Abstract
In 2006 and 2007 beef and pork carcass swabs from provincially inspected abattoirs in Alberta, Canada were tested to determine the levels of total aerobic bacteria, coliform bacteria, and generic Escherichia coli, and the prevalence of Salmonella spp., Campylobacter spp., and Shiga toxin-producing E. coli (STEC). Swabs from beef and pork carcasses from 48 and 34 facilities, respectively, were analyzed. All samples tested were positive for aerobic bacteria with 99.8% of beef and 96.0% of pork samples, having total counts of ≤ 100 000 CFU/cm2. Coliform bacteria were isolated from 22.4% and 42.0% of beef and pork carcass samples, respectively. Generic E. coli were recovered from 14.6% of beef and 33.7% of pork carcass samples. For beef carcasses, positive tests were obtained for 0.1% of 1036 samples tested for Salmonella spp., 1.5% of 1022 samples tested for Campylobacter spp. and 5.4% of 1018 samples tested for STEC. For pork carcasses, positive tests were obtained for 1.6 % of 1076 samples tested for Salmonella spp., 8.8% of 1070 samples tested for Campylobacter spp. and 4.8% of 1067 samples tested for STEC.
Résumé
Étude de référence microbiologique des carcasses de bovins et de porcs dans les abattoirs inspectés par les autorités provinciales de l’Alberta, au Canada. En 2006 et en 2007, des écouvillons des carcasses de bovins et de porcs provenant des abattoirs inspectés par les autorités provinciales de l’Alberta, au Canada, ont été testés pour déterminer les quantités de bactéries aérobies totales, de bactéries coliformes et d’Escherichia coli générique et la prévalence de Salmonella spp., de Campylobacter spp. et d’E. coli produisant de la shiga toxine (STEC). Des écouvillons de carcasses de bovins et de porcs provenant de 48 et de 34 établissements, respectivement, ont été analysés. Tous les échantillons testés étaient positifs pour des bactéries aérobies et 99,8 % des échantillons de bovins et 96,0 % des échantillons de porcs présentaient des numérations totales ≤ 100 000 CFU/cm2. Des bactéries coliformes ont été isolées dans 22,4 % et 42,0 % des échantillons des carcasses de bovins et de porcs, respectivement. E. coli générique a été récupéré dans 14,6 % des échantillons de carcasses de bovins et dans 33,7 % des échantillons de carcasses de porc. Pour les carcasses de bovins, des tests positifs ont été obtenus dans 0,1 % des 1036 échantillons testés pour Salmonella spp., dans 1,5 % des 1022 échantillons testés pour Campylobacter spp. et dans 5,4 % des 1018 échantillons testés pour STEC. Pour les carcasses de porcs, des tests positifs ont été obtenus dans 1,6 % de 1076 échantillons testés pour Salmonella spp., dans 8,8 % des 1070 échantillons testés pour Campylobacter spp. et dans 4,8 % des 1067 échantillons testés pour STEC.
(Traduit par Isabelle Vallières)
Introduction
Consumers, regulatory agencies, governments, and the food industry all consider food safety to be of the utmost importance. The implementation of Hazard Analysis Critical Control Point (HACCP) programs, Good Manufacturing/Production Practices, and various interventions by slaughter and meat processing facilities play a large role in enhancing the safety of meat products. Baseline studies to determine microbial levels of commensal organisms and pathogen prevalence can be used to assess the effectiveness of these programs and interventions. The United States Department of Agriculture, Food Safety and Inspection Service (USDA/FSIS) conducted several baseline studies of meat and poultry products from federally inspected facilities to determine the prevalence and levels of bacteria of public health concern (1–3). The national baseline data collection programs conducted from June 1997 to May 1998 included testing 1881 sponge samples from beef carcasses and 2127 from pork carcasses for levels of generic Escherichia coli and the prevalence of Salmonella spp. (2,3). In Canada, the Ontario Ministry of Agriculture, Food and Rural Affairs (OMAFRA) conducted baseline studies of poultry, beef, and pork carcasses from provincially inspected abattoirs from 1999 to 2002 to determine the counts of total aerobic bacteria, coliform bacteria, and E. coli, the prevalence and levels of Salmonella spp. and Campylobacter spp., and the prevalence of Listeria monocytogenes and E. coli (4–6).
