Abstract
The mammalian target of rapamycin (mTOR) is a central regulator of cell growth and proliferation in response to growth factor and nutrient signaling. Consequently, this kinase is implicated in metabolic diseases including cancer and diabetes, so there is great interest in understanding the complete spectrum of mTOR-regulated networks. mTOR exists in two functionally distinct complexes, mTORC1 and mTORC2, and whereas the natural product rapamycin inhibits only a subset of mTORC1 functions, recently developed ATP-competitive mTOR inhibitors have revealed new roles for both complexes. A number of studies have highlighted mTORC1 as a regulator of lipid homeostasis. We show that the ATP-competitive inhibitor PP242, but not rapamycin, significantly down-regulates cholesterol biosynthesis genes in a 4E-BP1–dependent manner in NIH 3T3 cells, whereas S6 kinase 1 is the dominant regulator in hepatocellular carcinoma cells. To identify other rapamycin-resistant transcriptional outputs of mTOR, we compared the expression profiles of NIH 3T3 cells treated with rapamycin versus PP242. PP242 caused 1,666 genes to be differentially expressed whereas rapamycin affected only 88 genes. Our analysis provides a genomewide view of the transcriptional outputs of mTOR signaling that are insensitive to rapamycin.
Keywords: sterol regulatory element-binding protein-2, eIF4E-binding protein-1 phosphorylation, 3-hydroxy-3-methylglutaryl-CoA reductase, thioredoxin interacting protein, expression array
The mammalian target of rapamycin (mTOR) is a serine/threonine kinase that belongs to the family of PI3K-related kinases. A nodal point in cell signaling, mTOR integrates growth factor signaling with nutrient availability to directly regulate protein translation and affect cell growth and proliferation (1, 2). mTOR hyperactivation caused by mutation or dysregulation of mTOR upstream effectors is frequently observed in human cancers (3, 4). The natural product rapamycin was instrumental in the discovery of mTOR and has provided a pharmacological tool for studying mTOR function in settings in which genetic approaches have been unsuccessful, as mTOR loss of function mutations result in an embryonic lethal phenotype in mice (5). mTOR exists in two distinct complexes, mTOR complex 1 (mTORC1) and complex 2 (mTORC2), of which only the former is sensitive to rapamycin inhibition (6, 7). mTORC1 regulates protein synthesis by phosphorylating both p70 S6 kinase 1 (S6K1) and the eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) to control ribosomal protein translation and cap-dependent translation (8). mTORC2 has been less well characterized because of the lack of a specific inhibitor. The only known mTORC2 substrates are Akt and serum- and glucocorticoid-induced kinase (SGK), which it phosphorylates on their C-terminal hydrophobic motifs at Ser473 and Ser422 respectively, to contribute to full kinase activation (9, 10).
Recently developed mTOR inhibitors bind to the active site rather than the rapamycin binding site, thus allowing for pharmacological interrogation of both mTOR complexes. Importantly, active site mTOR inhibitors have also revealed a surprising distinction between the two known mTORC1 substrates, S6K1 and 4E-BP1. Although rapamycin is classified as an mTORC1 inhibitor, it potently inhibits only S6K1 and not 4E-BP1 phosphorylation and cap-dependent translation, whereas ATP-competitive inhibitors PP242, Torin1, Ku-0063794, and WYE-354 completely block both mTORC1 outputs (11–14). The ATP-site inhibitors also completely block cell growth and proliferation in cell lines in which rapamycin has only a partial effect. More recently, phosphoproteomic studies have compared the new generation of mTOR inhibitors with rapamycin (15, 16). In general, ATP-competitive mTOR inhibitors could elucidate new rapamycin-resistant outputs of mTOR.
mTOR signaling has recently been linked to lipid metabolism (17, 18). Fatty acids and cholesterol are not only integral membrane components, but cholesterol and its biosynthetic intermediates serve as precursors for the generation of steroid hormones and other signaling molecules. Moreover, dysregulation of lipid homeostasis is the fundamental basis of disorders such as obesity, diabetes, and cardiovascular disease. Thus, there is great interest in understanding the mechanisms which regulate lipid homeostasis.
