Abstract
Interactions of proteins with low-molecular-weight ligands, such as metabolites, cofactors, and allosteric regulators, are important determinants of metabolism, gene regulation, and cellular homeostasis. Pharmaceuticals often target these interactions to interfere with regulatory pathways. We have developed a rapid, precise, and high-throughput method for quantitatively measuring protein-ligand interactions without the need to purify the protein when performed in cells with low background activity. This method, differential radial capillary action of ligand assay (DRaCALA), is based on the ability of dry nitrocellulose to separate the free ligand from bound protein–ligand complexes. Nitrocellulose sequesters proteins and bound ligand at the site of application, whereas free ligand is mobilized by bulk movement of the solvent through capillary action. We show here that DRaCALA allows detection of specific interactions between three nucleotides and their cognate binding proteins. DRaCALA allows quantitative measurement of the dissociation constant and the dissociation rate. Furthermore, DRaCALA can detect the expression of a cyclic-di-GMP (cdiGMP)-binding protein in whole-cell lysates of Escherichia coli, demonstrating the power of the method to bypass the prerequisite for protein purification. We have used DRaCALA to investigate cdiGMP signaling in 54 bacterial species from 37 genera and 7 eukaryotic species. These studies revealed the presence of potential cdiGMP-binding proteins in 21 species of bacteria, including 4 unsequenced species. The ease of obtaining metabolite-protein interaction data using the DRaCALA assay will facilitate rapid identification of protein-metabolite and protein-pharmaceutical interactions in a systematic and comprehensive approach.
Keywords: whole-cell assay
Interactions of low-molecular-weight ligands with protein receptors are critical in biological signaling both between cells and within individual cells. Examples of intercellular signaling mediated by small molecules include quorum signaling in bacteria, hormone and neurotransmitter responses in endocrine systems of animals, and auxin and abscisic acid regulation in plants (1). Intracellular signaling also involves regulatory protein-binding molecules, such as calcium and cyclic nucleotides [e.g., cAMP, cGMP, cyclic-di-GMP (cdiGMP)] (2, 3). In fact, nucleotide receptors are often targets for therapeutic intervention (4). Thus, these protein-small ligand interactions have important implications in modern drug design and use. Considering that many protein-ligand interaction pairs represent potential targets of pharmaceutical intervention in disease or agriculture, there is an urgent need to collect qualitative and quantitative data for such protein-ligand interactions in a high-throughput manner. Current efforts in metabolomics are directed at cataloging the presence of various metabolites through mass spectrometric analysis of biological samples (5–8). However, this approach lacks the ability to confirm interactions with protein partners, and therefore fails to reveal functional significance. Thus, the study of the interactions of a specific metabolite with all available cellular proteins, which we term “metabolite interactomics,” has been limited by the available assay systems. Current assays for specific protein-ligand interactions, including equilibrium dialysis, filter-binding assays, ultracentrifugation, isothermal calorimetry (ITC), surface plasmon resonance, and many other assays (9–15), are not high throughput because they are limited by sample processing time, equipment requirements, and assay-specific manipulations. Protein array technology requires purified proteins fixed on solid support (16). Although protein array technology is feasible and quite powerful (17–19), large-scale protein purification is limited by individual protein characteristics that often hinder isolation of functionally active proteins. Currently, protein array technology is limited to only a few laboratories capable of performing mass parallel purification of functional proteins and arraying them. To bypass such constraints of existing assays, we have developed a high-throughput differential radial capillary action of ligand assay (DRaCALA) that can be used to detect binding and to quantitate the fraction of a small-molecule ligand that is bound to a protein of interest. DRaCALA is rapid and quantitative, and it allows detection of protein-ligand interactions for both purified proteins and proteins expressed in whole cells, thus bypassing the requirement for protein purification. Unlike most comparable protein-ligand detection systems, DRaCALA does not require a wash step; thus, the total ligand available to protein is quantifiable, resulting in an accurate, simple, and precise measure of the fraction of ligand bound.
Results
Principle of DRaCALA.
