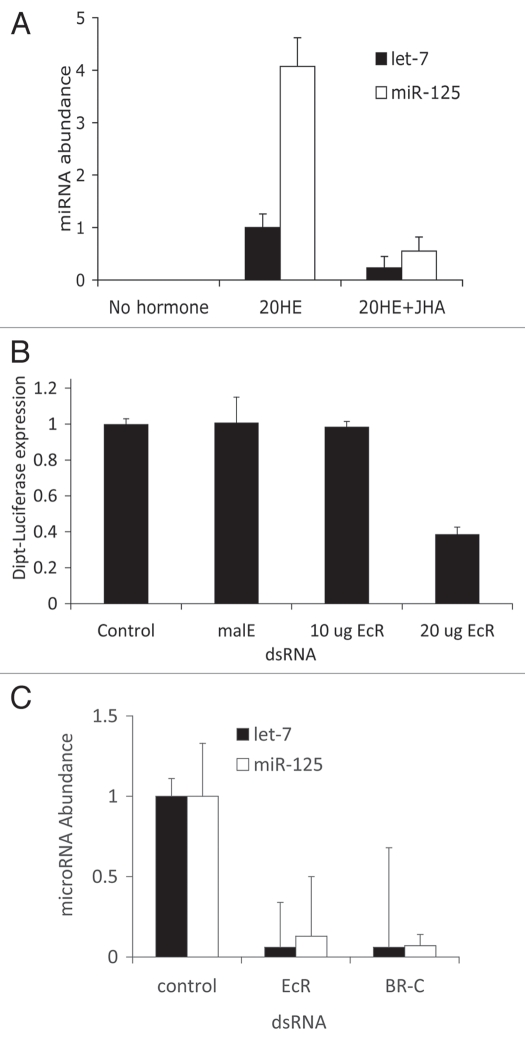
As measured by quantitative PCR, let-7 and mir-125 were robustly induced in S2 cells exposed to 20-HE for 24 hours (Fig. 1A), in agreement with previous reports.12–14 Likewise as reported by Sempere et al. (reviewed in ref. 14), the 20-HE-induced expressions of let-7 and mir-125 were repressed by the juvenile hormone analog methoprene (JHA) (Fig. 1A). To help resolve ambiguity among published reports as to whether the induction of microRNA by 20-HE required the ecdysone receptor, we transfected cells with EcR-dsRNA at a dose that effectively reduced EcR-mediated signaling (Fig. 1B). The levels of both let-7 and mir-125 were reduced in 20-HE-treated cells treated with EcR-dsRNA (Fig. 1C). As a positive control we also confirmed that transfection of BR-C-dsRNA reduced the induction of let-7 and mir-125 by 20-HE, as previously reported (Fig. 1C).13
Figure 1.
(A) Relative abundance of let-7 and miR-125 in S2 cells after 24 h exposure to 5 uM 20 hydroxy-ecdysone (20-HE) or to 5 uM 20-HE and 10 uM juvenile hormone analog (JHA) methoprene (with std). Cells without hormone treatment produced an undetectable quantity of miRNA; for both microRNA the difference between 20-HE and 20HE + JHA were significant, t-test, p < 0.001. Without hormone the mRNA of both let-7 and miR-125 were undetectable by RT-PCR (CT > 35). (B) Efficiency of EcR-dsRNA verified by repression of EcR-dependent induction of the dipt-Luciferase reporter within S2 cells exposed to peptidoglycan (with std). Control cells received no dsRNA. Cells treated with malE-dsRNa (negative control) or with 10 ug EcR-dsRNA did not reduce reporter expression. Treatment with 20 ug EcR-dsRNA was sufficient to reduce reporter activity relative to all other groups (Tukey-Kramer HSD test, p < 0.0001). (c) MicroRNA expression from S2 cells treated with 20-HE and dsRNA for EcR or BR-C. The negative control cells were treated with malE-dsRNa. In each condition, cells were treated with 20 ug dsRNA. For both microRNa: no significant difference between EcR and BR-C dsRNA treatment; significant differences between control and EcR or BR-C (Tukey-Kramer HSD test, p < 0.0001).