Lipid metabolic gene expression is controlled by master transcriptional regulators SREBP-1 and SREBP-2 (sterol regulatory element-binding protein-1 and -2), which, respectively, regulate fatty acid and cholesterol biosynthesis. SREBP proteins are synthesized as precursors that reside in the endoplasmic reticulum membrane, and, when cellular sterol levels are low, SREBP is processed in the Golgi apparatus to release the N-terminal fragment that acts as a transcription factor to control its own expression and the expression of lipid biosynthesis and uptake genes (19, 20). Recent studies that used rapamycin have established a role for mTOR in regulating SREBP-1 processing and the expression of fatty acid biosynthetic genes (21–23). However, conflicting data have been obtained by groups attempting to use rapamycin to study cholesterol biosynthesis. Although activation of Akt, an upstream activator of mTOR, induced the expression of SREBP1 and its target genes, the effect on SREBP2 was less pronounced (24). Moreover, in different cell types, rapamycin has opposing effects on SREBP-2 processing and the expression of 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMGCR), the rate-limiting step in cholesterol biosynthesis (25–27). Collectively, these data have led to the notion that mTOR may regulate SREBP-1 and SREBP-2 differently and that some regulation may be cell type-specific.
In this study, we use a combination of pharmacological and genetic tools to dissect the molecular basis of mTORC1 regulation of cholesterol biosynthesis. We find that cholesterol biosynthesis is coordinately regulated by 4E-BP1 and S6K1 and the relative contribution of each regulator varies between cell types. Finally, to provide an unbiased assessment of the rapamycin-resistant outputs of mTOR, we carried out genomewide comparative transcriptional profiling of rapamycin and ATP-competitive mTOR inhibitor PP242. Our gene expression profiling revealed a 20-fold increase in the number of PP242-sensitive genes compared with rapamycin. These findings represent a critical step toward our understanding of the complete transcriptional programs regulated through mTOR, which are only mildly or not at all sensitive to rapamycin.
Results
Rapamycin-Resistant Role for mTORC1 in Regulating Cholesterol Biosynthetic Gene Expression.
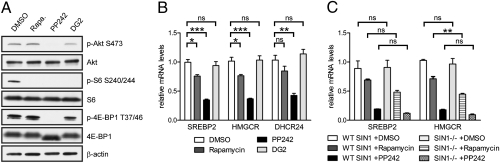
In murine NIH 3T3 cells, we confirmed by phosphospecific immunoblotting of known mTOR outputs that rapamycin and PP242 inhibit the phosphorylation of ribosomal protein S6 on Ser240/244, but PP242 additionally inhibits mTORC2 phosphorylation of Akt on Ser473 and mTORC1 phosphorylation of 4E-BP1 on Thr37/46 (Fig. 1A). The selected doses of rapamycin and PP242 represent the EC90 values for each drug as measured by antiproliferative activity (11).
Fig. 1.
mTORC1 regulates cholesterol biosynthetic gene expression. (A) Immunoblotting from NIH 3T3 cell lysates after 18 h treatment with 50 nM rapamycin (Rapa.), 2 μM PP242, or 20 μM DG2. (B) NIH 3T3 cells were treated as in A, and relative mRNA levels were analyzed by qRT-PCR. (C) Relative mRNA levels in WT and SIN1−/− MEFs after 18 h treatment with DMSO, 50 nM rapamycin, or 2 μM PP242 (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant).
We focused on regulation of cholesterol biosynthesis by using PP242 in hopes of identifying a rapamycin-resistant role for mTOR. Quantitative RT-PCR (qRT-PCR) analysis indicated that, in murine NIH 3T3 cells, PP242 was significantly more effective than rapamycin at inhibiting the expression of the transcriptional regulator SREBP2 and its target genes, including HMGCR, the rate-limiting enzyme, and 24-dehydrocholesterol reductase (DHCR24), one of the terminal enzymes in cholesterol biosynthesis (Fig. 1B). Culture in lipoprotein deficient serum (LPDS) did not significantly increase the expression of cholesterol biosynthesis genes in NIH 3T3 cells, so we turned to an LPDS-stimulatable HeLa cell system (28). We first validated that, in HeLa cells, culture in LPDS instead of FBS shows significant up-regulation of HMGCR and squalene epoxidase (SQLE), another gene in the cholesterol biosynthetic pathway (Fig. S1). In this LPDS-stimulated cell system, PP242, but not rapamycin, significantly blocks expression of cholesterol biosynthesis genes. We therefore demonstrate that the transcriptional effects of both classes of mTOR inhibitors are consistent between unstimulated and LPDS-stimulated cell systems.
To confirm that the observed transcriptional effects of PP242 were caused by mTOR inhibition and not off-target effects, we also showed that Ku-0063794, a structurally distinct mTOR inhibitor, similarly decreased cholesterol gene expression (Fig. S2). To confirm that PP242 was not a general inhibitor of transcription, we showed that PP242 did not significantly affect ribosomal protein gene RPS16 expression (Fig. S3). We also asked if PP242 regulated the proteolytic processing of SREBP-2; however, SREBP-2 was undetectable by immunoblotting in NIH 3T3 cells.