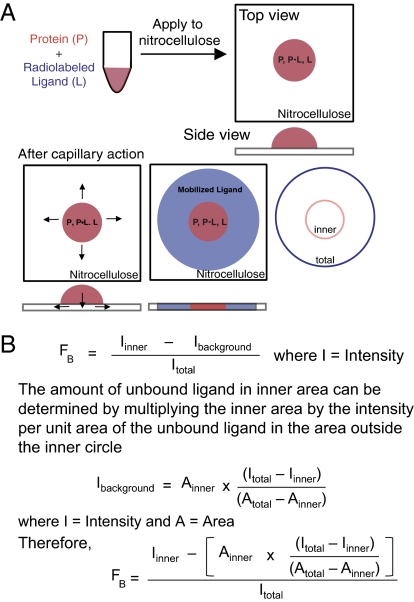
DRaCALA exploits the ability of nitrocellulose membranes to sequester proteins preferentially over small-molecule ligands. When a mixture of protein and radiolabeled ligand is spotted onto a dry nitrocellulose membrane, protein and bound ligand are immobilized at the site of contact, whereas free ligand is mobilized by capillary action with the liquid phase (Fig. 1A). DRaCALA is a rapid assay because the capillary action can be completed in less than 5 s. Because DRaCALA does not use a wash step, the pattern of ligand migration allows rapid detection of both the total ligand and the ligand sequestered by proteins. Because capillary action distributes the unbound ligand throughout the mobile phase, the calculation for the fraction bound (FB) must correct for this background (see below for edge effects at the solvent front and annulation of the protein). Therefore, FB is defined by the equation in Fig. 1B, where Iinner is the intensity of signal in the area with protein (inner circle) and Itotal is the intensity of total signal of the entire sample (outer circle). The Iinner signal consists of both ligand bound to protein and unbound ligand that has not mobilized beyond the area of the inner circle, which we define as Ibackground. Ibackground can be calculated by subtracting the Iinner from the total ligand Itotal and adjusting for the relative areas of the inner (Ainner) and outer (Atotal) circles (Fig. 1B). For free ligand alone, the signal for the ligand is not sequestered, and therefore has a baseline FB of 0.01 ± 0.04 (Fig. S1A), whereas protein alone does not mobilize on nitrocellulose (Fig. S1B). Herein, we demonstrate the uses of DRaCALA for rapid detection of metabolite binding to proteins and describe potential uses for identification of unknown interactions and pharmaceutical screening of protein agonists and antagonists.
Fig. 1.
Principle of DRaCALA. (A) Schematic representation of DRaCALA assay on application of protein-ligand mixture onto nitrocellulose and capillary action. Protein (P), ligand (L), and protein–ligand complex (P•L) distribution during the assay is shown. (B) Equations are used to analyze DRaCALA data for FB for purified proteins. An explanation of the apparent edge effect at the capillary migration front is provided in Fig. S3.
DRaCALA Detection of Protein-Ligand Interactions.
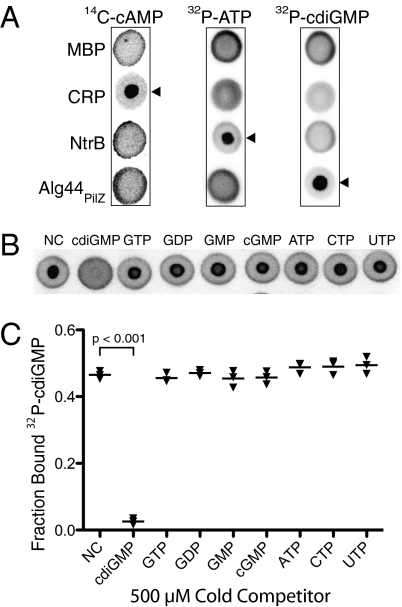
The principle of DRaCALA was illustrated by measuring ligand binding to known nucleotide binding proteins: Pseudomonas aeruginosa Alg44PilZ binds cdiGMP (20), Escherichia coli cyclic AMP receptor protein (CRP) binds cAMP (21, 22), and E. coli nitrogen regulator B (NtrB) binds ATP (23). Radiolabeled ligands were incubated with each of the proteins, and the mixtures were spotted on nitrocellulose. After spreading by capillary action, membranes were dried and quantitated using a phosphorimager. Maltose-binding protein (MBP), which does not bind to any of these small molecules, was used as a control. As expected, each of the radiolabel signals from MBP mixtures was distributed by capillary action (Fig. 2A). CRP specifically bound cAMP, as demonstrated by the sequestration of the signal, but it did not bind cdiGMP or ATP (21, 22) (Fig. 1A). NtrB bound ATP but not cdiGMP or cAMP (23) (Fig. 2A). Similarly, Alg44PilZ bound cdiGMP but not cAMP or ATP (20) (Fig. 2A). The specificity of Alg44PilZ binding to cdiGMP was further tested by competition with excess unlabeled nucleotides (400-fold molar excess relative to the Alg44PilZ protein). Alg44PilZ binding to 32P-cdiGMP was abolished by cdiGMP but not by cGMP, GMP, GDP, GTP, ATP, CTP, or UTP as was previously described (Fig. 2B). The FB for cdiGMP was 0.31 ± 0.07, which was reversed by competition with unlabeled cdiGMP to the background level of 0.04 ± 0.01 (Fig. 2C).
Fig. 2.
