Abstract
The proopiomelanocortin gene (POMC) is expressed in a group of neurons present in the arcuate nucleus of the hypothalamus. Neuron-specific POMC expression in mammals is conveyed by two distal enhancers, named nPE1 and nPE2. Previous transgenic mouse studies showed that nPE1 and nPE2 independently drive reporter gene expression to POMC neurons. Here, we investigated the evolutionary mechanisms that shaped not one but two neuron-specific POMC enhancers and tested whether nPE1 and nPE2 drive identical or complementary spatiotemporal expression patterns. Sequence comparison among representative genomes of most vertebrate classes and mammalian orders showed that nPE1 is a placental novelty. Using in silico paleogenomics we found that nPE1 originated from the exaptation of a mammalian-apparent LTR retrotransposon sometime between the metatherian/eutherian split (147 Mya) and the placental mammal radiation (≈90 Mya). Thus, the evolutionary origin of nPE1 differs, in kind and time, from that previously demonstrated for nPE2, which was exapted from a CORE-short interspersed nucleotide element (SINE) retroposon before the origin of prototherians, 166 Mya. Transgenic mice expressing the fluorescent markers tomato and EGFP driven by nPE1 or nPE2, respectively, demonstrated coexpression of both reporter genes along the entire arcuate nucleus. The onset of reporter gene expression guided by nPE1 and nPE2 was also identical and coincidental with the onset of Pomc expression in the presumptive mouse diencephalon. Thus, the independent exaptation of two unrelated retroposons into functional analogs regulating neuronal POMC expression constitutes an authentic example of convergent molecular evolution of cell-specific enhancers.
Keywords: shadow enhancer, retroposition, retrotransposition, obesity, satiety
Transcriptional regulation in multicellular organisms is orchestrated by complex molecular mechanisms that were shaped throughout evolution to control multistage developmental programs and cell-specific gene expression. The current modular architecture view of gene expression regulation is based on the idea that genes have several transcriptional enhancers scattered along intergenic and/or intronic regions, each of them carrying a unique cis-transcriptional code to drive gene expression with distinct spatiotemporal resolution (for recent reviews see refs. 1–3). A unique level of complexity has been suggested recently, on the basis of the discovery that several developmental genes contain not just one but two enhancers capable of guiding similar spatiotemporal expression patterns (4–6). Examples include invertebrate genes, such as brinker (5), sog (5), snail (6), and shavenbaby (4) from Drosophila melanogaster and cog-1 from Caenorhabditis elegans (7), as well as the vertebrate developmental genes Shh (8) and Sox10 (9). The more distal enhancers of the aforementioned D. melanogaster genes reproduced the expression pattern driven by the proximal enhancers in reporter gene assays and were named, therefore, shadow enhancers (5).
It has been proposed that shadow enhancers could play important roles throughout development to ensure precise and reproducible patterns of gene expression during embryogenesis (5). In fact, two recent works performed in transgenic flies challenged the idea that shadow enhancers are just redundant sequences and suggested instead that they are critical elements that provide phenotypic robustness by buffering environmental perturbations that would otherwise imperil normal development (4, 6). These studies showed that, under optimal experimental conditions, the sole presence of either a primary or secondary enhancer is sufficient for normal gene expression and fly development but, in contrast, the simultaneous presence of both partners is required by the fly developmental program to overcome the challenges imposed by critical environmental constraints (4, 6). Although the adaptive advantage of having two functionally similar enhancers became apparent from these two studies, it still remains to be determined whether shadow enhancers originated from the duplication of an ancestral sequence or by independent and convergent evolutionary mechanisms.
The proopiomelanocortin gene (POMC) is expressed in a relatively small group of neurons that originate in the developing ventral hypothalamus of all jawed vertebrates (10, 11). Pomc-expressing neurons mature in the arcuate nucleus of the hypothalamus, integrate into neuronal circuits that become fully functional after weaning, and continue to actively express Pomc throughout the life span (12). POMC encodes a prohormone that is enzymatically processed to produce several peptides that play fundamental roles in the central control of food intake and energy balance (13). The physiological relevance of POMC-derived peptides in the brain can be readily appreciated in mice lacking central Pomc expression, which are hyperphagic and display early-onset severe obesity (14). Transcriptional regulation of Pomc in neurons is conveyed by two upstream distal enhancers, named nPE1 and nPE2, which are highly conserved in mammals (15). Expression studies in transgenic mice showed that nPE1, as well as nPE2, is sufficient to independently drive reporter gene expression to POMC neurons when placed upstream of a minimal heterologous promoter (15). Moreover, only the concurrent removal of nPE1 and nPE2 from transgenic constructs leads to the loss of reporter gene expression in POMC hypothalamic neurons (15). However, whether both enhancers drive distinctive expression patterns within the arcuate nucleus still remains unknown.
