Abstract
Horizontal gene transfer (HGT) can radically alter the genomes of microorganisms, providing the capacity to adapt to new lifestyles, environments, and hosts. However, the extent of HGT between eukaryotes is unclear. Using whole-genome, gene-by-gene phylogenetic analysis we demonstrate an extensive pattern of cross-kingdom HGT between fungi and oomycetes. Comparative genomics, including the de novo genome sequence of Hyphochytrium catenoides, a free-living sister of the oomycetes, shows that these transfers largely converge within the radiation of oomycetes that colonize plant tissues. The repertoire of HGTs includes a large number of putatively secreted proteins; for example, 7.6% of the secreted proteome of the sudden oak death parasite Phytophthora ramorum has been acquired from fungi by HGT. Transfers include gene products with the capacity to break down plant cell walls and acquire sugars, nucleic acids, nitrogen, and phosphate sources from the environment. Predicted HGTs also include proteins implicated in resisting plant defense mechanisms and effector proteins for attacking plant cells. These data are consistent with the hypothesis that some oomycetes became successful plant parasites by multiple acquisitions of genes from fungi.
Keywords: osmotrophy, pseudofungi, lateral gene transfer
Horizontal gene transfer (HGT) involves the transfer of genetic material between reproductively isolated lineages (1, 2) and has been an important factor in prokaryotic evolution (3–5). By contrast, the role of HGT between eukaryotic genomes is less clear (2, 6) and seems to have occurred at a lower frequency, with some reports suggesting that HGT between eukaryotic kingdoms is rare (7). Conversely, HGTs originating from prokaryotic genomes constitute a significant, but relatively small, proportion of the total inventory of genes in phagotrophic protists (8–12) and some osmotrophic fungi (13).
Fungi branch with animals on the tree of life (14) and encompass an extremely diverse group of organisms, including species adapted to forming intimate relationships with plants, including mutualistic and parasitic associations, as well as saprophytic growth (15–17). To explore the frequency of HGT between distinct eukaryotic groups that form parasitic associations with plants, we previously used the predicted protein-encoding sequences from the genome of a fungal plant parasite, Magnaporthe oryzae (18), as a BLASTp search seed and identified four HGTs from fungi to oomycetes with strong phylogenetic support (19). The oomycetes are distant relatives of fungi and branch within the stramenopile radiation, which includes a diversity of photosynthetic microbes possessing plastid organelles of secondary endosymbiotic ancestry (20). Oomycetes, however, are not photosynthetic and display filamentous growth, closely resembling fungi in many aspects of their biology. Both fungi and oomycetes, for instance, feed exclusively by osmotrophy, secreting depolymerizing enzymes to break down complex biological materials in the extracellular environment, followed by transport of broken-down metabolic units into the cell. Fungi and oomycetes also cause many of the world's most serious plant diseases. Sudden oak death is caused by the oomycete Phytophthora ramorum, for example, whereas the Irish potato famine of the 19th century was caused by the late blight parasite Phytophthora infestans. Important crop diseases caused by fungi include the devastating rice blast disease caused by M. oryzae and the rusts, smuts, and mildews that affect wheat, barley, and maize. In this study we report that HGT between fungi and oomycetes has occurred to a far greater degree than hitherto recognized (19). Our previous analysis suggested four strongly supported cases of HGT, but by using a whole-genome, gene-by-gene phylogenetic analysis we now reveal a pattern of 34 transfers and propose that these transfers have been fundamental to the evolution of plant parasitic traits within the oomycetes.
Results and Discussion
Identifying and Testing the Pattern of HGT Between Fungi and Oomycetes.