Detection of specific protein-ligand interactions by DRaCALA. (A) DRaCALA images of interactions between purified proteins (20 μM) incubated with 500 nM 14C-cAMP, 4 nM [32P]ATP, or 4 nM 32P-cdiGMP. Protein-ligand mixtures were spotted on nitrocellulose and allowed to dry before imaging using a Fuji FLA7100 PhosphorImager. Cognate protein-nucleotide combinations are indicated by arrowheads. MBP was used as a negative control. (B) DRaCALA images of competition assays assessing the ability of 1 mM indicated cold nucleotides to compete with binding interactions between 4 nM 32P-cdiGMP and 2.5 μM HisMBP-Alg44PilZ. (C) Graph of FB for each sample in Fig. 1B, with averages indicated by a horizontal bar. NC, no competitor. P values were determined by a Student t test for significant differences compared with the NC control for three independent experiments. The Itotal of each DRaCALA spot in Fig. 2 A and B is provided in Tables S1 and S2, respectively.
Use of DRaCALA to Quantitate Protein-Ligand Interactions.
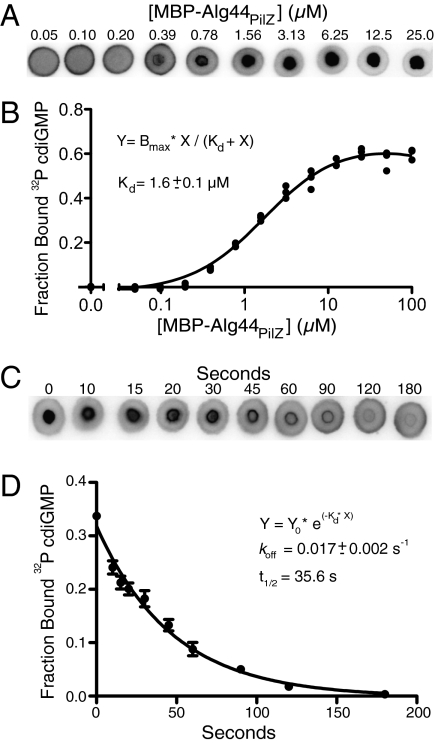
In addition to qualitative assessments of specific protein-ligand interactions, DRaCALA is useful for quantitating biochemical parameters, including the dissociation constant (Kd) and the dissociation rate (koff). The Kd can be measured by altering either the protein or ligand concentration in titration experiments; because DRaCALA detects only the ligand mobility, ligand concentrations are always held constant. As an example, the Kd of Alg44PilZ binding to cdiGMP was determined by analyzing mixtures of 4 nM 32P-cdiGMP with 0.006–100 μM Alg44PilZ (Fig. 3A). At concentrations of protein above the Kd, the FB approaches saturation; this binding decreases as the Alg44PilZ concentration is decreased, reaching a level indistinguishable from background at the lowest protein concentrations. Analysis of this binding curve indicated that Kd = 1.6 ± 0.1 μM, which is in reasonable agreement with our previously reported value of 5.6 μM determined by ITC (20) (Fig. 3B). Application of identical samples to the dot blot apparatus for a vacuum-mediated filter-binding assay resulted in problems associated with high protein concentrations and, as a consequence, difficulty in assessing saturation of binding (Fig. S2). Because the assay is completed in less than 5 s, DRaCALA can also be used to determine the koff for those protein–ligand complexes with slower off rates. The koff was determined for cdiGMP and Alg44PilZ by spotting at the indicated time points after the addition of 1 mM unlabeled cdiGMP to a preincubated mixture of Alg44PilZ and 32P-cdiGMP (Fig. 3C). The fractions bound were plotted against time and analyzed by nonlinear regression, which yielded a koff of 0.017 ± 0.002 s−1, corresponding to a t1/2 of 35.6 ± 10.7 s (Fig. 3D). The binding of 32P-cdiGMP to Alg44PilZ was completely competed away by 1 mM unlabeled cdiGMP within 90 s. Occasionally, we observed an increased signal at the leading edge of the capillary action and in the protein portion of the DRaCALA spot. Both edge and annulation effects are explained in Fig. S3. The edge effect is attributable to evaporation of the solvent during the time of the experiment, and it is dependent on the humidity of the local environment around the nitrocellulose support. The evaporation results in a smaller total area (Atotal observed in Fig. S3A) and leads to an increased value in the calculated Ibackground. As a secondary correction for the edge effect, the FB determined for spotted ligand in the absence of protein can be subtracted from all samples in parallel (Fig. S3B). The annulation effect does not alter the FB calculation (Fig. S3C). Results from these experiments demonstrate the utility of DRaCALA for rapid and precise quantitation of biochemical parameters.
Fig. 3.
Determination of Kd and koff by DRaCALA. (A) DRaCALA images were used for Kd determination for the interaction of Alg44PilZ and cdiGMP. His-MBP-Alg44PilZ was varied from 100 μM to 6 nM, and the 32P-cdiGMP was held constant at 4 nM. Representative images of six sets of DRaCALA experiments are shown for 40 nM to 25 μM Alg44 protein. (B) FB from data in A were plotted as a function of [MBP-Alg44PilZ], and the best-fit line was determined by nonlinear regression using the indicated equation. A no-protein control was also plotted. The fitting program varied both Kd and Bmax to obtain the best fit, indicated by the solid line. (C) koff was determined by spotting protein-ligand mixtures onto nitrocellulose at various times after the addition of 1 mM cold cdiGMP to a mixture of 4 nM 32P-cdiGMP and HisMBP-Alg44PilZ. (D) Time course of decrease of FB from analysis of the data in C was fitted to a single exponential decay, indicating koff. The Itotal of each DRaCALA spot in Fig. 3 A and C is provided in Tables S3 and S4, respectively.