20-HE has multiple effects on cultured Drosophila cells, and often with some delay. At least 18 hours of 20-HE treatment was required before S2 cells could immunologically respond to peptidoglycan,17 and 25 hour of 20-HE treatment was needed before S2 and Kc167 cells induced let-7 and mir-125.12,13 The delay in these responses may occur for several nonexclusive reasons. The responses may be a by-product of cell differentiation and S2 cells do change shape and adhesion after 24 hours exposure to 20-HE.20 Independent of differentiation, the responses to 20-HE may be substantially indirect, requiring induction and translation of multiple intervening factors. Thirdly, by the standard protocol for these experiments, cells were exposed to hormones in an abrupt and monotonic manner, unlike the graded and periodic presentation of 20-HE and JH that is expected within developing flies. Any of these factors could produce the observed differences between cultured cells and pupating animals in the timing of microRNA with respect to hormones.
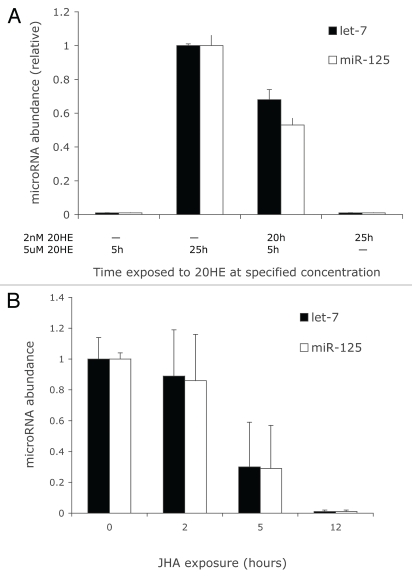
To investigate the third alternative we assessed whether S2 cells exposed to 20-HE at nanomolar concentrations, as occurs in the physiological context of the early prepupae, might prime cells to later robustly respond to 20-HE at micromolar concentrations, as occurs during the peak of pupation.21 As a control, S2 cells were exposed to 5 µM 20-HE for 5 h. These cells did not induce let-7 or mir-125 in contrast to the robust expression of both microRNA after 25 h exposure with 5 µM 20-HE (Fig. 2A). However, let-7 and mir-125 were readily induced in cells exposed to 5 µM 20-HE for 5 h after they were first exposed to 2 nM 20-HE for 20 h. Since nanomolar concentration of 20-HE alone was not sufficient to elevate lit-7 or mir-125 or to produce morphological change in the cells, we conclude that a priming exposure to nanomolar 20-HE is required before micromolar concentrations can induce microRNA. While extended exposure to 20-HE is required before peptidoglycan can induce antimicrobial peptides in S2 cells, JHA represses this response without delay.17 We found a similar asymmetric dynamic applied to microRNA induction. In cells first exposed to 5 uM 20-HE for 25 h, induced microRNA declined within 5 h of JHA treatment (Fig. 2B). JHA, unlike 20-HE, has an immediate effect on the transcription or stability of let-7 and mir-125 RNA.
Figure 2.
(A) Relative abundance of miRNA induced in S2 cells exposed to 20HE first at 2 nM and then at 5 uM for specified durations. For both microRNa: no significant difference among means of 5 uM 20-HE for 5 h and 2 nM 20-HE for 25 h; each remaining treatment differed significantly from all other groups (Tukey-Kramer HSD test, p < 0.01). (B) Duration of JHA (10 uM) exposure required to suppress miRNA expressed in S2 cells previously exposed to 20-HE (5 uM) for 25 hours. For both microRNA: no significant difference among means of zero and two hours JHA exposure; means from 5 hour JHA exposure were significantly less than those of zero hour exposure; means from 12 hour exposure were significantly less than those of all other treatments (Tukey-Kramer HSD test, p < 0.01).