Among the best methods for identifying HGT is to identify a gene phylogeny that places a taxonomic group (recipient) within the branches of a distantly related group (donor) in direct contradiction to the known phylogenetic relationships of the respective taxa (2, 6). To identify all potential gene transfers between fungi and oomycetes, we selected the predicted proteomes of the oomycete species Phytophthora ramorum, Phytophthora sojae, Phytophthora infestans, and Hyaloperonospora parasitica (also named Hyaloperonospora arabidopsidis) (21–23). We processed each proteome separately, first excluding all putative transposable elements (SI Materials and Methods). Then we clustered the genes from each genome into closely related cluster groups (24) (SI Materials and Methods). Cluster groups that BLASTp demonstrated were exclusively found only in the oomycete genomes were then removed (SI Materials and Methods). This process left 3,014, 3,018, 3,233, and 2,169 cluster groups from P. ramorum, P. sojae, P. infestans, and H. parasitica, respectively, totaling 11,434 gene clusters (Table 1). We then used a gene-by-gene phylogeny pipeline (7) to generate fast maximum-likelihood trees for all 11,434 gene family groups, using a database of 795 (173 eukaryotic and 622 prokaryotic) genomes (Table S1). These data were searched manually for trees that demonstrated putative fungi/oomycete gene transfer. Gene families that produced unresolved tree topologies were reprocessed by running the tree-building pipeline again but using different gathering thresholds (SI Materials and Methods).
Table 1.
Comparative gene-by-gene phylogenomic analysis and identified HGTs
| Species | P. ramorum | P. sojae | P. infestans | H. parasitica |
| Total no. of predicted proteins | 15,743 | 19,027 | 22,658 | 13,240 |
| Total proteins used in phylogenetic analysis | 6,871 | 6,079 | 7,093 | 3,365 |
| No. of gene cluster groups used for phylogenetic analyses [after OrthoMCL (24) clustering] | 3,014 | 3,018 | 3,233 | 2,169 |
| No. of HGT gene families strongly supported by all methods | 19 | 17 | 11 | 4 |
| No. of “gray zone” HGT gene families (Table 2 and Table S3) | 11 | 10 | 7 | 5 |
| Total putative ORFs from fungal-derived HGTs | 143 | 117 | 48 | 21 |
| Total secreted proteins in genome identified by both WoLFPSORT and SignalP | 1,326 | 1,586 | 1,123 | 411 |
| Total no. of proteins analyzed with phylogeny predicted to be secreted | 521 | 630 | 351 | 121 |
| No. of fungal HGT-derived proteins with N-terminal secretion motif | 101 | 89 | 30 | 13 |
| % Total predicted secretome derived from fungi by HGT | 7.61 | 5.61 | 2.67 | 3.16 |
| % Proteins analyzed that are predicted to be secreted and are derived by HGT | 19.38 | 14.13 | 8.55 | 10.74 |
All of the predicted fungi/oomycete HGTs were subject to reevaluation of taxon sampling using additional database searches and manual alignment improvement, followed by recalculation of the phylogenetic tree, combined with bootstrap analyses (see SI Materials and Methods and Table S2 for details of phylogenetic analysis). These analyses identified multiple cases in which oomycete genes (the recipient group) branched within a clade of fungal genes (the donor group) and a single case demonstrating the opposite relationship. To confirm the results of these phylogenetic analyses, we used alternative topology tests to test whether it was possible to reject monophyly of the donor group. Where taxon sampling allowed, we used a variety of different topology constraints, corresponding to different relationships within the fungi (15, 25). This approach enabled us to partially polarize the ancestry of the HGT event relative to the donor group (Table S3). Taken together, our analyses identified 20 gene families predicted to have been transferred from within the fungi to the oomycetes and one case in which the transfer occurred from the oomycetes to the fungi (Fig. 1 and Figs. S1.1–S1.21).
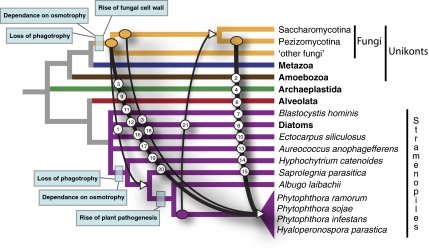
Fig. 1.