The aim of the present study was to further understand the evolutionary mechanisms that shaped the neuronal-specific control of Pomc expression in mammals using two different enhancers. To this end we investigated the evolutionary history of the neuronal Pomc enhancer nPE1 in relation to that of nPE2 and, in addition, explored whether these two enhancers control identical or complementary spatiotemporal expression patterns from early to adult stages of mouse brain development.
Results
Functional and Evolutionary Dissection of the POMC Neuronal Enhancer nPE1.
To investigate the evolutionary history of nPE1, we performed BLAST searches in all available vertebrate genomes deposited in the Ensembl database and the trace archives of the National Center for Biotechnology Information. Using human and mouse nPE1 sequences as queries, we identified nPE1 orthologs in the genomes of all available placental mammalian orders (Eutheria), except Xenarthra (armadillo and sloth), probably because of insufficient sequencing coverage (Fig. S1). We failed to identify nPE1 sequences from the marsupials (Metatheria) short-tailed opossum (Monodelphis domestica) and wallaby (Macropus eugenii), as well as from the monotreme (Prototheria) egg-laying platypus (Ornithorhynchus anatinus). In addition, we failed to identify nPE1 orthologs in all examined nonmammalian vertebrates, including the green lizard (Anolis carolinensis), the chicken (Gallus gallus), the frog Xenopus tropicalis, and several teleost fishes. On the basis of these findings, we conclude that nPE1 is a placental mammalian novelty that originated in an ancestor to extant placental mammals after their split from the lineage leading to marsupials, 147 Mya (16). Thus, nPE1 appeared as an upstream neuronal enhancer of Pomc at least 20 million years later than nPE2, which originated from the exaptation of a CORE-short interspersed nucleotide element (SINE) retroposon in the lineage leading to basal mammals (17), more than 166 Mya (16).
In previous work we defined nPE1 as a 630-bp human-mouse-dog conserved sequence able to direct reporter gene expression to hypothalamic POMC neurons of transgenic mice (15). Because the critical functional regions of cell-specific enhancers are usually under stronger selective constraint than neighboring nonfunctional regions, we performed an evolutionary divergence analysis to calculate the substitution rate along nPE1 sequences in every branch of a phylogenetic tree constructed from all available mammalian species (Fig. 1 A and B). A sliding-window plot based on the alignment and phylogenetic tree of 16 nPE1 sequences showed two highly conserved fragments in the central region with substitution rates lower than 4 nt per site (Fig. 1B). These two fragments are separated by a diversity peak generated primarily by a rodent-specific deletion (Fig. 1D). Altogether this area, that we have named nPE1core, constitutes 144 and 156 bp of mouse and human nPE1, respectively (Fig. 1C). The aligned sequences upstream and downstream of nPE1core are increasingly divergent, indicative of more relaxed evolutionary constraint (Fig. 1B). A ClustalW alignment of nPE1core sequences using 16 different species representing most mammalian orders revealed a remarkably high overall conservation, with the most divergent sequences being from the tenrec (Echinops telfairi) and the elephant (Loxodonta africana) (Fig. 1D).
Fig. 1.
nPE1 evolutionary divergence in placental mammals. (A) The tree represents phylogenetic relationships among the orthologous nPE1 enhancers of mammals. (B) Evolutionary divergence sliding-window plot of nPE1. The histogram shows the number of substitutions per site along each branch of the phylogeny displayed in A, estimated every 25 bases in 50-base intervals. Each layer of the plot represents a branch or node of the tree (numbers 1–13), and colors are maintained with respect to the tree. Substitutions were estimated from the best-fit maximum-likelihood model, which incorporates unequal equilibrium nucleotide frequencies and unequal rates of transitional and transversional substitutions (HKY85). The histograms are stacked so that the total height represents the density of substitutions over the entire phylogeny. (C) Schematic of nPE1 showing the most conserved region nPE1core in yellow. (D) Sequence alignment of nPE1core enhancer, among representative placental mammals. The shading of the alignment is based on the identity of residues and shows percentage of conservation within each column. One hundred percent identical aligned nucleotides are shaded in black. More than 80% conservation is depicted in dark gray. Columns with less than 80% and more than 60% conservation are shaded in light gray, whereas nucleotides with less than 60% are not shaded.