DRaCALA Detection of Ligand-Binding Proteins in Whole Cells.
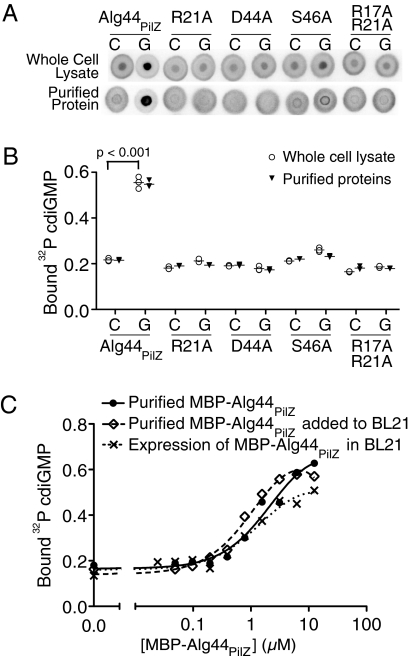
A major limitation of most biochemical assays is the requirement for purified protein. We asked whether DRaCALA could be applied to crude extracts to overcome this limitation. Alg44PilZ binding to cdiGMP requires a number of conserved residues, including R17, R21, D44, and S46, in the PilZ domain of the protein (20, 24). E. coli BL21(DE3) expressing Alg44PilZ and variants with R21A, D44A, S46A, and R17A/R21A substitutions were lysed and tested for binding to cdiGMP using DRaCALA. Protein extracts from E. coli expressing MBP alone did not bind cdiGMP (Fig. S1 C and D). Only the whole-cell lysates from E. coli expressing WT Alg44PilZ sequestered 32P-cdiGMP (Fig. 4A). Specificity of 32P-cdiGMP sequestration in the background of all other cellular macromolecules was demonstrated by competition with 1 mM unlabeled specific competitor cdiGMP or the nonspecific competitor GTP. A significant difference between the bound fractions for cdiGMP or GTP competition experiments was detected for the WT Alg44PilZ but not for the PilZ domain mutants (Fig. 4 A and B). The results from whole-cell lysates are in agreement with the results obtained with purified proteins (20) (Fig. 4B). The sensitivity of DRaCALA detection of MBP-Alg44PilZ binding to cdiGMP was tested by testing serial dilutions of purified protein alone or in the presence of BL21(DE3) whole-cell lysates. The results show that the binding of cdiGMP by MBP-Alg44PilZ is not affected by the presence of cellular proteins (Fig. 4C). Furthermore, serial dilution of extracts from BL21(DE3) cells expressing MBP-Alg44PilZ also resulted in a similar binding curve for comparable levels of MBP-Alg44PilZ proteins (Fig. 4C and Fig. S4). A common problem during expression of heterologous protein in a foreign host is that the protein is often insoluble and forms inclusion bodies. Expression of both Alg44PilZ and PelD without the MBP tag resulted in insoluble proteins (Fig. S5A). We tested the whole-cell extracts with soluble and insoluble proteins and found that either form of the protein can specifically sequester cdiGMP by DRaCALA (Fig. S5 B and C). Serial dilution of the whole-cell extracts reduced cdiGMP sequestration to background levels for BL21 whole-cell extracts (Fig. S5D). The ability to detect protein-ligand interactions in whole-cell lysates makes DRaCALA amenable to high-throughput analysis of whole-cell lysates for the presence of ligand-binding proteins.
Fig. 4.
Detection of specific protein-ligand interaction in whole-cell lysates by DRaCALA. (A) Images of Alg44PilZ interaction with 4 nM 32P-cdiGMP and either 1 mM cold cdiGMP or GTP with purified proteins or when expressed in E. coli BL21(DE3). C, cdiGMP; G, GTP. (B) Graph of 32P-cdiGMP binding by whole-cell lysate samples (○) and purified proteins (▼) in Fig. 4A, with the average indicated by a horizontal bar. P values were determined by a Student t test for significant differences compared with the no-competitor control for three independent experiments. The Itotal of each DRaCALA spot in Fig. 4B is provided in Tables S5 and S6. (C) Graph of 32P-cdiGMP binding by purified MBP-Alg44PilZ, purified MBP-Alg44PilZ added to BL21 whole-cell lysates, and whole-cell lysates of BL21(DE3) overexpressing MBP-Alg44PilZ. Protein concentrations were determined by separation on SDS/PAGE and staining with Coomassie blue (Fig. S4).