Pattern of HGTs between fungi and oomycetes, demonstrating that the majority of the fungal-derived gene transfers are into/or retained by the plant parasitic oomycetes. Using the results of the phylogenetic analysis in combination with alternative topology tests, it was possible to estimate the earliest point of transfer for each of the 21 strongly supported HGTs (Table S3). By comparing the taxonomic distribution of the HGTs it was then possible to identify the putative primary point of acquisition. Incomplete phylogenetic resolution and incomplete taxon sampling may cause these estimates to misplace the HGT events. We also note that the figure is a based on a hypothetical cladogram and does not identify the pattern of transfer relative to either phylogenetic distance or time. Additional genome sampling will enable improved resolution of these transfers. The number labels on each transfer event refer to phylogenetic data in Figs. S1.1–S1.21. Major events in cell evolution are marked to polarize HGTs in relation to evolutionary history of these microbes.
Our pipeline analysis also identified four gene families that were exclusively found in oomycete and fungal genomes and not present in any additional taxonomic groups analyzed. We checked to confirm that the taxon sampling was robust by comparison with the GenBank nr database, using both BLASTp and psi-BLAST with five iterations (26), and by interrogating the GenBank EST and the Taxonomically Broad EST database using tBLASTn (27). Because of the taxon distribution it was not possible to perform phylogenetic analysis to test an HGT hypothesis directly, because the taxon sampling included only oomycetes and fungi. However, punctate taxon distribution of a gene family can be used to suggest HGT because alternative explanations involving ancient acquisition of a gene in the last common ancestor, coupled to gene loss, are less parsimonious (see ref. 6 for description of different HGT scenarios). On the basis of these criteria, we have included the additional four gene families among the putative fungi–oomycete HGTs (Table 2 and Table S3).
Table 2.
Summary of support for each HGT and gene annotation in brief
| HGT ID 1–34 | GenBank accession no./Joint Genome Institute database protein ID | Support for HGT: phylogeny (P), bootstrap analysis (B), topology comparison tests (Ct), and/or taxon distribution (Td) | Conserved introns between donors and recipients (Table S4) | Annotation of putative function | Reported by Richards et al. (19) |
| 1 (Fig. S1.1) | EEY57756 | P, B, Ct, Td | 1 shared intron | MFS transporters similar to saccharide monomer transporters | Yes (named AraJ) |
| 2 (Fig. S1.2) | 82760 (P. ramorum) | P, B, Ct | Extracellular esterase/lipase | Yes (named esterase/lipase (ref. 19, Fig. S3B) | |
| 3 (Fig. S1.3) | AAM48174.1 | P, B, Ct | Aldose 1-epimerase | Yes (named GalM) | |
| 4 (Fig. S1.4) | 71178 (P. ramorum) | P, B, Ct, Td | α-ketoglutarate dependent xanthine dioxygenase (XanA) | ||
| 5 (Fig. S1.5) | EEY56552 | P, B, Ct | Dehydrogenase/reductase family protein | ||
| 6 (Fig. S1.6) | 72257 (P. ramorum) | P, B, Ct, Td | Extracellular quercetin 2,3-dioxygenase | ||