The importance of baseline information on pathogen prevalence and levels of commensal bacteria as a benchmark for measuring improvements in provincially inspected abattoirs was also recognized in Alberta. The Food Safety and Animal Health Division of Alberta Agriculture and Rural Development initiated baseline studies of poultry, beef, and pork carcasses in provincially inspected abattoirs starting in November 2004. Poultry carcass rinses, beef, and pork carcasses were tested to determine counts of total aerobic bacteria, coliform bacteria and E. coli, and the prevalence of Salmonella spp., Campylobacter spp., and Shiga toxin-producing E. coli (STEC). The results of the baseline study of poultry carcass rinses were recently reported (7). The results of the baseline study for beef and pork carcass sponges collected from provincially inspected Alberta abattoirs are reported herein.
Materials and methods
Experimental design
Samples were collected from provincially inspected cattle and hog slaughter facilities between May 16, 2006 and April 20, 2007. The slaughter facilities were categorized into two strata (high or low numbers of animals slaughtered) based on the annual slaughter numbers from the previous year (2005). The cut-off was a slaughter volume of < 1000 or ≥ 1000 for cattle and < 8000 or ≥ 8000 for hogs. Samples were tested from a total of 50 facilities. Many of the facilities slaughtered both cattle and hogs. Beef carcasses were tested from 35 low-volume and 13 high-volume abattoirs. Pork carcasses were tested from 31 low-volume and 3 high-volume abattoirs.
Sample collection
Beef and pork carcass sponges were collected according to the USDA/FSIS sample collection guidelines (8). Convenience samples were taken based on accessibility of the carcasses in the coolers. Three sterile, pre-moistened sponges (Qualicum Scientific, Ottawa, Ontario) were used to swab adjacent areas on the same side of each carcass. One sponge was used for enumeration of total aerobic bacteria, coliform bacteria, and generic E. coli. The second sponge was used for the isolation of Salmonella spp. and STEC. The third sponge was used for the isolation of Campylobacter spp. The carcass sponges were collected in sterile plastic bags, put on ice in coolers and sent to the laboratory for testing. Samples received at the laboratory at temperatures > 10°C or more than 48 h after collection were rejected.
Culture methods
All media used were Difco brand (Difco Laboratories, Becton Dickinson Microbiology Systems, Sparks, Maryland, USA) unless otherwise indicated. Total aerobic bacteria, coliform bacteria, and generic E. coli were enumerated using the pour plate technique. Fifteen milliliters of 0.1% peptone water were added to the sponge samples and mixed for 2 min using a stomacher (Seward Stomacher 400; Seward Laboratory, London, UK). Ten-fold serial dilutions of carcass sponge diluent were made using 0.1% peptone water, 1-mL aliquots were dispensed into sterile Petri dishes, and molten agar added. Total aerobic bacteria were enumerated on plate count agar incubated at 35°C for 48 h. Coliform bacteria and E. coli were enumerated on Violet Red Bile agar with MUG (4-methylumbelliferyl-β-D-glucuronide) after incubation at 35°C for 24 h. Coliform bacteria were confirmed by the production of gas after incubation in 2% Brilliant Green Bile broth at 35°C for 48 h. Generic E. coli were confirmed by the production of gas and fluorescence after incubation in EC medium with MUG at 45°C for 48 h. Incubated EC medium with MUG that had gas production but no fluorescence was streaked onto Columbia Blood Agar plates (BAP; Oxoid, Nepean, Ontario), incubated at 35°C for 24 h and the identity of at least 1 colony determined by a Vitek instrument (bioMéreiux Canada, St. Laurent, Quebec) using a GNI+ card.