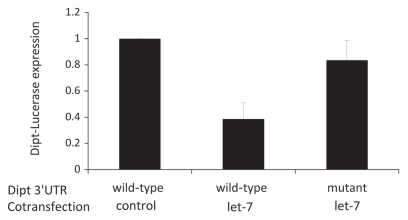
20-HE thus appears to induce microRNA and to facilitate the expression of antimicrobial peptides. To address if 20-HE independently affects microRNA and antimicrobial peptides or regulates these peptides via microRNA, we asked whether the 3′UTR sequences of Drosophila antimicrobial peptides contain potential microRNA binding sites. We used the PITA database (http://genie.weizmann.ac.il/pubs/mir07/index.html) of predicted conserved microRNA binding sites32 to search for candidate targets in the 3′UTR of genes listed in Genbank as encoding Drosophila antimicrobial peptides. PITA predicts a stem-loop configuration of the let-7 target at position 156–164 relative to the stop codon within diptericin (CTA TCT CAA ATG CCA TCA; bases expected to complement the dme-let-7 seed region shown in bold). Of the quarried antimicrobial peptides only drocomycin also showed any candidate miRNA sites, again for let-7 (data not shown). To evaluate whether let-7 can regulate diptericin from this site, we cloned the diptericin 3′UTR into a firefly luciferase translation reporter vector (MT-fLuc-dpt-3′UTR) and cotransfected this into S2 cells with a vector expressing either let-7 microRNA sequence or the microRNA sequence of mir-92b, which is not expected to affect diptericin mRNA. Cells cotransfected with MT-fLuc-dpt-3′UTR and control mir-92b vectors strongly expressed reporter luciferase (Fig. 3). Luciferase expression was markedly repressed in cells cotransfected with MT-fLuc-dpt-3′UTR and let-7 vectors. To verify that the identified let-7 sequence of the diptericin 3′UTR was responsible for its repression, we generated a MT-fLuc-dpt-3′UTRmutant where 20 nucleotides including the candidate let-7 binding site were deleted. This mutant luciferase reporter was not repressed in cells cotransfected with the let-7 vector (Fig. 3). Overall, these results demonstrate that the 3′UTR of antimicrobial peptide gene diptericin contains an operational binding site from which let-7 can repress translation.
Figure 3.
Expression of luciferase reporter cloned to diptericin 3′UTR with wild-type sequence or sequence mutated to remove the candidate let-7 target site. These S2 cells were cotransfected with vectors to express a negative control microRNa (mir-92b) or let-7 microRNA. Relative to mir-92b control, reporter expression was significantly less in UTR-wild-type/let-7 treatment (t = 4.27, p < 0.01); expression of UTR-mutant/let-7 was significantly greater than that of wild-type/let-7 (t = 2.97, p < 0.05).
Discussion
Our results may help resolve whether 20-HE can modulate expression of let-7 and mir-125 in Drosophila. In previous work, conditional reduction of the ecdysone receptor within larvae did not impair expression of these microRNA.12 We now show that EcR can be required for let-7 and mir-125 expression in S2 cells, similar to previous work with cells where BR-C was required for 20-HE to induce these microRNA.14 Thus, at least for cell culture there is consistent evidence that ecdysone signaling positively modulates microRNA expression. Reduction of EcR by transiently expressed dsRNA in animals might not reveal this dependence for several reasons. Larvae, unlike S2 and Kc167 cells, might not have the potential for 20-HE to regulate microRNA. Alternatively, the transient nature of the heat shock induced EcR-dsRNA in larvae used by Bashirullah et al.12 may have been able to repress early targets of ecdysone signaling but the gene knock-down was not able to repress indirect or delayed targets of ecdysone signaling. Although we cannot fully distinguish between these explanations we see that the capacity for 20-HE to induce microRNA depends on past exposure to the hormone. And the history of past 20-HE exposure is likely to have differed among the methods of the original reports analyzing the relationship of 20-HE and microRNA. We now show that treating cells with nanomolar levels of 20-HE primes cells to rapidly induce microRNA when exposed to 20-HE in micromolar concentration. If late third instar larvae express 20-HE at nanomolar concentrations and thus prime animals to rapidly induce let-7 and mir-125 when 20-HE increases at pupation, their response of microRNA to 20-HE would appear to be sudden, as seen in the original studies using cell culture. Interestingly, one potential mechanism for such priming might involve how 20-HE induces transcription of its own receptor, EcR;22,23 a small initial concentration of 20-HE may thereby increase the abundance of the receptor prior to the time when hormone titers are strongly elevated. Studies with cycloheximide likewise suggest there are intervening factors between the initial exposure of 20-HE and immune response since protein synthesis is required for 20-HE to induce the expression of the immunity-associated Hemolin of Hyalophora cecropia.28 Finally, we note that the regulation of EcR of Drosophila appears to be negatively regulated by microRNA mir-14,24 while the mammalian estrogen-bound receptor ERα was found to downregulate expression of microRNA;29 microRNA may thus play a broad role in the overall modulation of ecdysone hormone or steroid hormone action.