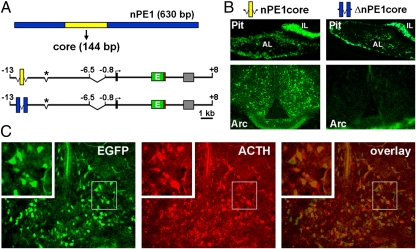
We next tested the hypothesis that nPE1core retained the critical functional sequences for neuronal enhancer activity of nPE1 by performing a transcriptional reporter expression analysis in transgenic mice. To this end, we constructed nPE1core-PomcEGFP, a transgene carrying nPE1core as the only source of nPE1 sequences cloned upstream of a mouse Pomc proximal promoter driving EGFP expression (Fig. 2A). In addition, we constructed nPE1Δcore-PomcEGFP, a transgene that carries a deletion version of mouse nPE1 in which the conserved 144 bp core of nPE1 was entirely removed. As an internal control for reporter expression, both transgenes included the mouse Pomc proximal promoter known to drive expression to POMC cells of the pituitary gland but unable to function independently in hypothalamic neurons (15, 17). The ability of these transgenes to drive EGFP expression to the brain and pituitary was determined in coronal sections taken from founder transgenic mice at postnatal day 1. We detected expression of EGFP in hypothalamic arcuate neurons in 9 of 11 independent nPE1core-PomcEGFP transgenic founder mice (Fig. 2B). Importantly, the only two nPE1core-PomcEGFP transgenic mice that did not show hypothalamic expression also failed to show expression in pituitary cells, probably as a consequence of transgene insertion into transcriptionally silent heterochromatin. In contrast, six independent transgenic founder mice carrying nPE1Δcore-PomcEGFP did not show a single EGFP-positive neuron in the arcuate nucleus, despite showing expression in the intermediate and anterior pituitary lobes (Fig. 2 B and C). The difference in arcuate expression between the two transgenes was significant (P < 0.005, two-tailed Fisher's exact test). Together, these results indicate that mouse nPE1core is both necessary and sufficient to drive reporter gene expression to POMC arcuate neurons, supporting the idea that these 144 bp contain the essential cis-acting code that determines neuronal-specific Pomc expression.
Fig. 2.
Functional dissection of the neuronal POMC enhancer nPE1 in transgenic mice. (A) Two structurally similar transgenes were constructed to express EGFP (green box) under the transcriptional control of the 144-bp core region of nPE1 (yellow box) or a deleted core version of nPE1 (blue boxes). The deletion of nPE2 is indicated by an asterisk, and the three mouse Pomc exons are boxed. (B) Expression analysis in the anterior (AL) and intermediate (IL) lobes of the pituitary (Pit) and the hypothalamic arcuate nucleus (Arc) in coronal sections of P1 founder transgenic mice carrying nPE1corePomc-EGFP (Left) or nPE1ΔcorePomc-EGFP (Right). (C) A coronal hypothalamic section of an nPE1corePomc-EGFP transgenic mouse shows native EGFP expression (Left) and immunolabeled ACTH neurons (red, Center). A superimposed image (Right) shows that the large majority of EGFP signal coexpresses within POMC neurons.
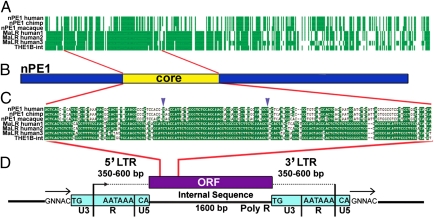
To trace the evolutionary origin of nPE1, we pursued an in silico paleogenomics strategy (17) based on BLAST searches for nPE1 paralogs in representative mammalian genomes. Using human nPE1core sequence as a query and our own modification of the BLAST software (SI Materials and Methods), we found in the human genome (Build 37 genome database reference GRCh37) 18 significant high identity hits in addition to the anticipated 100% hit at the POMC locus. Fifteen of these hits corresponded to repetitive sequences annotated by Repeat Masker in the human genome as derived from LTR retrotransposons of the mammalian-apparent LTR retrotransposons (MaLR) family (18), all located in noncoding regions. Two of the remaining three high-scoring hits are located in noncoding conserved regions (5′ upstream of PRK and SAMD12), and the third one within a conserved region of untranslated exon 2 of OLIG2. The location and conservation level of these three sequences suggest the possibility that they play regulatory functions. We obtained a reliable sequence alignment between the MaLR-derived hits and the human nPE1core sequence (Fig. 3A). The alignment further extended to the complete 630 nt of nPE1 is also shown (Fig. 3A). The highest identity score was found between nPE1core and the internal sequence of the MaLR THE1B (RepBase; www.girinst.org), one of the most abundant MaLRs in primate genomes (Fig. 3 A–C) (18, 19). Interestingly, the second-highest identity of nPE1 is MLT1, a member of the oldest group of MaLRs described in mammals to date (18). MaLRs are LTR-like retrotransposons that originated approximately 80–100 Mya, before the radiation of eutherians (18). This timing matches with our finding that nPE1 is a placental novelty. The identity between human nPE1core and a consensus THE1B-int sequence taken from RepBase is 62%, with 40 nt of the central region of nPE1core showing the highest identity (80%; Fig. 3C). This level of identity is similar to those found among orthologous nPE1 sequences from distantly related mammals, such as the elephant and the tenrec (Table S1), and to other recently reported cases of mobile elements exapted into functional enhancers (17, 20, 21).