| 7 (Fig. S1.7) | EEY52979 | P, B, AU test is borderline | Extracellular α-l-rhamnosidase (glycosyl hydrolase 78) | ||
| 8 (Fig. S1.8) | EEY63463 | P, B, Ct | Lactonohydrolase/gluconolactonase | ||
| 9 (Fig. S1.9) | EEY64355 | P, B, Ct | Extracellular glucooligosaccharide oxidase | ||
| 10 (Fig. S1.10) | EEY59160 | P, B, Ct | Extracellular unsaturated rhamnogalacturonyl hydrolase (glycosyl hydrolase 88) | ||
| 11 (Fig. S1.11) | 83543 (P. ramorum) | P, B, Ct | Transcription factor | ||
| 12 (Fig. S1.12) | 85044 (P. ramorum) | P, B, Ct, Td | 1 shared intron | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase/Phenylphosphate carboxylase family | |
| 13 (Fig. S1.13) | EEY53137 | P, B, Ct | 1 shared intron | Phosphatidylinositol transfer protein | |
| 14 (Fig. S1.14) | 133521 (P. sojae) | P, B, Ct | Extracellular protein similar to prokaryote lipases | ||
| 15 (Fig. S1.15) | 142730 (P. sojae) | P, B, Ct | 1 shared intron | Esterase/lipase protein domain family | |
| 16 (Fig. S1.16) | EEY68514 | P, B, Ct | Xylitol dehydrogenase/sorbitol dehydrogenase | ||
| 17 (Fig. S1.17) | EEY55544 | P, B, Ct, Td | 2 shared introns | Extracellular arabinan-endo-1,5-α-l-arabinosidase (glycosyl hydrolase 43) | |
| 18 (Fig. S1.18) | EEY59913 | P, B, Ct, Td | Transporter for purines and pyrimidines | Yes (named CodB) | |
| 19 (Fig. S1.19) | 141189 (P. sojae) | P, B, Ct | Extracellular protein with an esterase/lipase domain | ||
| 20 (Fig. S1.20) | EEY58144 | P, B, Ct, Td | Putative member of the NPP1 (necrosis and ethylene-inducing peptide 1 proteins) | ||
| 21 (Fig. S1.21) | EEY64154 | P, B, Ct, Td | 1 shared intron | Conserved hypothetical protein with similarity to a prokaryotic antibiotic biosynthesis monooxygenase | |
| 22 (Fig. S1.22) | EEY62062 | P, Td | Pectate lyase | ||
| 23 (Fig. S1.23) | EEY67135 | P, Td | Extracellular intradiol dioxygenase | Yes (named PcaH) | |
| 24 (Fig. S1.24) | ABB22031 | P, Td | Extracellular endoglucanase (glycosyl hydrolase 12) | ||
| 25 (Fig. S1.25) | EEY67552 | P, B | 1 shared intron | Extracellular histidine phosphatase domain protein | |
| 26 (Fig. S1.26) | EEY65395 | P, B | 2 shared intron | Extracellular endo-1,4-β-xylanase (glycosyl hydrolase 10) | |
| 27 (Fig. S1.27) | 75147 (P. ramorum) | P, B | Member of Zinc metalloprotease proteins which include archaemetzincin | ||
| 28 (Fig. S1.28) | EEY56384 | P, B | Extracellular arabinogalactan endo-1,4-β-galactosidase (glycosyl hydrolase 53) | ||
| 29 (Fig. S1.29) | 77558 (P. ramorum) | P, B | Aliphatic nitrilase (CN hydrolase family) | ||
| 30 (Fig. S1.30) | EEY68947 | P, Td | Extracellular α-l-rhamnosidase (glycosyl hydrolase 78) | ||
| 31 | 76863 (P. ramorum) | Td | LysM domain protein | ||
| 32 | EEY55495 | Td | Extracellular conserved hypothetical protein | ||
| 33 | 134308 (P. sojae) | Td | NmrA, a negative transcriptional regulator | ||
| 34 | EEY58177 | Td | Conserved hypothetical protein |
Sequences for P. ramorum and P. sojae are available from the Department of Energy, Joint Genome Institute website. Protein sequences can be obtained using the protein ID and searching at http://genome.jgi-psf.org/pages/search-for-genes.jsf?organism=Phyra1_1 for P. ramorum and http://genome.jgi-psf.org/pages/search-for-genes.jsf?organism=Physo1_1 for P. sojae.