Salmonella spp. were identified using the USDA/FSIS method with modifications (8). Briefly, 50 mL of Buffered Peptone Water (BPW) were added to the carcass sponge. The sample was mixed thoroughly and incubated at 35°C for 24 h ensuring the sponge was immersed in the pre-enrichment broth. One milliliter and 0.1 mL of BPW pre-enrichment culture were transferred to 10 mL of tetrathionate (TT) broth and 10 mL of Rappaport Vassiliadis (RV) broth (EM Science, Merck, Darmstadt, Germany), respectively, and incubated at 42°C for 24 h. One hundred and fifty microliters of each selective enrichment broth (TT and RV) were combined for DNA extraction followed by real-time polymerase chain reaction (PCR) (9). Culture was continued on those samples that were positive by real-time PCR. Enrichment cultures (TT and RV) from positive samples were streaked onto Brilliant Green Sulfa agar (BGS), Xylose-Lysine-Tergitol 4 agar (XLT4) and Rambach agar (Merck, Darmstadt, Germany) and incubated at 35°C. Plates were examined after 24 h of incubation and suspect colonies were selected for biochemical testing. Plates were incubated for an additional 24 h and reexamined for additional suspect colonies.
Suspect colonies were streaked on BAP and MacConkey agar plates and inoculated into Urea agar, Triple Sugar Iron agar and Lysine Iron agar slants. Plates were examined for purity and reactions of the slants and MacConkey agar plates were noted. Isolates that appeared to be Salmonella spp. were further tested using polyvalent O and O1 antisera (Denka Seiken, Tokyo, Japan). Samples that were positive by the real-time PCR assay but negative by the culture method previously described were retested using immunomagnetic separation (IMS). One milliliter each of BPW, TT and RV enrichment broths were mixed with immunomagnetic beads (Dynabeads® anti-Salmonella; Dynal, Lake Success, New York, USA) on a semi-automated magnetic particle processor (KingFisher™ mL, Fisher Scientific, Nepean, Ottawa) according to the manufacturer’s instructions. The bead-bacteria complexes were inoculated into TT, incubated and then plated onto XLT4 and Rambach agar plates and examined for typical Salmonella colonies. Typical colonies were tested using biochemical and serological tests as previously described.
Campylobacter spp. were isolated and enumerated using a modification of the FDA method (10). The sample sponge was added to 90 mL of Campylobacter Enrichment Broth, Bolton formula (Oxoid) with laked horse blood. The sample was placed under microaerobic conditions in an anaerobic jar filled with 10% CO2, 5% O2, 85% N2 and incubated at 42°C for 48 h. After 24 and 48 h of incubation, the enrichment broth was plated onto Campy-line agar (CLA; Dalynn Biologicals, Calgary, Alberta) and Modified Campylobacter Blood-Free Selective agar (CCDA; Oxoid) plates using a 10 μL loop and the plates were incubated microaerobically at 42°C. Plates were examined for typical colonies after 24 h and 48 h incubation. Typical smooth, convex, translucent, colorless to cream colonies on CCDA plates and deep red to magenta, smooth, shiny and convex colonies with a defined edge or flat with an irregular edge on CLA plates were streaked onto BAP. The BAP was incubated microaerobically at 42°C for 24 h. Campylobacter species were identified by wet mount, oxidase, catalase, hippurase, and naladixic acid/cephalothin susceptibility tests.
Shiga toxin-producing E. coli (STEC) were identified following the method cited in Renter et al (11). Briefly, DNA was extracted from the BPW enrichment broths that were used for the isolation of Salmonella spp. Extracted DNA was tested for the presence of the genes for Shiga toxin 1 (stx1) and Shiga toxin 2 (stx2) with the use of PCR in a multiplex assay. Enrichment broths positive for the stx1 and/or stx2 genes were streaked onto MacConkey and Violet Red Bile Agar with MUG and incubated at 42°C for 24 h. Twenty isolated colonies from each plate were stab inoculated onto a MacConkey plate and incubated at 35°C for 24 h. Deoxyribonucleic acid was extracted from the isolates and tested by PCR for the stx1 and stx2 genes. If no positive isolates were identified, the enrichment broth was restreaked onto agar plates and an additional 40 colonies were tested. Only samples in which a positive isolate could be identified were reported as positive. STEC isolates were characterized for O157 antigen using Prolex™ E. coli O157 latex test kit (Pro-Lab Diagnostics, Richmond Hill, Ontario) and verotoxin (Shiga toxin) production using GLISA Duopath™ Verotoxin test kit (EM Science, Merck).