Because rapamycin is able to effectively block S6K1 activity but not cholesterol gene expression, we hypothesized that these transcriptional outputs were not regulated through S6K1. We took a pharmacological approach to examine the effect of the selective S6K1 inhibitor DG2 on the expression of cholesterol biosynthesis genes. In NIH 3T3 cells, DG2 effectively inhibited ribosomal protein S6 phosphorylation (Fig. 1A) but did not inhibit the expression of SREBP2 and its target genes (Fig. 1B). These results confirmed that cholesterol biosynthetic gene expression was not regulated through S6K1 in NIH 3T3 cells.
To determine which mTOR complex was responsible for the differential gene expression induced by PP242, we used SIN1-knockout (SIN1−/−) primary mouse embryonic fibroblasts (MEFs), which lack a unique and integral component of mTORC2 (29). PP242 down-regulated SREBP2 and HMGCR similarly in the WT and SIN1−/− MEFs, demonstrating that regulation of the cholesterol pathway did not require intact mTORC2 and was therefore controlled by mTORC1 (Fig. 1C).
mTORC1 Effector 4E-BP1 Regulates Expression of SREBP2 and Its Target Genes.
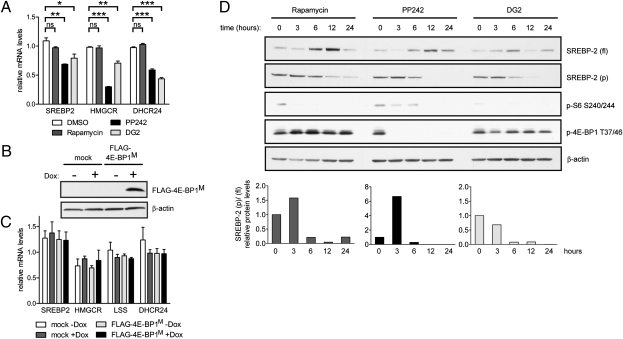
A previous study in breast cancer cell lines showed that siRNA knockdown of eukaryotic initiation factor 4E inhibits SREBP-1 processing and that expression of a nonphosphorylatable 4E-BP1 mutant inhibits transcriptional activity of a fatty acid biosynthesis reporter gene (23). We asked if 4E-BP1 regulated the expression of SREBP2 and its target genes. We first exploited a cell line that we discovered to be selectively resistant to PP242-mediated inhibition of 4E-BP1 phosphorylation. In SW620 colorectal carcinoma cells, PP242 prevents S6K1 phosphorylation of ribosomal protein S6 at Ser240/244 and mTORC2 phosphorylation of Akt at Ser473, but does not effectively inhibit 4E-BP1 phosphorylation at Thr37/46, whereas in HCT15 colorectal carcinoma cells, PP242 effectively blocks all three mTOR outputs (Fig. 2A). In HCT15 cells PP242 significantly decreases the expression of SREBP2 and target genes HMGCR and DHCR24. Compared with HCT15 cells, SW620 cells express lower basal levels of SREBP2 and DHCR24; nevertheless, HCT15 cells are sensitive to PP242-mediated down-regulation of cholesterol biosynthetic genes whereas SW620 cells are resistant (Fig. 2B). Incomplete resistance to PP242 inhibition of HMGCR and DHCR24 may be the result of a small fraction of unphosphorylated 4E-BP1 in PP242-treated SW620 cells (Fig. 2A).
Fig. 2.
mTORC1 effector 4E-BP1 regulates the expression of SREBP2 and its target genes. (A) Western blot of lysates from HCT15 and SW620 colorectal carcinoma cells treated with varying doses of PP242 for 4 h. (B) Relative mRNA levels in HCT15 and SW620 cells after 18 h treatment with DMSO or 2 μM PP242. (C) NIH 3T3 cells were cotransfected with FLAG-4E-BP1M and Tet activator expression constructs and induced with 1 μg/mL doxycycline, and cell lysates were prepared for immunoblotting. (D) NIH 3T3 cells were transfected as in C and mRNA levels were analyzed by qRT-PCR. (E) 4E-BP1 immunoblot analysis from four different cell lines. (Right) Protein levels are quantified and expressed as normalized ratios (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant).