Our analysis also identified an additional nine gene families that demonstrated phylogenetic support for the oomycete gene branching within the fungi to the exclusion of all other taxa, but the approximately unbiased (AU) alternative topology tests proved inconclusive, suggesting that the case for HGT is not definitive given available data (Table S3). In four of these cases (Figs. S1.22–S1.24 and S1.30) our analyses demonstrated that the gene family was present in a mosaic scattering of prokaryote, fungi, and oomycete taxa, with the oomycetes branching within the fungal clade but with weak bootstrap support. These data sets implicate HGT (on the basis of taxonomic distribution of the gene family with phylogenetic analyses suggesting fungi-to-oomycete HGT), but alternative topology tests proved inconclusive. The remaining five gene families also suggested fungi-to-oomycete HGT, placing the oomycete gene within the fungi, separate from other eukaryotic taxa, with moderate to strong bootstrap support but with alternative topology tests again proving inconclusive (Figs. S1.25–S1.29 and Table S3). We therefore find strong evidence for 21 gene transfers and tentative evidence for a further 13 transfers, suggesting an important and pervasive pattern of HGT between these distantly related groups (Table S3).
Theoretically, the presence of conserved introns in both recipient and donor taxa would suggest that any HGT is the product of a direct eukaryotic-to-eukaryotic transfer. To test for evidence of such a pattern, we searched all 30 HGT gene families supported by phylogenetic analysis (Figs. S1.1–S1.30) for evidence of a conserved intron present in the donor and recipient taxa. We found eight such cases (Table 2 and Table S4), providing additional data to support the hypothesis that in these cases the transfers are eukaryotic-to-eukaryotic HGT events.
Distribution of HGTs Relative to Evolution of Parasitic Traits in the Oomycetes.
To provide additional data for polarizing the ancestry of these HGT events, we sequenced the genome of Hyphochytrium catenoides using a combination of DNA and RNA (transcribed as cDNA) sequencing using both the Roche 454 GS FLX titanium and Illumina GAIIx paired-end methods (SI Materials and Methods). H. catenoides forms a sister branch to the oomycetes and, like the oomycetes, was originally classified as a fungus because of its fungal-like morphology and osmotrophic feeding habit (20, 28). However, molecular phylogenetics have since demonstrated that oomycetes and hyphochytriomycetes are sister groups and branch separately from fungi within the stramenopile radiation (20, 28). Analysis of this draft genome assembly revealed that all of the 34 oomycete/fungi HGTs are absent from the Hyphochytrium genome (SI Materials and Methods). Of the 34 putative HGTs identified, we noted that only 2 were present in the genome assembly of the oomycete fish parasite Saprolegnia parasitica (Figs. S1.1 and S1.21), one of which is an HGT from the oomycete lineage to the fungi (Fig. 1). To further test this pattern we compared the 34 HGTs with 13,807 proteins from the oomycete Albugo laibachii genome available in GenBank (29). Albugo forms a deeper branch sister to the Phytophthora/Hyaloperonospora clade and possesses a distinct repertoire of effector proteins compared with other oomycete plant pathogens (29). Comparative analysis suggests that only one of the HGTs was present in the Albugo assembly (HGT 34; Table 2 and Table S3). However, all 34 HGTs were present in one or more of the Phytophthora/Hyaloperonospora plant parasitic oomycete species. The majority of the HGTs therefore seem to have been acquired specifically within the oomycete plant parasite clade, although this result will need retesting as more oomycete genomes become available.