Fig. 3.
nPE1 is an exapted retroposon of the MaLR family. (A) Sequence alignment of the human, chimpanzee, and macaque nPE1 enhancer sequences, three representative human instances of the LTR-like retrotransposon MaLR, and the THE1B internal consensus sequence from RepBase (THE1B-int). Green filled vertical bars and white spaces represent conserved and nonconserved residues, respectively, in each column of the alignment. (B) Diagram shows nPE1 and nPE1core. (C) nPE1core region of the alignment is shown in detail. (D) Diagram shows MaLR functional regions according to ref. 18.
Comparative Spatiotemporal Expression Profiles of nPE1 and nPE2.
To investigate the degree of overlap of enhancer function between nPE1 and nPE2, we performed a spatiotemporal transgenic expression study from early mouse hypothalamic development to the adult brain. We designed two structurally similar transgenes carrying either red (tomato) or green (EGFP) fluorescent protein coding sequences under the transcriptional control of nPE1 or nPE2, respectively (Fig. 4A). For each transgene we selected one pedigree displaying the highest level of eutopic reporter gene expression in POMC neurons for further detailed analysis. Immunolabeled POMC arcuate neurons coexpressed EGFP in coronal brain sections from nPE2Pomc-EGFP transgenic mice (Fig. 4B). In turn, the expression of tomato in POMC neurons was determined by crossing nPE1Pomc-tomato transgenic mice with Pomc-lacZ knockin mice (Fig. 4C), because of technical difficulties in detecting tomato in immunolabeled POMC neurons. To compare the expression domains driven by nPE1 and nPE2 throughout mouse brain development, we collected compound nPE1Pomc-tomato.nPE2Pomc-EGFP transgenic embryos at embryonic day (e) 9.5, e10.5, e12.5, and e18.5, because these ages encompass the earliest detection of Pomc expression at e10.5 in a small population of differentiating neurons in the prospective ventral hypothalamus (17), the birth of most additional POMC neurons between e10.5 and e12.5, and the subsequent migration and accumulation of Pomc-expressing neurons in the mantle zone of the arcuate nucleus. In e9.5 compound transgenic embryos, we did not detect any red or green fluorescent cells (Fig. S2A). In contrast, e10.5 embryos revealed the presence of a compact population of neurons at the base of the presumptive hypothalamus that coexpressed tomato and EGFP (Fig. 4D, Bottom, and Fig. S2B). Thus, the onset of nPE1 and nPE2 transgene expression coincided in time and space within the same neurons and matched the developmental onset of mouse Pomc expression (22). Analysis of sagittal sections of e12.5 compound transgenic mice showed coexpression of both reporter genes in all labeled cells (Fig. S3A). Analysis of e18.5 mouse embryos (1 d before birth) showed a larger number of fluorescent neurons along the ventromedial hypothalamus (Fig. S3B, Left). At this stage we found that the level of coexpression of both fluorescent markers was almost complete, with more than 97% of the neurons showing the presence of tomato and EGFP (Fig. S3B, Right). Similarly, expression analysis of adult compound transgenic mice revealed a remarkably high level of coexpression of both fluorescent proteins (>85%) along the entire anteroposterior and mediolateral axes of the arcuate nucleus (Fig. 4D, Top and Middle; further details in Fig. S4). Altogether, the level of coexpression of both reporters throughout development and in adult mice indicates that nPE1 and nPE2 drive gene expression to a nearly identical population of neurons in the ventromedial hypothalamus. The independent origin of these two neuronal POMC enhancers displaying similar function reveals a convergent evolutionary process that initiated in the lineage leading to monotremes, more than 166 Mya (16), and concluded after the marsupial/placental split dated approximately 147 Mya (16).