We also tested if overexpression of an unphosphorylatable dominant-negative 4E-BP1 mutant in which all five mTORC1-regulated phosphorylation sites have been mutated to alanine (FLAG-4E-BP1M) could replicate the effect of PP242 on cholesterol gene transcription. NIH 3T3 cells were cotransfected with a tetracycline-inducible FLAG-4E-BP1M expression construct and Tet activator expression construct. Expression of FLAG-4E-BP1M was confirmed by immunoblot analysis, and although expression was induced several fold by doxycycline, low expression levels were detected even in the absence of doxycycline (Fig. 2C). FLAG-4E-BP1M expression caused a decrease in SREBP2, DHCR24, and lanosterol synthase (LSS) gene expression (Fig. 2D). Interestingly, even low levels of FLAG-4E-BP1M expressed in the absence of doxycycline are sufficient to decrease transcript levels of some but not all SREBP2 target genes. As a negative control, FLAG-4E-BP1M overexpression had no significant effect on ACTIN mRNA levels (Fig. S4). In contrast to our findings in NIH 3T3 cells, FLAG-4E-BP1M expression had no significant effect on cholesterol biosynthetic gene expression in HCT15 and SW620 cells (Fig. S5). We asked if these cell types differed in their endogenous levels of 4E-BP1. We found that NIH 3T3 cells express nearly twice the level of phospho-4E-BP1 as HCT15 and SW620 cells, potentially confounding the effects of the dominant-negative mutant in the latter cell types (Fig. 2E).
mTORC1 Effector S6K1 Controls SREBP-2 Processing and Cholesterol Biosynthetic Gene Expression in Hepatocellular Carcinoma Cells.
A recent study in MEFs expressing constitutively active mTOR showed that siRNA knockdown of S6K1 causes a decrease in SREBP-1 processing and the expression of fatty acid biosynthesis genes, suggesting that fatty acid biosynthesis is controlled through S6K1 (22). In contrast, studies in retinal pigment epithelial cells and MEFs showed that siRNA knockdown and genetic deletion of S6K1 had no effect on SREBP-1 processing (30). We asked if cell type dictated whether lipid metabolism was regulated mostly through S6K1 or 4E-BP1 and chose to examine hepatocellular carcinoma (HCC) cells because they are derived from the liver and therefore specialized in lipid metabolism.
In HCC cells, PP242 and DG2, but not rapamycin, significantly reduced cholesterol biosynthetic gene expression (Fig. 3A). Although expression of FLAG-4E-BP1M did not affect SREBP2 and its target gene transcript levels (Fig. 3 B and C), HCC cells do not contain high levels of phospho-4E-BP1 and thus may not be sensitive to FLAG-4E-BP1M overexpression (Fig. 2E). We next examined the relative amounts of full-length SREBP-2 [SREBP-2 (fl)] and processed SREBP-2 [SREBP-2 (p)] upon inhibitor treatment over a 24-h time course. Phosphospecific immunoblotting confirmed that all three inhibitors effectively block ribosomal protein S6 phosphorylation whereas only PP242 blocks 4E-BP1 phosphorylation. Whereas PP242 and DG2 fully block SREBP-2 (p) formation within 12 h, rapamycin is an incomplete inhibitor (Fig. 3D). The time-dependent disappearance of SREBP-2 (p) correlates with an accumulation of full-length protein. Consistent with previous work that used siRNA knockdown of S6K1 in TSC2-null cells, we suggest that, in HCC cells, S6K1, but not 4E-BP1, is a key regulator of cholesterol biosynthesis (22).
Fig. 3.
mTORC1 effector S6K1 controls SREBP-2 processing and cholesterol biosynthetic gene expression in HCC cells. (A) HCC cells were treated as indicated in Fig. 1B, and relative mRNA levels were assessed by qRT-PCR. (B) HCC cells were transfected as described in Fig. 2C, followed by immunoblot analysis. (C) HCC cells were transfected as in B, and mRNA levels were analyzed by qRT-PCR. No differences were statistically significant. (D) HCC cells were treated with the inhibitors described in A over a 24-h time course. One hour before lysis, cells were treated with 25 μg/mL ALLN and cell lysates were prepared for Western blotting. (Bottom) Protein levels were quantified, and the ratio of processed to full-length SREBP-2 is shown throughout the time course (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant).
PP242 but Not Rapamycin Causes Global Transcriptional Rewiring.