In 2006 Richards et al. (19) provided preliminary evidence of a pattern of HGT between fungi and oomycetes. These data were based on a BLAST survey of a single fungal genome and provided strong evidence for four gene transfers from fungi to oomycetes, with an additional four candidates for which the tree topologies could not be resolved but could potentially indicate HGT. We used these data to propose that HGT led to transfer of osmotrophic characteristics from fungi to oomycetes and was an important step in the evolution of the fungal-like biology of oomycetes and possibly their sisters (e.g., hyphochytridiomycetes), which together form the “Pseudofungi” (20). The data reported here were based on a comprehensive gene-by-gene phylogeny approach and confirmed five of these HGTs [including the four strongly supported HGTs reported previously (19); Table 2 and Table S5]. The remaining three HGTs, which were listed as tentative in the 2006 article, are not supported by this analysis (Table S5). Using additional genome sampling including a de novo assembly of H. cateniodes, we were then able to test the hypothesis that fungal-to-oomycete HGTs were important for the transition from a phagotrophic algal form to an osmotrophic filamentous form deep within the oomycete/hyphochytrium clade. Contrary to our previous suggestion (19), the results of this study support an alternative hypothesis, with the pattern of HGT seeming to have occurred much later, with the majority of the HGTs specifically retained by oomycete plant parasites, a pattern consistent with the putative function of these proteins, discussed below (Fig. 2, Table 2, and Table S5). Furthermore, the pattern demonstrates that the HGTs occurred after the last common ancestor of the oomycetes and the hyphochytriomycetes had lost the capacity for phagotrophy and had instead evolved an osmotrophic/filamentous lifestyle (Fig. 1), demonstrating that the absence of phagotrophy, at least for the oomycetes, is not a barrier to HGT.
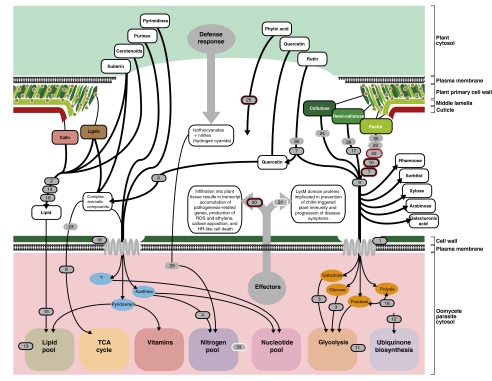
Fig. 2.
Schematic diagram demonstrating the oomycete functional proteome derived from fungi by HGT and its putative function regarding host attack. The majority of functions are related to attacking and/or feeding on plant cell tissue. HGTs were represented on the figure when it was possible to identify a putative function (these are illustrated in numbered gray oblongs, with the numbers referring to the 34 HGTs identified; Table 2 and Table S5), with strongly supported HGTs confirmed by alternative topology tests) enclosed within a black line (Figs. S1.1–S1.21). Oblongs with additional red borders represent genes shown to be up-regulated in P. infestans microarray data (22) after 5 d infection of plant tissue (Table S5).
The pattern of fungal-derived HGTs implicates the gene acquisitions as being important in the evolution of plant parasitism. To test this idea, we examined the HGTs in detail. First, we found that of the 34 HGTs, 21 demonstrated evidence of gene duplication after transfer. In some cases the number of gene duplication events was extensive, such that the 33 fungi-to-oomycete HGTs contributed a total of 329 predicted genes, including 143 genes to P. ramorum, 117 to P. sojae, 48 to P. infestans, and 21 to H. parasitica (Table 1).
Putative Function of HGTs in the Oomycetes.
We set out to determine the proportion of the HGT candidates that were predicted to encode secreted proteins (SI Materials and Methods). Both fungi and oomycetes secrete large numbers of proteins, including plant cell wall-degrading enzymes, attachment factors, and effector proteins to subvert host defenses. We used a combination of SignalP and WolfPSORT to identify HGT-derived genes that putatively encoded proteins with N-terminal secretion signals (Table S5). This analysis showed that between 62% and 76% of all the HGT-derived genes in each of the four oomycete genomes investigated possess putative N-terminal secretion signals (Table 1). This is particularly striking in P. ramorum and P. sojae, where 71% and 76% of the HGT-derived genes putatively possess N-terminal signal peptides, respectively. The fungal-derived HGTs therefore account for between 2.7% (P. infestans) and 7.6% (P. ramorum) of the total predicted secretome of these plant parasites (Table 1).