DRaCALA Detection of cdiGMP-Binding Proteins in Diverse Prokaryotic and Eukaryotic Organisms.
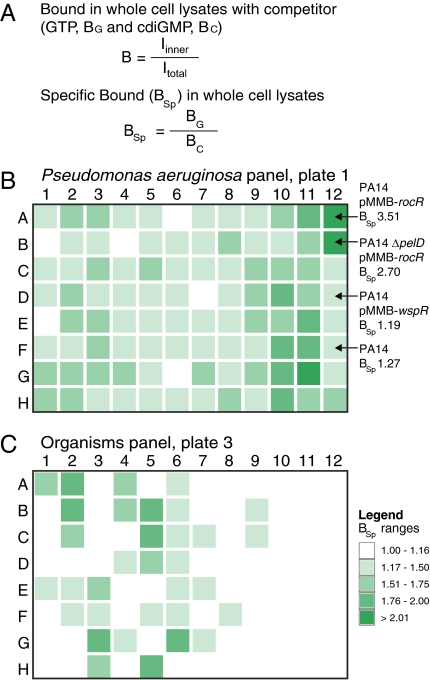
The applicability of DRaCALA to high-throughput metabolite interactomics was demonstrated by screening for binding proteins of an important secondary signaling dinucleotide, cdiGMP. Recent findings have identified cdiGMP as the signaling molecule that controls biofilm formation, motility, and a number of other bacterial functions (2, 3, 25–27). Although the enzymes known to synthesize and degrade cdiGMP are restricted to bacteria, there are questions as to which bacterial species express cdiGMP-binding proteins. cdiGMP has also proven to be useful as an adjuvant during immunization to enhance the mammalian immune response (28), which suggests that there may be cdiGMP-binding proteins in higher eukaryotes. We used DRaCALA to test 191 strains of P. aeruginosa and a panel of 61 other species in a 96-well plate format. As a control for specificity, each extract was tested for binding to the labeled ligand by competition with the unlabeled specific or nonspecific ligand. As in the example of whole-cell lysates of E. coli, unlabeled GTP competitor was used to detect specific 32P-cdiGMP binding [bound during GTP competition (BG)] and unlabeled cdiGMP competitor was used to detect nonspecific 32P-cdiGMP binding [bound during cdiGMP competition (BC)] (Fig. 5A). The ratio of BG to BC is called specific binding (BSp). The limit of nonspecific binding was calculated by adding 2 SDs to the average BC, resulting in a conservative cutoff value for a positive BSp value of 1.17 (Fig. 5A and Table S7). Of the 191 P. aeruginosa isolated from various sources, 184 (96%) displayed a positive BSp value greater than 1.17 (96 samples shown in Fig. 5B and all data presented in Table S7). These results suggest that most P. aeruginosa strains express detectable levels of cdiGMP-binding proteins. When strains isolated from different sources were analyzed for cdiGMP binding, all groups had an average BSp value greater than 1.48, suggesting that cdiGMP signaling is retained (Fig. S6 and Table S7). The range of cell lysate concentrations required for consistent signal detection was tested by diluting cell extracts to 10–60 absorbance (280 nm) units in intervals of 10. Each lysate dilution yielded similar BSp values, suggesting that this range of cell lysate concentration provides a reliable readout for the detection of cdiGMP binding (Fig. S7).
Fig. 5.
Analysis of cdiGMP-binding proteins in various organisms. BSp values of whole-cell lysates are shown as a heat map using the range indicated in the legend. (A) Equations used to analyze DRaCALA data for BSp for whole-cell lysates or tissue extracts. (B) Plate 1 is the analysis of cdiGMP binding by lysates from P. aeruginosa isolates. Specific strains discussed in the text are indicated by arrows. Sources of all strains in plates 1 and 2, as well as the raw data for each lysate, are shown in Table S7. (C) Plate 3 is the analysis of cdiGMP binding by lysates from various organisms. Plate numbers, column numbers, and row letters correspond to the strains and organisms listed in Tables S7 and S8.
One potential complicating factor is the effect of cdiGMP metabolism and endogenous cdiGMP levels on the DRaCALA readout. Extracts of the laboratory P. aeruginosa strain PA14 overexpressing either the phosphodiesterase RocR (29) or the diguanylate cyclase (DGC) WspR (30) were tested for their ability to bind cdiGMP. WT PA14 showed a BSp value of 1.27 indicating that cdiGMP-binding proteins are not fully occupied by endogenous cdiGMP consistent with what is expected for signaling systems (F12 of plate 1 of Fig. 5B and Table S7). Increasing the cellular cdiGMP concentration through WspR overexpression decreased the BSp value to 1.19 (D12 of Fig. 5B and Table S7). Reducing cellular cdiGMP through RocR overexpression increased the BSp value to 3.51 (A12 in Fig. 5B and Table S7). The amount of cdiGMP sequestered by a whole-cell extract should reflect the amount and affinity of the binding proteins present. This was tested by overexpression of RocR in the PA14ΔpelD background, which lacks the cdiGMP-binding protein PelD (31). Without PelD, the BSp value was reduced to 2.70 (B12 in Fig. 5B and Table S7), indicating that PelD is an important binding protein for cdiGMP and that other proteins also bind cdiGMP (20, 31). These results indicate that endogenous cdiGMP metabolism affects but does not abolish the ability of DRaCALA to detect cdiGMP-binding proteins.