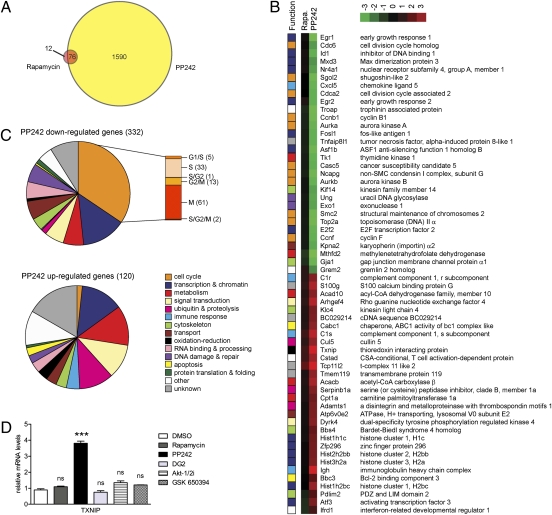
Given the differential abilities of rapamycin and PP242 to regulate the cholesterol biosynthetic gene program, we wanted to compare the transcriptional outputs of rapamycin and PP242 in an unbiased manner. Microarray analysis in NIH 3T3 cells revealed that, at a B statistic (log-odds) cutoff of B > 0, PP242 caused 1,666 genes to be differentially expressed whereas rapamycin affected a mere 88 genes (Fig. 4A). Moreover, only 12 of these genes were uniquely rapamycin-sensitive, and for most of the 76 shared targets, PP242 caused a greater magnitude of change in gene expression (Fig. S6). The shared targets included SREBP2 (Srebf2), which was down-regulated 1.4-fold by rapamycin and 1.7-fold by PP242. A previous study had identified more than 500 rapamycin-sensitive genes in human lymphoma cells (31). To address this discrepancy, we filtered our data according to the previously published statistical parameters and observed 236 and 3,892 genes differentially regulated by rapamycin and PP242, respectively, thus bringing our rapamycin analyses in line with previous transcriptional analyses of this drug. Both studies show similar functional classes of genes being targeted by rapamycin inhibition of mTOR. Regardless of the statistical methods used, we see an approximately 20-fold greater number of genes affected by ATP-site inhibition compared with rapamycin inhibition of mTOR.
Fig. 4.
PP242 causes greater changes in gene expression than rapamycin. (A) NIH 3T3 cells were treated with DMSO, 50 nM rapamycin, or 2 μM PP242 for 18 h, RNA was harvested, and cDNA was prepared for transcriptional profiling. Comparison of genes differentially expressed after rapamycin and PP242 treatment (B > 0). (B) List of the 30 most up-regulated and 30 most down-regulated genes that are uniquely PP242-sensitive but rapamycin-insensitive (FDR < 0.005). The differential gene expression shown represents the mean log2-based fold change from 3 replicate samples, and each gene is functionally annotated in the leftmost column according to the legend in Fig. 4C. (C) Functional annotation of all 452 PP242-sensitive genes (FDR < 0.005). Cell cycle genes are subcategorized by cell cycle phase; the number of genes in each subcategory is shown in parentheses. (D) qRT-PCR analysis of TXNIP mRNA levels in NIH 3T3 cells after 18 h treatment with DMSO, 50 nM rapamycin, 2 μM PP242, 20 μM DG2, 10 μM Akt-1/2i, or 10 μM GSK 650394.
We next used a more stringent false discovery rate (FDR) cutoff of less than 0.005 to identify a subset of 452 genes that displayed a statistically significant difference in gene expression between PP242 and rapamycin treatment (Table S1). These uniquely PP242-sensitive genes ranged from 6.5-fold down-regulated to 12-fold up-regulated, and we functionally annotated the most strongly up- and down-regulated genes according to the Gene Ontology database (Fig. 4B). The 30 most down-regulated genes were particularly enriched in transcriptional regulation (e.g., E2F transcription factor 2 and early growth response 1) and cell cycle control (e.g., cyclin F and aurora kinase A), which is consistent with the ability of PP242, but not rapamycin, to fully inhibit cell growth and proliferation. Genes involved in biosynthetic processes (e.g., thymidine kinase 1) and molecular transport (e.g., importin α2) were also strongly down-regulated. The 30 most up-regulated genes spanned a broad range of functional categories, including catabolic enzymes (e.g., acetyl CoA carboxylase β) and proteins involved in ubiquitin-mediated proteolysis (e.g., cullin 5).
Functional annotation of all 452 PP242-sensitive genes indicated that genes involved in transcription, metabolism, and signal transduction were widely represented in both datasets (Fig. 4C). An overwhelming 35% of down-regulated genes were involved in the cell cycle, which is consistent with the cytostatic effect of PP242. Although genes from all phases of the cell cycle were affected, more than half were involved in mitosis. Transport and DNA damage and repair genes were mostly down-regulated whereas proteolytic, apoptotic, redox, and immune response genes were more prominently up-regulated. Overall, PP242 induces a significantly broader transcriptional response compared with rapamycin, thus providing a rich source of mTOR-regulated pathways previously obscured by the limited effects of rapamycin.