Our observations also reveal a novel function for microRNA in Drosophila. Besides its established role in life stage transitions,15 we have explored microRNA in the context of innate immunity. We show that the 3′UTR of antimicrobial peptide diptericin contains sequence for let-7 binding and that expression of let-7 can repress translation of protein associated with this 3′UTR. let-7 thus appears to be a direct negative regulator of diptericin. This arrangement is particularly interesting because mRNA of both diptericin and let-7 are upregulated directly or indirectly by 20-HE via the ecdysone receptor, at least in S2 cells. As has been proposed for microRNA to function as feedback loop regulators of thresholds,25 we suggest that let-7 may be co-regulated with diptericin to set a limiter or governor on the antimicrobial peptide. In this view 20-HE would sensitize cells to transcribe diptericin mRNA when exposed to bacteria but at the same time activate a translational regulator of the antimicrobial peptide via its induction of let-7. JH appears to turn off both aspects of this dynamic since it represses the ability of 20-HE to sensitize expression of diptericin mRNA and reduces the translational repressor let-7. Such regulation of fly innate immunity may be important because expression of anti-microbial peptides entail costs in terms of reduced fecundity and long-term survival.19,35
microRNA may play an analogous role in the innate immunity/inflammatory response of mammals. Repression of TNFα translation from its 3′UTR is released when cells are exposed to pathogen-associated lipopolysaccharides.26 Bioinformatic analysis has predicted binding sites for mir-125a and mir-125b within the 3′UTR of TNFα and exposure to lipopolysaccharides represses mir-125b.27 We thus hypothesize from these observations and our current data that specific steroid hormones might potentiate the induction of the innate immune/inflammatory response in mammals but simultaneously induce a limiter in the form of microRNA to prevent unwarranted or excess activation of the system. Upon infection, specific molecular signals or pathogen-derived cues could reduce the microRNA and thereby elevate the innate immune/inflammatory state. microRNA in this context may play a conserved role in the homeostasis of innate immunity and inflammatory responses.
Materials and Methods
S2* cell culture.
All cells were Drosophila Schneider-2-star cells (S2*) and these cells were stably transfected with a firefly-luciferase reporter vector driven by the promoter of diptericin (S2* Dpt-luc).30 Cells were cultured in Schneider's media with 10% fetal bovine serum (FBS) and passed every five days. New flasks were seeded three days before dsRNA or transfection experiments.
dsRNA.
3 × 106 S2* Dpt-luc cells per well were plated in 1.1 mL of Schneider's Cell Media (Sigma) without fetal bovine serum (FBS) in a six-well plate. Cells were treated with 60 µl at a concentration of 1 ug/µL of EcR-dsRNA, BR-C-dsRNA or MalE-dsRNA (maltose binding protein, a negative control). 2.2 mL of Schneider's Cell Media with 10% FBS was added to each well after 1 hr incubation. When indicated, cells were treated with hormone 24 hr after dsRNA treatment.
Hormone treatment.
S2* Dpt-luc cells (3 × 103 cells) in 3 µL of media per well were treated with 5 µM 20-HE (Sigma) or with 2 nM 20-HE as noted, and with 10 µM juvenile hormone analog (JHA) methoprene (Sigma) as indicated. All hormones were diluted in ethanol, and an equivalent volume of ethanol was added to control wells.
Luciferase assay.
Luciferase activity was measured following the protocol and reagents from the Brite-Glo Luciferase Assay Kit (Promega). Cells from each incubation-well were aliquoted into 6 to 10 wells of a 96-well plate (black/clear bottom) (3 × 103 cells). Luciferase was quantified with a SpectraMax M5 (Molecular Devises). For translation reporter plasmids, firefly and Renilla luciferase activity was measured from the same well using the Dual-Glo Luciferase Assay Kit (Promega).
qRT-PCR.
Total RNA was isolated from S2* cells using Trizol (Invitrogen). TaqMan (Applied Biosystems) probes for quantitative Real-Time PCR was used to assay mirR-125, let-7 and miR-2 microRNA expression in S2* cells after treatment with ecdysone and dsRNA. Analysis was conducted on an ABI 7300 and normalized relative to miR-277.
S2* cell transfection.