cdiGMP signaling occurs in a wide variety of bacterial species but is not known to be present in Eukarya (32). We tested 54 bacterial species from 37 genus and 7 eukaryotic species, including protozoa, fungi, nematodes, plants, and mammals. Of the 82 tested bacteria strains, 31 (38%) displayed a BSp value greater than 1.17. Included in the 31 positive samples are 21 species for which functional cdiGMP signaling has yet to be demonstrated, of which 4 species, Serratia marcescens, Pseudomonas alcaligenes, Pseudomonas diminuta, and Brevundimonas vesicularis, have not yet been sequenced (Fig. 5C and Table S8). We tested 6 bacterial species with sequenced genomes but do not have annotated DGCs, and all of them failed to sequester cdiGMP above the threshold. We also tested 8 eukaryotic species, none of which have annotated DGCs. Whole-cell extracts of protozoa, fungi, and nematodes displayed a BSp value below 1.17, indicating that cdiGMP-binding proteins are absent or below the limit of detection. Mammalian tissue extracts from rodent and human cell lines displayed high nonspecific binding with BC values greater than 3 SDs above the average BC value (>0.233; F6, E12, G12, and H12 in Fig. 5C and Table S8). Furthermore, the nonspecific binding was eliminated after three twofold dilutions of these tissue extracts, indicating that mammalian tissues may contain receptors with low affinity or low abundance. Only positive BSp results from DRaCALA can be interpreted for utilization of cdiGMP signaling. As a result, DRaCALA is most effective in whole-cell extracts with low nonspecific binding. Utilization of DRaCALA in a high-throughput format has expanded our knowledge of the bacterial organisms harboring cdiGMP-binding proteins and confirmed the absence of abundant high-affinity cdiGMP-binding proteins in eukaryotes.
Discussion
In this work, we have developed and characterized DRaCALA as a rapid and precise method for quantitatively measuring protein-ligand interactions. We show the utility of DRaCALA by using the example of cdiGMP binding to Alg44PilZ as a proof of principle. The dissociation constant of 1.6 μM obtained by DRaCALA is similar to those obtained in previous studies using filter binding, ITC, and surface plasmon resonance assays (20, 31). Previous studies of the dissociation rate of cdiGMP from Alg44PilZ were based on saturating the protein with radiolabeled cdiGMP and separating the protein–ligand complex from unbound cdiGMP over a Sephadex column. The t1/2 of the complex was estimated by filter binding as 5 min, in contrast to 35.6 ± 10.7 s as detected by DRaCALA. This discrepancy is likely attributable to two key differences between the two assays. First, DRaCALA is able to quantify the total signal directly in each sample. Because of the various separation steps required for the filter-binding assay, the total ligand in each sample is just assumed to be equivalent. For DRaCALA, the total signal of labeled ligand is known for each individual sample, therefore eliminating the need to assume that the total signal is equivalent. The ability to detect the total signal and FB significantly increases the precision of the measurement and reduces the error incurred from pipetting and other physical manipulations. Second, the processing times of the assays are dramatically different. The filter assay involved binding, separation of bound ligand from free ligand, filter binding, and the associated wash time, requiring at least 5 to 10 min of processing time. DRaCALA directly assays the binding without prior processing or the subsequent wash steps. As a result, DRaCALA can be completed within 5 to 30 s, depending on the volume of the sample spotted. Because all binding interactions have off-rates, the speed of the assay is critical to capture accurate data. Other techniques for determining biochemical interaction are also available, such as ITC or surface plasmon resonance; however, these techniques require dedicated specialized instrumentation and individual processing of samples, resulting in longer assay time and lower throughput. An important feature of DRaCALA is that it will make biochemical approaches accessible to molecular and cellular biologists interested in precise and simple measurements of interactions between protein-ligand pairs of interest. The ability to determine koff , in addition to Kd, allows calculation of the on-rate. Differences in the koff can be useful in understanding biological processes, because interactions with similar affinities can result in distinct biological outputs.