Because PP242 inhibits both mTOR complexes, we expected genes identified in an unbiased screen to be uniquely sensitive to PP242 and not rapamycin to include genes downstream of mTORC2. Thioredoxin interacting protein (TXNIP) was one of the most strongly up-regulated genes, so we assessed its dependence on mTORC2. Txnip is a negative regulator of thioredoxin, and thioredoxin not only maintains cellular redox homeostasis, it also interacts with several transcription factors and signaling molecules including phosphatase and tensin homologue, which signals upstream of Akt and mTOR (32, 33). As a result of the broad cellular roles of thioredoxin, Txnip is implicated in diverse cellular processes including proliferation, apoptosis, and differentiation (34). A recent study identified TXNIP as an upstream regulator of mTOR activity (35), but here we report mTOR as a regulator of TXNIP. TXNIP was significantly up-regulated by PP242 but not by rapamycin or DG2 (Fig. 4D). In contrast to our results in NIH 3T3 cells, TXNIP was unresponsive to PP242 in WT and SIN1−/− MEFs, so we turned to pharmacological inhibition of mTORC2 substrates as an alternative means to distinguish between mTORC1 and mTORC2 dependence. Inhibition of mTORC2 effectors Akt and SGK by Akt-1/2i and GSK 650394, respectively, did not affect TXNIP expression, suggesting that TXNIP expression is not regulated through mTORC2.
Discussion
The emerging class of ATP-competitive mTOR inhibitors has revealed rapamycin-resistant outputs of mTOR and made pharmacological interrogation of the complete spectrum of mTOR functions possible. Meanwhile, mTORC1 has emerged as a regulator of lipid biosynthesis, although its role in cholesterol metabolism has been less well defined than for fatty acid metabolism as a result of the limited effects of rapamycin. By using ATP-site mTOR inhibitor PP242, we confirmed a role for mTORC1 in cholesterol biosynthesis.
In NIH 3T3 cells, PP242 is much more effective than rapamycin at inhibiting the expression of cholesterol biosynthesis genes, which we attribute to a unique regulatory role of mTORC1 effector 4E-BP1. 4E-BP1 may regulate SREBP-2 activation by controlling the cap-dependent translation of one or more proteins involved in SREBP-2 processing. Alternatively, cholesterol biosynthetic gene transcription is coordinately regulated by SREBP-2 and other transcriptional cofactors, and these auxiliary cofactors may undergo highly cap-dependent translation. Another possibility is that 4E-BP1 regulates the degradation of SREBP2 transcript. It will be interesting to elucidate the exact mechanism by which 4E-BP1 controls transcript levels of SREBP2 and its target genes to control cholesterol metabolism.
mTORC1 regulates not only SREBP2 transcription but also its posttranslational processing. Whereas 4E-BP1 is a primary regulator of cholesterol biosynthetic gene expression in NIH 3T3, HCT15, and SW620 cells, S6K1 regulated SREBP-2 processing and the expression of its target genes in HCC cells. Interestingly, rapamycin fully inhibited ribosomal protein S6 phosphorylation but was an incomplete inhibitor of SREBP-2 processing, suggesting that SREBP-2 processing is independent of S6 phosphorylation and may be mediated through another S6K1 substrate. Collectively, our data suggest that cholesterol biosynthesis is a complex process that is controlled by multiple mTOR outputs. We propose that S6K1 and 4E-BP1 coordinately regulate cholesterol metabolism and that the relative contribution of each mTORC1 effector varies between cell types.
We hypothesized that there were additional transcriptional consequences of ATP-competitive mTOR inhibition and evaluated this by gene expression profiling in NIH 3T3 cells. Consistent with previous studies showing active site mTOR inhibition to be more effective than rapamycin inhibition of cell proliferation and cap-dependent protein synthesis (11–14), we found that PP242 differentially regulated approximately 20 times as many genes as rapamycin. Follow-up analysis of one of the most differentially expressed genes, TXNIP, confirmed that this rapamycin-resistant expression profile does not represent the gene targets of mTORC1 effector S6K1 and further suggests that the expression profile does not represent the transcriptional outputs of mTORC2 through Akt or SGK, two known kinases activated by mTORC2. It will be interesting to use the genetic and pharmacological tools outlined in the present study to determine which mTOR complex and effector controls the expression of each of the PP242-sensitive genes.