Fig. 4.
Spatiotemporal reporter gene expression driven by nPE1 and nPE2 in compound transgenic mice. (A) Two structurally similar transgenes were constructed to express either the red fluorescent protein tomato under the transcriptional control of nPE1 or EGFP by nPE2. (B) Immunofluorescence using an antibody against ACTH (red, Left) and EGFP natural fluorescence (green, Center) detected in coronal sections of nPE2Pomc-EGFP adult transgenic mice at the level of the medial basal hypothalamus at a lower (Upper) and higher (Lower) magnification. Right: Superimposed images demonstrate an almost complete penetrance of eutopic expression of the transgene in POMC (ACTH immunolabeled) neurons. White arrows denote rare ACTH immunolabeled neurons with no detectable EGFP expression. (C) Immunohistochemistry using an antibody against tomato in coronal brain slices of knockin mutant mice expressing lacZ from the Pomc locus reveals high penetrance of eutopic expression of nPE1Pomc-tomato (brown reaction product) in POMC neurons visualized by X-gal staining (blue). (D) Expression of the fluorescent proteins tomato (Left) or EGFP (Center) in coronal brain sections of adult (Top and Middle) and sagittal sections of e10.5 (Bottom) compound transgenic mice carrying nPE1Pomc-tomato and nPE2Pomc-EGFP. Right: Superimposed images demonstrate an almost complete level of cellular coexpression of both fluorescent reporter proteins. White arrows indicate rare neurons expressing tomato but not EGFP, whereas a gray arrow points to a neuron expressing EGFP but not tomato.
Rate of Molecular Evolution of nPE1 and nPE2.
The existence of two different neuronal Pomc enhancers with overlapping and apparently redundant function suggests that one of them could be relatively less constrained to evolve (5, 6). To test this hypothesis we compared the evolutionary rates of nPE1 and nPE2 in two different scenarios: a long-scale interspecies’ comparison of mammalian orthologs that allows calculating the sequence variation along the last 100 million years, and a short-scale intraspecies analysis based on a population genetics study in humans. First, we compared the nucleotide substitution rate using a relative ratio test that indicated that nPE1 is evolving 2.64 times faster than nPE2 (Table S2). Interestingly, the functionally active portion of nPE1, nPE1core, is also evolving faster than nPE2 [relative ratio (RR) = 1.73], although at a slower rate than the entire 630 bp of nPE1. nPE1 is also evolving faster than exon 3 sequences coding for the bioactive POMC peptides adrenocorticotropic hormone (ACTH), melanocyte stimulating hormones (MSH) α and β (α-MSH and β-MSH), and β-endorphin (RR = 1.70; Table S2), whereas nPE2 and the exon 3 coding region are evolving at a similar rate (RR = 0.523; Table S2).
To gain insight into the genetic variation of nPE1 and nPE2, we resequenced DNA samples from a global human diversity panel of 90 individuals representing most ethnic groups (Table S3). We found no polymorphic sites at nPE2 and three polymorphic sites at nPE1. Interestingly, two of these SNPs are located within the nPE1core and have not been previously reported (Fig. S5 and Table S3). As an internal comparison we resequenced the coding region of POMC exon 3 and detected four polymorphic sites, with one of them representing a unique SNP (Table S3). Our data indicate that at the human population level nPE1 and nPE2 are evolving more slowly than coding exon 3, although the large majority of exon 3 SNPs do not code for amino acids present in bioactive POMC peptides but rather in linker regions (Fig. S5 and Table S3). Altogether, our interspecies and intraspecies sequence comparisons indicate that nPE2 is under stronger purifying selection than nPE1.
Discussion
In this study we provide evolutionary and functional evidence supporting the independent exaptation of two unrelated retroposons into neuronal-specific enhancers that dictate Pomc expression in the arcuate nucleus of the hypothalamus of placental mammals. The example of nPE1 and nPE2 constitutes an authentic demonstration of cell-specific enhancer analogs that originated by convergent molecular evolution. Convergent evolution occurs when unrelated ancestors give rise to similar forms or functions. There are several morphological and functional examples of convergent evolution, such as the development of wings in birds and bats, eyes in cephalopods and mammals, and immune systems in jawed and jawless fishes (23). Some examples of convergent molecular evolution at the protein coding level have also been reported, including the origin of visual pigment genes in fishes and humans (24) and protein phosphatases in animals and plants (25). In the case reported here, a MaLR and a CORE-SINE retroposon independently evolved into functional arcuate-specific enhancers of POMC in mammals. Convergent evolution of these two enhancer modules was a long-lasting evolutionary process that occurred over at least 20 million years, from the earlier fixation of nPE2 before the Prototheria split, 167 Mya (16), to the later exaptation of a MaLR retroposon into nPE1 after the Metatheria/Eutheria split (147 Mya), as depicted in Fig. 5. Bioinformatic analysis revealed three nucleotide motifs and eight putative transcription factor binding sites shared by nPE1core and nPE2 (Fig. S6). Functional assays will be necessary to unravel the common neuronal-specific transcriptional code used by these two unrelated enhancers.