The principle of DRaCALA should be universally applicable to any system in which the ligand can be mobilized by capillary action in conjunction with a solid support capable of sequestering the macromolecule. Thus, the choice of the support, the solvent composition of the mobile phase, and the specific properties of the ligand can be altered to enhance the effectiveness of DRaCALA for various protein-ligand systems. In this paper, we have described the application of DRaCALA only for radiolabeled ligands; however, the principle of the assay should also apply to fluorescently labeled ligands. The solid support could also be changed to sequester other macromolecules based on their unique chemical properties. For example, nucleic acids can be retained by DEAE-cellulose (12). The solvent composition could also be changed to alter the chemical properties of the mobile phase. Detergents, salts, and other agents may be added to alter the relative behavior of the protein and ligand on the solid support.
Studies of numerous systems involving low-molecular-weight biological ligands and receptor macromolecules can benefit from DRaCALA, including nucleotide derivatives, amino acid derivatives, metal ions, sugars, and other small signaling molecules. The function of biological ligands can be specific to subset of organisms that produce or use these molecules. To identify biological samples that may be enriched in a specific ligand-binding protein, a similar approach to the screen for cdiGMP-binding proteins (Fig. 5C) can be taken to identify model organisms for study of the ligand of interest. Alternatively, expression of ligand-binding proteins may be regulated. DRaCALA provides a method for rapidly screening a single organism grown in different conditions, as in phenotypic arrays, and a test for ligand-binding activity.
The systematic identification of protein receptors for each of these small molecular ligands will allow for a comprehensive understanding of the biological effect of these signaling molecules, and DRaCALA offers a high-throughput platform that should greatly facilitate this process. Furthermore, the ability of DRaCALA to detect ligand binding in whole-cell extracts should allow for systematic screening of whole-genome ORF libraries (ORFeomes) for proteins that bind various small molecules. Although the binding proteins have been overexpressed, the key parameter for detection is that the cellular concentration of expressed protein is greater than the Kd for the ligand. For proteins with low affinities, these interactions will be difficult to detect in whole cells similar to purified systems. DRaCALA is scalable, and it has been performed using both a standard single-channel pipette and an eight-channel multichannel pipette with equal precision and accuracy. Thus, DRaCALA could be easily adapted for high-throughput applications by using a 96-well pin tool in combination with standard robotics. The volume required for the DRaCALA assay can be further reduced to allow for screening using the 384-well format. A current limitation of the whole cell-based DRaCALA system is the requirement to identify a cell that has low background expression of proteins that bind the ligand of interest. This problem can be solved using protein array technology (16–18, 33, 34). However, high-volume parallel protein purification requires specialized equipment, and it is difficult to assess the stability and functionality of all proteins on the chip. Nonetheless, DRaCALA and protein arrays can be viewed as complementary technologies. This is analogous to the identification of protein-protein interactions using the genetic/molecular biology approach of the yeast two-hybrid assay (35), which is complementary to the biochemical approach of mass spectrometric identification of coprecipitated proteins. An important advantage of DRaCALA is that insoluble proteins appear to have a similar behavior as soluble protein in whole-cell lysates, thus avoiding purification problems associated with insoluble proteins.
Development of DRaCALA as a high-throughput assay for the detection of protein-ligand interactions will be useful for identifying new targets for pharmaceutical intervention. To this end, DRaCALA might be used initially as a screening tool to identify new interaction pairs and then in a second round of DRaCALA to identify inhibitors that prevent the interaction. Labeling of the identified specific inhibitor would then allow for rapid sequential screening for even more potent molecules that can displace the original inhibitor. Our results show that DRaCALA can be developed as a platform to enable critical advances in metabolite interactomics and therapeutic intervention.
Materials and Methods
Protein Purification.
The arabinose inducible CRP expression plasmid (pBAD-CRP) was a generous gift from Sankar Adhya (National Institutes of Health, Bethesda, MD), and purified NtrB was a generous gift from Richard Stewart (University of Maryland, College Park, MD). Additional information is provided in SI Materials and Methods.
DRaCALA.
Protein or whole-cell lysates in 1× cdiGMP binding buffer (20 μL) were mixed with 4 nM radiolabeled nucleotide and allowed to incubate for 10 min at room temperature. Radiolabeled nucleotide was competed away by cold nucleotides in concentrations and for times indicated. Purified proteins were tested in technical replicates. Whole-cell lysates in Fig. 4 and Fig. S7 were tested in biological triplicates. Whole-cell lysates in Fig. 5 were tested in technical replicates. These mixtures were pipetted (2.5–5 μL) onto dry untreated nitrocellulose (GE Healthcare) in triplicate and allowed to dry completely before quantification. An FLA7100 Fujifilm Life Science PhosphorImager was used to detect luminescence following a 5-min exposure of blotted nitrocellulose to phosphorimager film. Data were quantified using Fujifilm Multi Gauge software v3.0.
Whole-Cell Lysate Preparation.