mTOR inhibition caused global changes in the expression of genes involved in the cell cycle, metabolism, transcription, signal transduction, and many other cellular processes. Several of the genes most differentially expressed by PP242 have previously reported roles in tumor biology and metabolic dysfunction. In NIH 3T3 cells, Txnip, an oxidoreductase that maintains cellular redox state, was up-regulated almost fourfold more by PP242 than rapamycin (Table S1), and this gene not only has tumor suppressor activity, a naturally occurring nonsense mutation in the TXNIP gene is associated with hyperlipidemia (36, 37). RRAS was up-regulated 1.8-fold by PP242, and activated forms of R-Ras have been shown to inhibit cell cycle progression (38). SUV39h2, which encodes the H3-K9 histone methyltransferase, was down-regulated 2.5-fold by PP242. A SNP in the SUV39h2 gene has been associated with an increased risk for lung cancer (39). The broad transcriptional profile revealed by ATP-site mTOR inhibition offers unique insight into medically relevant pathways controlled through mTOR, and detailed analysis of this gene set may contribute toward delineating new regulatory networks downstream of mTOR.
Materials and Methods
Inhibitors and Plasmids.
Rapamycin, Akt-1/2i, and N-acetyl-leucyl-leucyl-norleucinal (ALLN) were purchased from Calbiochem. PP242 (40) and DG2 (41) were synthesized as described previously. Ku-0063794 was a gift from Y. Liu and P. Ren (Intellikine, La Jolla, CA) and GSK 650394 was provided by D. Pearce (University of California, San Francisco, San Francisco, CA). FLAG-4E-BP1M and rtTA Tet activator expression constructs were described previously (42).
Immunoblotting.
Anti-SREBP-2 (1C6) antibody was purchased from BD Biosciences and anti-FLAG (anti-FLAG M2) antibody was from Sigma. All other antibodies were from Cell Signaling Technology. Lysate preparation and immunoblotting are detailed in SI Materials and Methods.
qRT-PCR.
Cells were treated with DMSO or inhibitor for 18 h. RNA was harvested, reverse-transcribed, and analyzed by qRT-PCR as described in SI Materials and Methods. All qRT-PCR data were analyzed by using one-way ANOVA with multiple comparisons or Student t test when appropriate, and error bars in figures represent the SEM of three biological replicates. Primer sequences are listed in Table S2.
Transfection.
Cells were transfected using the Nucleofector transfection system (Amaxa) according to the manufacturer's protocol. Cells (1 × 106) were cotransfected with 5 μg of FLAG-4E-BP1M and 0.5 μg of Tet activator expression constructs. One to 2 d after transfection, cells were induced with 1 μg/mL doxycycline and harvested 24 h later.
Microarray Analysis.
NIH 3T3 cells were treated with DMSO, 50 nM rapamycin, or 2 μM PP242 for 18 h. Each treatment was done in triplicate. Sample preparation, labeling, array hybridizations, and statistical analyses are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank B. Su, R. S. Warren, A. Goga, Y. Liu, and P. Ren; and D. Pearce, and D. Ruggero for providing reagents. We thank D. Fiedler and D. C. Gray for synthesizing DG2 and F. R. Papa and K. R. Yamamoto for use of their thermocyclers. We thank M. E. Feldman for manuscript advice and A. G. Lerner and D. Han for experimental guidance. This work was supported by the University of California Cancer Research Coordinating Committee, Samuel Waxman Cancer Research Foundation, Accelerate Brain Cancer, Multiple Myeloma Translational Institute, Sandler Asthma Basic Research Center Functional Genomics Core Facility, and National Institutes of Health/National Center for Research Resources Grant UL1 RR024131 (University of California, San Francisco, Clinical and Translational Science Institute).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE27784).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103746108/-/DCSupplemental.