Transient transfection of S2* cells with reporter or expression plasmids followed the protocol of Burgler (ref. 33): 2.0 × 106 of S2* cells were plated in 1.0 mL of medium without FBS into each well of a 6-well plate. Cells were transfected with 2 µg microRNA expression plasmid (pAct-let-7 or pAct-miR-92b) or empty pGEM vector (control), 50 ng fLuc translation reporter plasmid (MT-fLuc-Ttk 3′UTR or MT-fLuc-dpt-3′UTR) and 10 ng control Renilla reporter plasmid (MT-rLuc). All plasmids were diluted in 0.5 ml Schneider's Drosophila Medium without FBS (Sigma). 5 µl Cellfectin (Invitrogen) was diluted into 0.5 ml Schneider's Drosophila Cell Medium (Sigma) and incubated for 5 min and then mixed with the plasmid media. Cells were resuspended in the mixture after 45 min of co-incubation. Five hours after transfection, 0.5 ml of Schneider's Media with 30% FBS (Gibco) was added to cell culture. Forty-five hours after transfection, 700 mM CuSO4 was added to induce metallothionein (MT) promoters in reporter and control plasmids. Six hours after CuSO4 induction, cells were harvested and rLuc and fLuc levels were assayed. To normalize for transfection efficiency and cell viability across treatments, within each replicate fLuc levels were standardized against rLuc levels. Each treatment was conducted in two replicates and each experiment was performed three times.
Vectors.
Okabe (ref. 34) originally derived the metallothionein construct (MT-fLuc-Ttk-3′UTR) from the pRmHa-3 vector with Phontinus luciferase GL-3 (Promega) sequence followed by the Tramtrack (Ttk)-3′UTR, and likewise the vector MT-rLuc-Adh-3′UTR with the Drosophila metallothionein promoter driving Renilla luciferase followed by alcohol dehydrogenase (Adh) 3′-UTR. These reporter vectors, and the pAct-let-7 and pAct-mir-92b expression vectors (Burger and Macdonald, ref. 33) were generously provided by Fergal O'Ferrell (Department of Natural Sciences, Sodertorns Hogskola, Huddinge, Sweden).31 We subsequently constructed the MT-fLuc-dpt-3′UTR vector by amplifying the first 230 nt of genomic 3′UTR from Drosophila diptericin using primers Fwd: CAT TAG GGA TCC AAC and Rev: CAT TAG TCT AGA CGA TTC ATC ATT TTA CAA GGT CA and inserting the resulting sequence in place of the Ttk-3′-UTR at the BamHI/XbaI site of MT-fLuc-Ttk-3′UTR. To construct the MT-fLuc-dpt-3′UTRmutant vector, the MT-fLuc-dpt-3′UTR vector was PCR amplified using mutagenic primers to produce a 20 bp deletion that included the predicted let-7 target site (Fwd: CAA CGC CAA GGA CAT AAA TTA TGG TCA GGT ATG C; Rev: GCA TAC CTG ACC ATA ATT TAT GTC CTT GGC GTT GCAA CGC CAA GGA CAT AAA TTA TGG TCA GGT ATG C).
Acknowledgements
For providing vectors we thank Fergal O'Ferrell, Department of Natural Sciences, Sodertorns Hogskola, Huddinge, Sweden. We thank Thomas Flatt for advice and discussion. This work was supported in part by awards to Marc Tartar from the Ellison Medical Foundation, the Paul F. Glenn Foundation and NIH R01 AG031152.