BL21(DE3) cells expressing pVL847 (MBP), pVL882 (MBP-Alg44), or Alg44 point mutations were grown in LB at 30 °C and induced for overexpression with 100 μM isopropyl-β-d-thiogalactopyranoside. All Pseudomonas strains from Fig. 5A and Table S7 were grown for 16 h in LB broth at 37 °C with 200 rpm shaking. Growth conditions and sample preparation of all samples in Fig. 5B and Table S8 are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank R. Stewart for providing purified NtrB and S. Adhya for providing pBAD-CRP. We thank Drs. R. Stewart, D. Mosser, D. Stein, K. McIver, K. Ramamurthi, A. Records, and J. Kahn for useful discussions. We thank L. Cathcart, L. Zimmerman, L. Hause, R. Suresh, J. German-Shipley, Dr. J. Culver, Dr. V. Briken, Dr. K. McIver, and Dr. D. Stein for various strains and organism samples. V.T.L. was funded by a University of Maryland Seed Grant and National Institutes of Health Grant AI096083.
Footnotes
Conflict of interest statement: A provisional patent has been filed by the University of Maryland.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018949108/-/DCSupplemental.
References
- 1.Lewin B. Cells. Sudbury, MA: Jones and Bartlett Publishers; 2007. p. xix. [Google Scholar]
- 2.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 3.Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 4.Jacobson KA, Boeynaems JM. P2Y nucleotide receptors: Promise of therapeutic applications. Drug Discov Today. 2010;15:570–578. doi: 10.1016/j.drudis.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feist AM, Herrgård MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol. 2009;7:129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kafsack BF, Llinás M. Eating at the table of another: Metabolomics of host-parasite interactions. Cell Host Microbe. 2010;7:90–99. doi: 10.1016/j.chom.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horai H, et al. MassBank: A public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 8.Saito K, Matsuda F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol. 2010;61:463–489. doi: 10.1146/annurev.arplant.043008.092035. [DOI] [PubMed] [Google Scholar]
- 9.Connors KA. Binding Constants: The Measurement of Molecular Complex Stability. New York: Wiley; 1987. p. xiv. [Google Scholar]
- 10.Nirenberg M, Leder P. RNA codewords and protein synthesis. The effect of trinucleotides upon the binding of sRNA to ribosomes. Science. 1964;145:1399–1407. doi: 10.1126/science.145.3639.1399. [DOI] [PubMed] [Google Scholar]
- 11.Northup JK, et al. Purification of the regulatory component of adenylate cyclase. Proc Natl Acad Sci USA. 1980;77:6516–6520. doi: 10.1073/pnas.77.11.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: Application to protein-nucleic acid interactions. Proc Natl Acad Sci USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sturtevant JM. Some applications of calorimetry in biochemistry and biology. Annu Rev Biophys Bioeng. 1974;3:35–51. doi: 10.1146/annurev.bb.03.060174.000343. [DOI] [PubMed] [Google Scholar]
- 14.Alfthan K. Surface plasmon resonance biosensors as a tool in antibody engineering. Biosens Bioelectron. 1998;13:653–663. doi: 10.1016/s0956-5663(98)00020-7. [DOI] [PubMed] [Google Scholar]
- 15.Vuignier K, Schappler J, Veuthey JL, Carrupt PA, Martel S. Drug-protein binding: A critical review of analytical tools. Anal Bioanal Chem. 2010;398:53–66. doi: 10.1007/s00216-010-3737-1. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Snyder M. Protein chip technology. Curr Opin Chem Biol. 2003;7:55–63. doi: 10.1016/s1367-5931(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 18.Zhu H, et al. Analysis of yeast protein kinases using protein chips. Nat Genet. 2000;26:283–289. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]
- 19.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 20.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 21.Emmer M, deCrombrugghe B, Pastan I, Perlman R. Cyclic AMP receptor protein of E. coli: Its role in the synthesis of inducible enzymes. Proc Natl Acad Sci USA. 1970;66:480–487. doi: 10.1073/pnas.66.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zubay G, Schwartz D, Beckwith J. Mechanism of activation of catabolite-sensitive genes: A positive control system. Proc Natl Acad Sci USA. 1970;66:104–110. doi: 10.1073/pnas.66.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ninfa AJ, Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amiot N, Heintz K, Giese B. New approach for the synthesis of cdiGMP and its analogues. Synthesis. 2006;24:4230–4236. [Google Scholar]
- 25.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boehm A, et al. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 27.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen W, Kuolee R, Yan H. The potential of 3′,5′-cyclic diguanylic acid (c-di-GMP) as an effective vaccine adjuvant. Vaccine. 2010;28:3080–3085. doi: 10.1016/j.vaccine.2010.02.081. [DOI] [PubMed] [Google Scholar]
- 29.Kulasekara HD, et al. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- 30.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee VT, et al. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: Bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knickerbocker T, Chen JR, Thadhani R, MacBeath G. An integrated approach to prognosis using protein microarrays and nonparametric methods. Mol Syst Biol. 2007;3:123. doi: 10.1038/msb4100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacBeath G. Protein microarrays and proteomics. Nat Genet. 2002;32(Suppl):526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 35.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.