To investigate the putative function of all 34 HGT gene families, we used a combination of BLAST and PFAM HMM homology searches to annotate these genes (Table 2, SI Materials and Methods, and Table S5). Strikingly, 13 of the 32 HGT gene families that could be annotated are predicted to function in the breakdown, transport, or remodeling of sugars (Fig. 2, Table 2, and Table S5). Indeed, a total of nine fungal-derived HGT gene families were predicted to encode extracellular polysaccharide depolymerizing enzymes and therefore putatively to break down rutin, hemicellulose, or pectin polysaccharides (Fig. 2, Table 2, and Table S5) sugars, which are unique to plants. The predicted fungi-to-oomycete HGTs also includes four esterase/lipase-encoding genes, three of which are predicted to be secreted, and a further three enzymes that are putatively involved in the degradation of complex aromatic polymers (Fig. 2, Table 2, and Table S5). The HGTs include examples of proteins that putatively degrade plant-specific structural components, including lignin, hemicellulose, pectin, and suberin. These structural compounds include key components of plant cell walls (e.g., hemicellulose and pectin) and the waxy epidermis (cutin), which together constitute the first barrier to plant infection (Fig. 2). Consequently, any acquisition of enzymes used to break down these structures would be advantageous to a plant parasite.
Additional HGTs include several gene products predicted to be involved in nutrient acquisition including a nucleotide transporter, which putatively functions in purine and/or pyrimidine uptake (Table S5). Potentially coupled to this adaptation, the fungal-derived HGTs include a putative xanthine dioxygenase involved in hydroxylation of the purine xanthine to uric acid (30, 31) (Fig. 2). A putative monosaccharide transporter gene was identified that would enable the cell to traffic sugar monomers (Fig. 2), whereas two additional fungal-derived HGT acquisitions were also identified that putatively function to process monosaccharides within the cell (Fig. 2). A nitrilase is present among the putative HGTs, predicted to break down toxic nitriles to carboxylic acids and ammonia. Such enzymes in plant parasites have been shown to break down the plant defense compound hydrogen cyanide into formamide (32). The list of fungal-derived HGTs also encompasses a putative histidine phosphatase domain protein, which putatively functions in breakdown of phytic acid and other organophosphate substrates (33). Phytic acid is the primary storage form of phosphate and inositol in plants (34). Consequently, this acquisition might provide an adaptation for uptake of phosphates from plant tissue.
Finally, the list of HGTs includes two proteins that in fungi have been directly implicated in plant parasitism. The LysM domain-containing gene family acquired by HGT from fungi has, for instance, been shown in fungal plant parasites to be linked with suppression of plant defenses. In Cladosporium fulvum, the causal agent of leaf mold of tomato, implicated in suppression of chitin-triggered plant immunity (35). Second, there is an example of a necrosis-inducing protein or (Nep1)-like protein (NLP) that is a candidate fungi-to-oomycete HGT (36). Infiltration of NPP1 protein into leaves of Arabidopsis thaliana results in transcript accumulation of plant defense-related genes, production of reactive oxygen species and ethylene, callose apposition, and localized cell death (36).
Conclusion
When considered together, our analysis demonstrates a pattern of at least 21 HGTs, and probably more than 34, between fungi and oomycetes, with the vast majority of predicted HGTs (33 in total) transferring gene functions from fungi to oomycetes. This equates to a small proportion of the total genome for which phylogenetic analysis was possible (0.5–1% of the genome analyzed; Table 1), but in many cases these gene transfers have subsequently undergone numerous gene duplications. A large fraction of the HGT-derived proteins are predicted to be secreted (Table 1), strongly suggesting that HGT from fungi has played a significant role in the evolution of the oomycete secretome. The observed pattern of transfer may have facilitated, or aided, the spread of oomycetes to plant hosts and their evolution into successful plant parasites. Comparative genomics demonstrates that these transfers do not seem to date back to the initial transition to an osmotrophic lifestyle (19) because they are, in the majority, absent in the animal parasitic oomycete Saprolegnia and the free-living filamentous osmotrophic Hyphochytrium. This suggests that osmotrophy, and the capacity to grow as strand-like hyphal cells, probably arose before diversification of these lineages and before the acquisition of fungal genes by HGT. Interestingly, fungal-derived HGTs may, however, represent specific acquisitions to life as a plant parasite. Our conclusion is consistent with the predicted functions of the HGT candidates aiding entry to plant cells, allowing efficient nutrient acquisition and leading to microbial proliferation in plant tissue.