Abbreviations
- 20-HE
20-hydroxy-ecdysone
- JH
juvenile hormone
- JHA
juvenile hormone analog
- 3′UTR
3′ untranslated region
- BR-C
broad-complex C
- EcR
ecdysone receptor
- dsRNA
double stranded RNA
- malE
maltose binding protein
- dpt
diptericin
- fLuc
firefly luciferase
- rLuc
renilla luciferase
- FBS
fetal bovine serum
Footnotes
Previously published online: www.landesbioscience.com/journals/fly/article/13008
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 5.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 6.Chen K, Rajewsky N. Deep conservation of microRNA-target relationships and 3′UTR motifs in vertebrates, flies and nematodes. Cold Spring Harb Symp Quant Biol. 2006;71 doi: 10.1101/sqb.2006.71.039. [DOI] [PubMed] [Google Scholar]
- 7.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 10.Lodish HP, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by micro. RNAsNat Rev Immunol. 2000;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 11.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bashirullah A, Pasquinelli AE, Kiger AA, Perrimon N, Ruvkun G, Thummel CS. Coordinate regulation of small temporal RNAs at the onset of Drosophila metamorphosis. Dev Biol. 2003;259:1–8. doi: 10.1016/s0012-1606(03)00063-0. [DOI] [PubMed] [Google Scholar]
- 13.Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The Expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Developmental Biology. 2002;244:170–179. doi: 10.1006/dbio.2002.0594. [DOI] [PubMed] [Google Scholar]
- 14.Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and Broad-Complex gene activity. Dev Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 15.Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and mir-125 heterochronic microRNAs. Curr Biol. 2001;11:2008. doi: 10.1016/j.cub.2008.06.020. 18:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meister M, Richards G. Ecdysone and insect immunity: the maturation of the inducibility of the diptericin gene in Drosophila larvae. Insect Biochem Molec Biol. 1996;26:155–160. doi: 10.1016/0965-1748(95)00076-3. [DOI] [PubMed] [Google Scholar]
- 17.Flatt T, Heyland A, Rus F, Porpiglia E, Sherlock C, Yamamoto R, et al. Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. J Exp Biol. 2008;211:2712–2724. doi: 10.1242/jeb.014878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler DE, Nijhout HF. A perspective for understanding the modes of juvenile hormone action as a lipid signaling system. BioEssays. 2003;25:9941001. doi: 10.1002/bies.10337. [DOI] [PubMed] [Google Scholar]
- 19.Zerofsky M, Harel E, Silverman N, Tatar M. Aging of the innate immune response in Drosophila melanogaster. Aging Cell. 2005;4:103–108. doi: 10.1111/j.1474-9728.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- 20.Baum B, Cherbas L. Drosophila cell lines as model systems and as an experimental tool. Methods Mol Biol. 2008;420:391–424. doi: 10.1007/978-1-59745-583-1_25. [DOI] [PubMed] [Google Scholar]
- 21.Riddiford LM. Hormone receptors and the regulation of insect metamorphosis. Receptor. 1993;3:203–209. [PubMed] [Google Scholar]
- 22.Gauhar Z, Sun LV, Hua S, Mason CE, Fuchs F, Li TR, et al. Genomic mapping of binding regions for the Ecdysone receptor protein complex. Genome Res. 2009;19:1006–1013. doi: 10.1101/gr.081349.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karim FD, Thummel CS. Temporal coordination of regulatory gene expression by the steroid hormone ecdysone. EMBO J. 1992;11:4083–4093. doi: 10.1002/j.1460-2075.1992.tb05501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varghese J, Cohen SM. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007;21:2277–2282. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen SM, Brennecke J, Stark A. Denoising feedback loops by thresholding—a new role for microRNAs. Genes Dev. 2006;20:2769–2772. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- 26.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-Rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 27.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNFalpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 28.Roxstrom-Lindquist K, Assefaw-Redda Y, Rosinska K, Faye I. 20-Hydroxyecdysone indirectly regulates Hemolin gene expression in Hyalophora cecropia. Insect Mol Biol. 2005;14:645–652. doi: 10.1111/j.1365-2583.2005.00593.x. [DOI] [PubMed] [Google Scholar]
- 29.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, et al. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–347. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Silverman N, Zhou R, Stöven S, Pandey N, Hultmark D, Maniatis TA. Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Farrell F, Esfahani SS, Engstrom Y, Kylsten P. Regulation of the Drosophila lin-41 homologue dappled by let-7 reveals conservation of a regulatory mechanism within the LIN-41 subclade. Dev Dyn. 2008;237:196–208. doi: 10.1002/dvdy.21396. [DOI] [PubMed] [Google Scholar]
- 32.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 33.Burgler C, Macdonald PM. Prediction and verification of microRNA targets by MovingTargets, a highly adaptable prediction method. BMC Genomics. 2005;6:88. doi: 10.1186/1471-2164-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okabe M, Cummins JM. Mechanisms of sperm-egg interactions emerging from gene-manipulated animals. Cell Mol Life Sci. 2007;64:1945–1958. doi: 10.1007/s00018-007-7037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Libert S, Chao Y, Chu X, Pletcher SD. Trade-offs between longevity and pathogen resistance in Drosophila melanogaster are mediated by NFκB signaling. Aging Cell. 2006;5:533–543. doi: 10.1111/j.1474-9726.2006.00251.x. [DOI] [PubMed] [Google Scholar]