Positive and negative controls were included with each set of samples tested. Positive controls were ATCC strains (S. Typhimurium ATCC 14028, C. jejuni ATCC 33250, and E. coli O157:H7 ATCC 35150) inoculated into the appropriate enrichment broths. Negative controls were uninoculated enrichment broths.
Enumeration data were recorded as colony forming units (CFU)/mL and then converted to CFU/cm2 by dividing the bacterial counts by 12. This was derived from:
Statistical analysis
The chi-squared (χ2) test was used to compare the proportion of positive samples for Salmonella spp. and Campylobacter spp. across the 2 strata for beef and pork samples. Similarly, two-sample Wilcoxon rank-sum (Mann-Whitney) tests were used to compare microbial counts. The statistical analyses were carried out using statistical software (Stata Statistical Software: version 10; StataCorp, College Station, Texas, USA).
Results
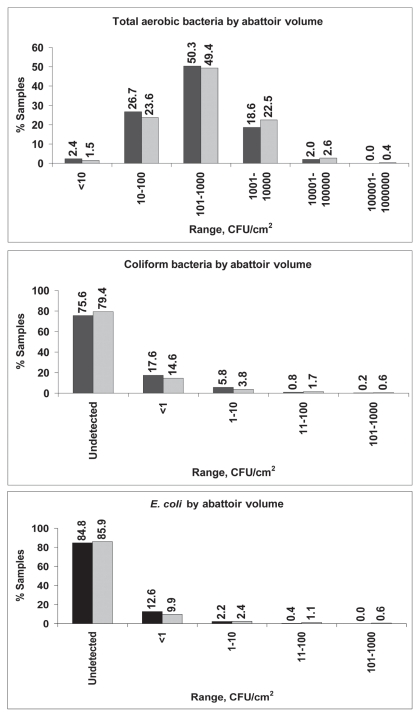
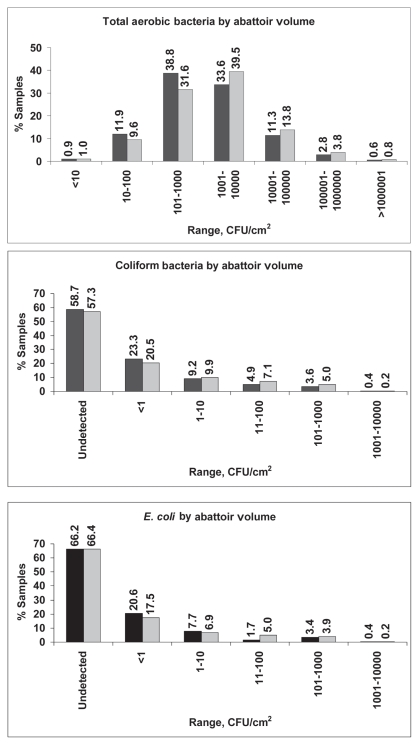
From May 16, 2006 to April 20, 2007, 2245 samples from 50 facilities were collected; however, not all samples met testing criteria. All beef and pork carcass samples tested for total aerobic bacteria (2084) were positive. Coliform bacteria were recovered from 22.4% of 1034 beef carcass samples and 42.0% of 1069 pork carcass samples (P < 0.01) and E. coli were recovered from 14.6% of beef carcasses and 33.7% of pork carcasses (P < 0.01). There was no significant difference between the positive rates for these organisms in high and low-volume abattoirs. Table 1 displays descriptive statistics for counts of commensal organisms on pork and beef carcasses, from both high and low volume abattoirs. The log means for total aerobic bacteria, coliform bacteria, and E. coli were significantly higher in pork carcass samples (all P < 0.01). The frequency distribution of samples according to the levels of these organisms is shown in Figures 1 and 2. Total aerobic bacteria were recovered from 100% of the samples tested, with 99.8% and 96.0% of beef and pork carcass samples, respectively, having counts of ≤ 100 000 CFU/cm2. Most (75.0%) of the beef carcass samples had counts in the range of 10 to 1000 CFU/cm2. Most (71.8%) of the pork carcass samples had aerobic bacteria counts in the range of 101 to 10 000 CFU/cm2. For coliform bacteria, most samples (98.4% for beef and 89.5% for pork) had levels of ≤ 10 CFU/cm2. Most samples (98.9% for beef and 92.7% for pork) had levels of E. coli of ≤ 10 CFU/cm2.