Fig. 5.
Evolutionary history of POMC, nPE1, and nPE2. The drawing represents the phylogeny of all mammalian orders (16, 35), vertebrate classes, and tunicates as an outgroup. POMC (black arrow) appeared before the radiation of all fishes including Agnathans, Chondrichthians, and Osteichthians (10). nPE2 (blue arrow) appeared in the lineage leading to mammals, and nPE1 (purple arrow) is found only in placental mammals.
The adaptive value of maintaining under purifying selection two functionally overlapping enhancers, instead of just one has been proposed for developmental genes (26) in light of the canalization theory independently proposed by Schmalhausen (27) and Waddington (28), who put forward the idea that the precise and stable spatiotemporal phenotypes normally observed during embryogenesis are assured by duplicated mechanisms that reduce variability by buffering environmental perturbations. Recently, two groups independently demonstrated for the D. melanogaster genes snail (6) and shavenbaby (4) that overlapping (shadow) enhancers provide adaptive robustness to overcome suboptimal environmental conditions during fly development (4, 6). More recently, it has been shown that a 6.5-kb deletion encompassing a distal retinal-specific shadow enhancer upstream of the human ATOH7 gene causes nonsyndromic congenital retinal nonattachment (NCRA; ref. 29). Although definitive proof that lack of this distal enhancer causes NCRA is still pending, the more parsimonious interpretation of the report challenges the semantics of the terms secondary or shadow for enhancers that could play fundamental roles even in normal conditions. Although Pomc expression starts at e10.5 in the mouse, it cannot be considered a classic developmental gene because all known functions of POMC-derived peptides are exerted after birth, and mice or humans lacking POMC do not exhibit overt developmental defects (30, 31). Thus, nPE1 and nPE2 are a unique example of overlapping enhancers acting in a gene primarily involved in postnatal development and adult physiology. For a gene like POMC, the adaptive value of having two apparently redundant enhancers may be related to the probability of increasing transcriptional rate and/or minimizing its variance. These two, not mutually exclusive, interpretations may be illustrated by a double-scull rowboat model that has the capacity to be propelled faster by both rowers when necessary but can keep going straight at basal speed if one rower wears out (acquisition of a deleterious mutation on one site). The processing of POMC prohormone in hypothalamic neurons generates α-MSH and β-endorphin, two potent neuropeptides that promote satiety and analgesia, respectively. Higher transcriptional efficiency of POMC provided by two overlapping enhancers could have played an adaptive role in evolution by increasing the ability of mammals to inhibit foraging behavior during risky environmental conditions (e.g., presence of predators) and to maintain long-lasting escapes after injury.
It has been proposed that shadow enhancers are duplicated copies of primary enhancers (5), although this hypothesis has not been rigorously tested. Our study shows, in contrast, that the overlapping enhancers nPE1 and nPE2 did not arise as a duplication event but rather as a convergent evolutionary mechanism. The independent emergence and fixation of two evolutionarily unrelated enhancers to control jointly neuronal Pomc expression may be viewed as a case of superfunctionalization of noncoding elements.
Retrotransposition is a fundamental driving force in evolution. Although originally considered genome parasites, some retrotransposon-derived sequences likely have gained advantageous functions (exaptation) for the host, because recent studies showed they are under purifying selection in evolutionarily related genomes (32, 33). Although it is conceivable that most conserved retroelement-derived sequences located in the vicinity of transcriptional units play some role in gene expression regulation, functional proof of exapted regulatory retroelements has been scarcely demonstrated. Indeed, nPE1 may be added to the incipient list of ancient retroposons coopted into cell-specific enhancers that includes three different types of SINEs: an LF-SINE exapted into an enhancer of Isl1 (20), a CORE-SINE exapted into the neuronal Pomc enhancer nPE2 (17), and an AmnSINE1 that evolved into a brain Fgf8 enhancer (21). Evidence that nPE1 derives from the exaptation of a MaLR is based on the high percentage of identity between human nPE1 and THE1B (62% along the 160 bp of nPE1core). The similarity is remarkably high within a 43-bp region of nPE1 (80% between nucleotides 41 and 83; Fig. 3C). This level of identity is similar to those reported for the opossum nPE2 and a consensus CORE-SINE [59% along 70 bp, with a central 45-bp region showing 71% identity (17)], the human ISL1 enhancer and consensus LF-SINE [61% along 55 bp (20)], and the consensus Amn-SINE and human Fgf8 enhancer [70% identity along 170 bp (21)]. Interestingly, a MaLR of the MSTB2 family was recently reported to have been exonized into an alternatively spliced exon of the IL22RA2 gene in primates (34).