References
- 1.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 2.Chiang GG, Abraham RT. Targeting the mTOR signaling network in cancer. Trends Mol Med. 2007;13:433–442. doi: 10.1016/j.molmed.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Gustin JP, et al. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc Natl Acad Sci USA. 2009;106:2835–2840. doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu ZH, et al. Mammalian target of rapamycin activator RHEB is frequently overexpressed in human carcinomas and is critical and sufficient for skin epithelial carcinogenesis. Cancer Res. 2010;70:3287–3298. doi: 10.1158/0008-5472.CAN-09-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakami M, et al. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 7.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 8.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 9.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 10.García-Martínez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 11.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Martínez JM, et al. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421:29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu K, et al. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 2009;69:6232–6240. doi: 10.1158/0008-5472.CAN-09-0299. [DOI] [PubMed] [Google Scholar]
- 15.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porstmann T, Santos CR, Lewis C, Griffiths B, Schulze A. A new player in the orchestra of cell growth: SREBP activity is regulated by mTORC1 and contributes to the regulation of cell and organ size. Biochem Soc Trans. 2009;37:278–283. doi: 10.1042/BST0370278. [DOI] [PubMed] [Google Scholar]
- 18.Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;19:R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 20.Sato R, et al. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J Biol Chem. 1996;271:26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- 21.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luyimbazi D, et al. Rapamycin regulates stearoyl CoA desaturase 1 expression in breast cancer. Mol Cancer Ther. 2010;9:2770–2784. doi: 10.1158/1535-7163.MCT-09-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porstmann T, et al. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene. 2005;24:6465–6481. doi: 10.1038/sj.onc.1208802. [DOI] [PubMed] [Google Scholar]
- 25.Ma KL, et al. Sirolimus inhibits endogenous cholesterol synthesis induced by inflammatory stress in human vascular smooth muscle cells. Am J Physiol Heart Circ Physiol. 2010;298:H1646–H1651. doi: 10.1152/ajpheart.00492.2009. [DOI] [PubMed] [Google Scholar]
- 26.Gueguen Y, et al. Compared effect of immunosuppressive drugs cyclosporine A and rapamycin on cholesterol homeostasis key enzymes CYP27A1 and HMG-CoA reductase. Basic Clin Pharmacol Toxicol. 2007;100:392–397. doi: 10.1111/j.1742-7843.2007.00066.x. [DOI] [PubMed] [Google Scholar]
- 27.Sharpe LJ, Brown AJ. Rapamycin down-regulates LDL-receptor expression independently of SREBP-2. Biochem Biophys Res Commun. 2008;373:670–674. doi: 10.1016/j.bbrc.2008.06.108. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura Y, Sakakibara J, Izumi T, Shibata A, Ono T. Transcriptional regulation of squalene epoxidase by sterols and inhibitors in HeLa cells. J Biol Chem. 1996;271:8053–8056. doi: 10.1074/jbc.271.14.8053. [DOI] [PubMed] [Google Scholar]
- 29.Jacinto E, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 30.Lewis CA, Griffiths B, Santos CR, Pende M, Schulze A. Genetic ablation of S6-kinase does not prevent processing of SREBP1. Adv Enzyme Regul. 2010;51:280–290. doi: 10.1016/j.advenzreg.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Peng T, Golub TR, Sabatini DM. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol Cell Biol. 2002;22:5575–5584. doi: 10.1128/MCB.22.15.5575-5584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishiyama A, et al. Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem. 1999;274:21645–21650. doi: 10.1074/jbc.274.31.21645. [DOI] [PubMed] [Google Scholar]
- 33.Hui ST, et al. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc Natl Acad Sci USA. 2008;105:3921–3926. doi: 10.1073/pnas.0800293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SY, Suh HW, Chung JW, Yoon SR, Choi I. Diverse functions of VDUP1 in cell proliferation, differentiation, and diseases. Cell Mol Immunol. 2007;4:345–351. [PubMed] [Google Scholar]
- 35.Jin HO, et al. TXNIP potentiates Redd1-induced mTOR suppression through stabilization of Redd1. Oncogene. 2011 doi: 10.1038/onc.2011.102. 10.1038/onc.2011.102. [DOI] [PubMed] [Google Scholar]
- 36.Han SH, et al. VDUP1 upregulated by TGF-beta1 and 1,25-dihydorxyvitamin D3 inhibits tumor cell growth by blocking cell-cycle progression. Oncogene. 2003;22:4035–4046. doi: 10.1038/sj.onc.1206610. [DOI] [PubMed] [Google Scholar]
- 37.Bodnar JS, et al. Positional cloning of the combined hyperlipidemia gene Hyplip1. Nat Genet. 2002;30:110–116. doi: 10.1038/ng811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komatsu M, Ruoslahti E. R-Ras is a global regulator of vascular regeneration that suppresses intimal hyperplasia and tumor angiogenesis. Nat Med. 2005;11:1346–1350. doi: 10.1038/nm1324. [DOI] [PubMed] [Google Scholar]
- 39.Yoon KA, et al. Novel polymorphisms in the SUV39H2 histone methyltransferase and the risk of lung cancer. Carcinogenesis. 2006;27:2217–2222. doi: 10.1093/carcin/bgl084. [DOI] [PubMed] [Google Scholar]
- 40.Apsel B, et al. Targeted polypharmacology: Discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat Chem Biol. 2008;4:691–699. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anand NK, et al. Preparation and structure activity of pyrazolo-pyrimidine derivatives as antitumor agents and kinase modulators. 2005 WO2005117909. [Google Scholar]
- 42.Hsieh AC, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.