Table 1.
Descriptive statistics of microbiological counts (CFU/cm2) of beef and pork carcasses from high- and low-volume abattoirs
| Beef
|
Pork
|
||||
|---|---|---|---|---|---|
| Bacteria | Summary statistics | Low volume | High volume | Low volume | High volume |
| Total aerobic bacteria | Geometric Mean | 240.9 | 317.2 | 1060.9 | 1525.3 |
| CIb Geometric Mean | 207.3 to 280.1 | 273.7 to 367.6 | 876.7 to 1283.8 | 1259.8 to 1846.7 | |
| Log10 Mean | 2.38 | 2.50 | 3.02 | 3.18 | |
| CI Log10 Mean | 2.32 to 2.45 | 2.44 to 2.56 | 2.94 to 3.11 | 3.10 to 3.27 | |
| Median Value | 225.0 | 291.7 | 916.7 | 1416.7 | |
| 10th percentile | 28.5 | 39.3 | 66.7 | 83.3 | |
| 90th percentile | 2416.7 | 2666.7 | 18333.3 | 28083.3 | |
| Coliform bacteriaa | Geometric Mean | 0.4 | 0.6 | 1.4 | 2.0 |
| CI Geometric Mean | 0.3 to 0.6 | 0.4 to 0.8 | 1.0 to 1.9 | 1.4 to 2.8 | |
| Log10 Mean | −0.37 | −0.23 | 0.14 | 0.29 | |
| CI Log10 Mean | −0.50 to −0.25 | −0.38 to −0.07 | 0.00 to 0.27 | 0.14 to 0.44 | |
| Median Value | 0.3 | 0.4 | 0.8 | 1.2 | |
| 10th percentile | 0.1 | 0.1 | 0.1 | 0.1 | |
| 90th percentile | 3.9 | 13.0 | 46.8 | 141.7 | |
| E. colia | Geometric Mean | 0.3 | 0.6 | 1.0 | 1.7 |
| CI Geometric Mean | 0.2 to 0.4 | 0.4 to 0.9 | 0.7 to 1.5 | 1.1 to 2.5 | |
| Log10 Mean | −0.54 | −0.23 | 0.01 | 0.23 | |
| CI Log10 Mean | −0.68 to −0.40 | −0.43 to −0.02 | −0.15 to 0.16 | 0.06 to 0.40 | |
| Median Value | 0.2 | 0.3 | 0.5 | 0.8 | |
| 10th percentile | 0.1 | 0.1 | 0.1 | 0.1 | |
| 90th percentile | 2.2 | 17.5 | 122.5 | 189.2 | |
For values above detection value.
95% confidence intervals.
Figure 1.
Frequency distribution of total aerobic bacteria, coliform bacteria, and generic E. coli from beef carcass sponge samples by abattoir volume. Low volume abattoirs are shown with black bars and high volume abattoirs are shown with grey bars.
Figure 2.
Frequency distribution of total aerobic bacteria, coliform bacteria, and generic E. coli from pork carcass sponge samples by abattoir volume. Low volume abattoirs are shown with black bars and high volume abattoirs are shown with grey bars.
For foodborne pathogens, the overall results [and 95% confidence intervals (CI)] were as follows: 0.8% (0.5% to 1.3%) of 2112 samples that were tested for Salmonella spp. were positive; 5.2% (4.3% to 6.2%) of 2092 samples that were tested for Campylobacter spp. were positive; and 5.1% (4.2% to 6.1%) of 2085 samples that were tested for STEC were positive. The prevalence of Salmonella spp. and Campylobacter spp. by carcass type is shown in Table 2. The difference between high- and low-volume abattoirs was not significant; therefore, aggregated results are reported. The difference in Campylobacter spp. and Salmonella spp. prevalence between beef and pork samples was statistically significant (P < 0.01 in both cases). This was not the case with STEC (P = 0.52). Only one of the STEC isolates from a beef carcass was determined by agglutination assays to be E. coli O157. Isolates from samples that were positive for Campylobacter spp. were further characterized to the species level with 84.4% of the isolates identified as C. coli, 13.8% as C. jejuni, and 1.8% of the isolates identified as C. lari. There were differences between beef and pork carcasses in the species of Campylobacter identified from positive samples. For beef carcass samples 73.3% and 2.7% of the positive isolates were identified as C. jejuni and C. coli, respectively. For pork carcass samples, 4.3% and 93.6% of the positive isolates were identified as C. jejuni and C. coli, respectively.