There are approximately 240,000 copies of MaLRs in the human genome (3.65% of the genome) (19). Sequence comparisons indicate that nPE1 is most similar to THE1B, a MaLR present in a high copy number particularly in the human, chimp, and macaque genomes but apparently absent from rodents (18). Thus, THE1Bs have probably been active until recently in the primate lineage (18), and their high copy number and level of sequence conservation in the three primate genomes analyzed were critical for the in silico paleogenomics discovery of nPE1 as an exapted MaLR. Because nPE1 is present in all placental mammalian orders, the exaptation event that created nPE1 probably occurred before the radiation of placental mammals and involved an ancient THE1B-like MaLR that was active in the lineage leading to this group. Because the evolutionary history of MaLRs has not been reexamined since 1993 (18), further phylogenetic studies of this large family of retroposon is necessary to definitively establish the identity of the MaLR exapted into nPE1 more than 90 Mya.
In summary, our study provides a unique evolutionary mechanism by which two distinct regulatory elements may be incorporated into a gene locus to control cell-specific expression. Given the large contribution of retropositional insertions to the total genome size in the human (45%), mouse (33%), and fly (15%), it is conceivable that convergent molecular evolution of analogous cell-specific enhancers by sequential exaptation of retroposons is a more generalized phenomenon than previously anticipated.
Materials and Methods
Sequences and Databases.
Blast searches were performed using nPE1 sequences against whole-genome assemblies from Ensembl (http://www.ensembl.org) and the Trace Archive (http://www.ncbi.nlm.nih.gov/Traces). Evolutionary rates among sites were modeled using the Gamma distribution, and equilibrium nucleotide frequencies were considered to be equal.
Transgenes and Reporter Gene Expression in Transgenic Mice.
Transgenes nPE1Pomc-tomato, nPE2Pomc-EGFP, nPE1corePomc-EGFP, and nPE1ΔcorePomc-EGFP were built as described in SI Materials and Methods. Mice from selected pedigrees were crossed among each other to obtain compound heterozygote pairs carrying nPE1-tomato and nPE2-EGFP. Spatiotemporal expression analysis of both reporter genes was performed at e9.5, e10.5, e12.5, e18.5, and in mature adults.
Human Population Sequence and Analysis.
A panel of 90 human samples from the Coriell Institute that broadly represents worldwide populations was used for resequencing. Chromatograms were aligned by the Sequencher software (Gene Codes Corporation), and polymorphisms were detected by direct visual inspection.
In Silico Paleogenomics.
Blast was modified incorporating Kimura's two-parameter evolutionary model to the scoring matrix calculation to search nPE1 paralogs in vertebrate genomes.
Detailed descriptions of all materials and methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Marta Treimun, Marisol Costa, and Vanina Rodríguez for excellent technical assistance and Anthony Coll for supplying Pomc-LacZ knockin mice. This work was supported by National Institutes of Health Grant DK068400 (to M.J.L. and M.R.), an International Research Scholar Grant of the Howard Hughes Medical Institute (M.R.), Agencia Nacional de Promoción Científica y Tecnológica, Argentina (to M.R. and L.F.F.), Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina, and Universidad de Buenos Aires. R.L.-L. received a doctoral fellowship from the Consejo Nacional de Investigaciones Científicas y Técnicas (Chile) and S.N. from the Agencia Nacional de Promoción Científica y Tecnológica (Argentina).
Footnotes
Conflict of interest statement: M.J.L., F.J.S.d.S., and M.R. have intellectual property and patent interests in the POMC neuronal-specific enhancers and have received income from the licensing of this intellectual property and related research material to financially interested companies.
Data deposition: SNP data have been deposited in www.ncbi.nlm.nih.gov/SNP.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104997108/-/DCSupplemental.