Table 2.
Proportion of beef and pork carcass sponge samples positive for Salmonella spp., Campylobacter spp., and Shiga toxin-producing E. coli (STEC)
| Number of samples (Number positive)
|
% Positive (95% CI)
|
|||||
|---|---|---|---|---|---|---|
| Beef | Pork | Total | Beef | Pork | Total | |
| Salmonella | 1036 (1) | 1076 (17) | 2112 (18) | 0.1a (0.0–0.5) | 1.6a (0.9–2.5) | 0.8 (0.5–1.3) |
| Campylobacter | 1022 (15) | 1070 (94) | 2092 (110) | 1.5a (0.8–2.4) | 8.8a (7.2–10.6) | 5.2 (4.3–6.2) |
| STEC | 1018 (55) | 1067 (51) | 2085 (106) | 5.4 (4.1–7.0) | 4.8 (3.6–6.2) | 5.1 (4.2–6.1) |
CI — 95% confidence intervals;
P < 0.01; STEC — Shiga toxin-producing E. coli.
Discussion
Microbiological studies are needed to assess the effectiveness of HACCP programs, Good Manufacturing/Production Practices, and interventions used in meat slaughter and processing facilities. In the absence of ongoing microbiological monitoring programs, such as those conducted in some European countries, baseline studies can serve as a means of benchmarking the current levels of hygiene in slaughter plants (12). Prior to this study there was no information on the levels of pathogens or commensal bacteria on beef and pork carcasses processed in Alberta provincially inspected meat facilities.
The percentages of carcass swab samples positive for total aerobic bacteria in this study were similar to those in the 1998 OMAFRA study (7,9), in which all the carcasses tested contained aerobic bacteria. The percentage of carcasses containing coliform bacteria and E. coli were slightly lower in Alberta than that reported in Ontario where 27.8% and 18.6% of beef carcasses and 61.3% and 39.5% of pork carcasses were positive for coliform bacteria and E. coli, respectively (4,6). The USDA baseline studies measured the levels of generic E. coli on beef and pork carcasses and these were very similar to those found in this study. The USDA reported that 98.9% of beef and 91.5% of pork carcasses contained ≤ 10 CFU/cm2 generic E. coli (13,14) and we found that 98.9 and 92.7% of beef and pork carcasses, respectively, had 10 or fewer CFU/cm2 E. coli.
In regards to the percentage of carcasses positive for food-borne pathogens, there was more variation among the different studies. In this study, Salmonella spp. were isolated from 0.1% and 1.6% of beef and pork carcasses, respectively, whereas the OMAFRA baseline found 1.6% of beef carcasses and 4.8% of pork carcasses positive (4,6). The USDA also reported higher results with 1.2% and 6.9% of beef and pork carcasses, respectively, positive for Salmonella spp. (2,3). However, a national survey in Australia in 2004 did not isolate any Salmonella spp. from 1147 chilled beef carcasses (15). Several other countries have also investigated the level of Salmonella spp. on pork carcasses and the reported results revealed that there is much variation in the percentage of positive carcasses. A study in Great Britain found that 5.3% were positive (13), a baseline of Swedish slaughterhouses revealed that no Salmonella spp. were isolated from pre-chilled carcasses (14), a study in Belgian abattoirs found an overall prevalence of 16.1% from carcasses sampled from the chill-room 2 to 4 h after slaughter (16), and a baseline study of pre-chill carcasses in Taiwan showed that 1.7% were positive for Salmonella spp. (17).
In Alberta, 1.5% of beef carcasses were positive for Campylobacter spp. and this is the same positive rate that was reported by OMAFRA (4). However, only 8.8 % of pork carcasses from Alberta contained Campylobacter spp. whereas Ontario reported a positive rate of 26.7% (6). The 2004 Australian national survey found that 0% of chilled beef carcasses were positive for Campylobacter spp. (13) which is similar to the low positive rate found in the Canadian studies. Baseline surveys of pre-chill pork carcasses in Sweden (14) and Taiwan (17) found that 1.0% and 13.8%, respectively, were positive for Campylobacter spp., which indicates wide variation in reported positive rates.
We determined that 5.4% of beef and 4.8% of pork samples were positive for STEC. However, only 1 of the isolates from a beef carcass (0.1%) was toxigenic E. coli O157. Other countries have also reported low positive rates for E. coli O157:H7. For beef carcasses, 0.1% were positive in Australia (15) and 2.7% were positive in Mexico (18). For pork carcasses, no E. coli O157:H7 were isolated from baseline studies in Sweden (14) or Taiwan (17) as was the case in this study. The OMAFRA study also identified pathogenic E. coli; however, they reported that only 0.3% of beef carcasses and 2.1% of pork carcasses were positive for verotoxigenic E. coli (4,6). The differences between the Alberta and Ontario results may be due to the different methodologies used. In this study PCR was used to identify STEC, whereas Ontario used a Vero cell assay to identify verotoxigenic E. coli.
Caution must be used when comparing results from different studies as variations in results may be due to differences in contamination rates, differences in laboratory methodologies, variations in sampling technique, time during the process when samples were obtained (for example, pre-chill, post-chill), study design, and seasonal or yearly effects.
The results of this baseline study indicate that improvements need to be made in Alberta’s provincially inspected abattoirs and/or other sections of the beef and pork production chain to reduce the prevalence of pathogenic bacteria and lower the levels of bacteria on beef and pork carcasses. Efforts are currently underway in Alberta to improve Good Manufacturing Practices and implement interventions in provincially inspected abattoirs. Subsequent studies will determine the effectiveness of these efforts in reducing microbial levels and pathogen prevalence.
Acknowledgments
The contributions of Claude Baker, Erin Benson, Jill Binder, Sylvia Checkley, Ken Dies, Ken Fahner, Ron Feniuk, Janice Futz, Becky Goulding, Sheila Hart, Robert Holowaychuk, Danielle Kneeland, Daryl Loback, Zelda Matthee, Denise Maxwell, Mark Miller, Marjan Morravej, Kate Sambey, Raminder Sandhu, Kevin Smith, Ross Smith, Tracy Stroud, Brenda Tchir, Mary VanderKop, Amanda Veldhuizen, Lea Wachowich, Kim Whitehead, Murray Yaschuk, and Amanda Zoefer are acknowledged. We thank John Coffin, Barbara Dakin, Ashwin Deo, Sandra Dyal, Suzanne Gibson, Kyla Kennedy, Robin King, Wayne Lazaroff, Denise Patterson, Cathy Sheppard, Catherine Taylor, and Cheryl Turner for their technical support. CVJ
Footnotes
Use of this article is limited to a single copy for personal study. Anyone interested in obtaining reprints should contact the CVMA office (hbroughton@cvma-acmv.org) for additional copies or permission to use this material elsewhere.
References
- 1.USDA. United States Department of Agriculture, Food Safety and Inspection Service, Nationwide Broiler Chicken Microbiological Baseline Data Collection Program, July 1994–June 1995. [Last accessed July 25, 2011]. Available from http://www.fsis.usda.gov/OPHS/baseline/broiler1.pdf.
- 2.USDA. Nationwide Sponge Microbiological Baseline Data Collection Program: Cattle, June 1997–May 1998. [Last accessed July 25, 2011]. Available from http://www.fsis.usda.gov/PDF/Baseline_Data_Cattle.pdf.
- 3.USDA. Nationwide Sponge Microbiological Baseline Data Collection Program: Swine, June 1997–May 1998. [Last accessed July 25, 2011]. Available from http://www.fsis.usda.gov/PDF/Baseline_Data_Swine.pdf.