References
- 1.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch M. The origins of eukaryotic gene structure. Mol Biol Evol. 2006;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- 3.Wilson MD, Odom DT. Evolution of transcriptional control in mammals. Curr Opin Genet Dev. 2009;19:579–585. doi: 10.1016/j.gde.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Frankel N, et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Meara MM, et al. Cis-regulatory mutations in the Caenorhabditis elegans homeobox gene locus cog-1 affect neuronal development. Genetics. 2009;181:1679–1686. doi: 10.1534/genetics.108.097832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- 9.Werner T, Hammer A, Wahlbuhl M, Bösl MR, Wegner M. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35:6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Souza FS, Bumaschny VF, Low MJ, Rubinstein M. Subfunctionalization of expression and peptide domains following the ancient duplication of the proopiomelanocortin gene in teleost fishes. Mol Biol Evol. 2005;22:2417–2427. doi: 10.1093/molbev/msi236. [DOI] [PubMed] [Google Scholar]
- 11.Raffin-Sanson ML, de Keyzer Y, Bertagna X. Proopiomelanocortin, a polypeptide precursor with multiple functions: From physiology to pathological conditions. Eur J Endocrinol. 2003;149:79–90. doi: 10.1530/eje.0.1490079. [DOI] [PubMed] [Google Scholar]
- 12.Low MJ. Role of proopiomelanocortin neurons and peptides in the regulation of energy homeostasis. J Endocrinol Invest. 2004;27(6 Suppl):95–100. [PubMed] [Google Scholar]
- 13.Lee M, Wardlaw SL. The central melanocortin system and the regulation of energy balance. Front Biosci. 2007;12:3994–4010. doi: 10.2741/2366. [DOI] [PubMed] [Google Scholar]
- 14.Smart JL, Tolle V, Low MJ. Glucocorticoids exacerbate obesity and insulin resistance in neuron-specific proopiomelanocortin-deficient mice. J Clin Invest. 2006;116:495–505. doi: 10.1172/JCI25243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Souza FS, et al. Identification of neuronal enhancers of the proopiomelanocortin gene by transgenic mouse analysis and phylogenetic footprinting. Mol Cell Biol. 2005;25:3076–3086. doi: 10.1128/MCB.25.8.3076-3086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bininda-Emonds OR, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 17.Santangelo AM, et al. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet. 2007;3:1813–1826. doi: 10.1371/journal.pgen.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smit AF. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 20.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki T, et al. Possible involvement of SINEs in mammalian-specific brain formation. Proc Natl Acad Sci USA. 2008;105:4220–4225. doi: 10.1073/pnas.0709398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Japón MA, Rubinstein M, Low MJ. In situ hybridization analysis of anterior pituitary hormone gene expression during fetal mouse development. J Histochem Cytochem. 1994;42:1117–1125. doi: 10.1177/42.8.8027530. [DOI] [PubMed] [Google Scholar]
- 23.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokoyama R, Yokoyama S. Convergent evolution of the red- and green-like visual pigment genes in fish, Astyanax fasciatus, and human. Proc Natl Acad Sci USA. 1990;87:9315–9318. doi: 10.1073/pnas.87.23.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moorhead GB, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–409. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- 26.Hobert O. Gene regulation: enhancers stepping out of the shadow. Curr Biol. 2010;20:R697–R699. doi: 10.1016/j.cub.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 27.Schmalhausen I. Factors of Evolution: The Theory of Stabilizing Selection. Chicago: Univ of Chicago; 1949. [Google Scholar]
- 28.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 29.Ghiasvand NM, et al. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat Neurosci. 2011;14:578–586. doi: 10.1038/nn.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Challis BG, et al. Mice lacking pro-opiomelanocortin are sensitive to high-fat feeding but respond normally to the acute anorectic effects of peptide-YY(3-36) Proc Natl Acad Sci USA. 2004;101:4695–4700. doi: 10.1073/pnas.0306931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krude H, et al. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat Genet. 1998;19:155–157. doi: 10.1038/509. [DOI] [PubMed] [Google Scholar]
- 32.Kamal M, Xie X, Lander ES. A large family of ancient repeat elements in the human genome is under strong selection. Proc Natl Acad Sci USA. 2006;103:2740–2745. doi: 10.1073/pnas.0511238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishihara H, Smit AF, Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piriyapongsa J, Polavarapu N, Borodovsky M, McDonald J. Exonization of the LTR transposable elements in human genome. BMC Genomics. 2007;8:291. doi: 10.1186/1471-2164-8-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy WJ, et al. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.