- 4.Johnson P, Mahdi A, Baker T, Odumeru J. Microbiological analysis of raw beef carcasses in Ontario abattoirs — A summary report. Food Inspection Branch, Ontario Ministry of Agriculture and Food; 2003. [Google Scholar]
- 5.Johnson P, Mahdi A, Baker T, Odumeru J. Microbiological analysis of raw chicken carcasses in Ontario abattoirs — A summary report. Food Inspection Branch, Ontario Ministry of Agriculture and Food; 2003. [Google Scholar]
- 6.Johnson P, Mahdi A, Baker T, Odumeru J. Microbiological analysis of raw pork carcasses in Ontario abattoirs — A summary report. Food Inspection Branch, Ontario Ministry of Agriculture and Food; 2003. [Google Scholar]
- 7.Bohaychuk VM, Checkley SL, Gensler GE, Romero Barrios P. Microbiological baseline study of poultry slaughtered in provincially inspected abattoirs in Alberta, Canada. Can Vet J. 2009;59:173–178. [PMC free article] [PubMed] [Google Scholar]
- 8.USDA. Appendix E — FSIS Sample Collection Guidelines and Procedure for Isolation and Identification of Salmonella from Raw Meat and Poultry Products. [Last accessed July 25, 2011];Federal Register. 1996 61(144) Rules and Regulations. Available from http://www.fsis.usda.gov/OA/fr/rule3.pdf#Appendix%20E. [Google Scholar]
- 9.Bohaychuk VM, Gensler GE, McFall ME, King RK, Renter DG. A real-time PCR assay for the detection of Salmonella spp. in a wide variety of food and food-animal matrices. J Food Prot. 2007;70:1080–1087. doi: 10.4315/0362-028x-70.5.1080. [DOI] [PubMed] [Google Scholar]
- 10.Hunt JM, Abeyta C, Tran T. Bacteriological Analytical Manual Online. U.S. Food and Drug Administration, Center for Food Safety and Applied Nutrition; 2001. [Last accessed July 25, 2011]. Isolation of Campylobacter Species from Food and Water. Available from http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/default.htm. [Google Scholar]
- 11.Renter DG, Bohaychuk VM, Van Donkersgoed J, King RK. Presence of shiga toxin-producing Escherichia coli in feces from feedlot cattle in Alberta and absence on corresponding beef carcasses. Can J Vet Res. 2006;2006;71:230–235. [PMC free article] [PubMed] [Google Scholar]
- 12.Anonymous. The Community Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in the European Union in 2008. EFSA Journal. 2010;8:1496. [Google Scholar]
- 13.Davies RH, Dalziel R, Gibbens JC, et al. National survey for Salmonella in pigs, cattle and sheep at slaughter in Great Britain (1999–2000) J Appl Microbiol. 2004;2004;96:750–760. doi: 10.1111/j.1365-2672.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- 14.Lindblad M, Lindmark H, Lambertz ST, Lindqvist R. Microbiological baseline study of swine carcasses at Swedish slaughterhouses. J Food Prot. 2007;70:1790–1797. doi: 10.4315/0362-028x-70.8.1790. [DOI] [PubMed] [Google Scholar]
- 15.Phillips D, Jordan D, Morris S, Jenson I, Sumner J. A national survey of the microbiological quality of beef carcasses and frozen boneless beef in Australia. J Food Prot. 2006;69:1113–1117. doi: 10.4315/0362-028x-69.5.1113. [DOI] [PubMed] [Google Scholar]
- 16.Delhalle L, De Sadeleer L, Bollaerts K, et al. Risk factors for Salmonella and hygiene indicators in the 10 largest Belgian pig slaughterhouses. J Food Prot. 2008;71:1320–1329. doi: 10.4315/0362-028x-71.7.1320. [DOI] [PubMed] [Google Scholar]
- 17.Yeh KS, Chen SP, Lin JH. One-year (2003) nationwide pork carcass microbiological baseline data survey in Taiwan. J Food Prot. 2003;68:458–461. doi: 10.4315/0362-028x-68.3.458. [DOI] [PubMed] [Google Scholar]
- 18.Varela-Hernandez JJ, Cabrera-Diaz E, Cardona-Lopez MA, et al. Isolation and characterization of Shiga toxin-producing Escherichia coli O157:H7 and non-O157 from beef carcasses at a slaughter plant in Mexico. Int J Food Microbiol. 2007;113:237–241. doi: 10.1016/j.ijfoodmicro.2006.06.028. [DOI] [PubMed] [Google Scholar]