Materials and Methods
Detailed descriptions of whole-genome gene-by-gene phylogenetic analysis, alternative topology tests, H. catenoides DNA and RNA preparation and sequencing, gene annotation, and identification of secreted proteins are provided in SI Materials and Methods. All potential HGTs found in a single oomycete genome were treated as possible cases of contamination. To test these, we conducted phylogeny of linked genes on DNA contigs. In all cases, we could confirm that the HGT was present within the oomycete genome (Table S6). An example of each HGT oomycete gene family is included as a combined FASTA file in Dataset S1.
Supplementary Material
Acknowledgments
T.A.R. received fellowship support from the Leverhulme Trust and funding from the Natural Environment Research Council (NERC) and the Biotechnology and Biological Sciences Research Council (BBSRC). M.D.M.J. is supported by NERC Grant NE/F011709/1. G.L. is supported by BBSRC Grant BB/G00885X/1.
Footnotes
The authors declare no conflict of interest.
Data deposition: The genome sequence data reported in this paper (Hyphochytrium) have been submitted to Sequence Read Archive for the raw data (SRP004821), and the assembled genome has been archived at the European Bioinformatics Institute (Genome Project: 61035).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105100108/-/DCSupplemental.
References
- 1.Doolittle WF. Lateral genomics. Trends Cell Biol. 1999;9:M5–M8. [PubMed] [Google Scholar]
- 2.Andersson JO. Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol. 2009;63:177–193. doi: 10.1146/annurev.micro.091208.073203. [DOI] [PubMed] [Google Scholar]
- 3.Doolittle WF, et al. How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Philos Trans R Soc Lond B Biol Sci. 2003;358:39–57. doi: 10.1098/rstb.2002.1185. , discussion 57–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher Y, et al. Lateral gene transfer and the origins of prokaryotic groups. Annu Rev Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. [DOI] [PubMed] [Google Scholar]
- 5.Jain R, Rivera MC, Moore JE, Lake JA. Horizontal gene transfer accelerates genome innovation and evolution. Mol Biol Evol. 2003;20:1598–1602. doi: 10.1093/molbev/msg154. [DOI] [PubMed] [Google Scholar]
- 6.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 7.Richards TA, et al. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell. 2009;21:1897–1911. doi: 10.1105/tpc.109.065805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doolittle WF. You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998;14:307–311. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- 9.Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 10.Archibald JM, Rogers MB, Toop M, Ishida K, Keeling PJ. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc Natl Acad Sci USA. 2003;100:7678–7683. doi: 10.1073/pnas.1230951100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loftus B, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 12.Andersson JO, Sjögren AM, Davis LAM, Embley TM, Roger AJ. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr Biol. 2003;13:94–104. doi: 10.1016/s0960-9822(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 13.Marcet-Houben M, Gabaldón T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 2010;26:5–8. doi: 10.1016/j.tig.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Burki F, Shalchian-Tabrizi K, Pawlowski J. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol Lett. 2008;4:366–369. doi: 10.1098/rsbl.2008.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James TY, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 16.Pirozynski KA, Malloch DW. The origin of land plants: A matter of mycotrophism. Biosystems. 1975;6:153–164. doi: 10.1016/0303-2647(75)90023-4. [DOI] [PubMed] [Google Scholar]
- 17.Wang B, Qiu YL. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza. 2006;16:299–363. doi: 10.1007/s00572-005-0033-6. [DOI] [PubMed] [Google Scholar]
- 18.Dean RA, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 19.Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ. Evolution of filamentous plant pathogens: Gene exchange across eukaryotic kingdoms. Curr Biol. 2006;16:1857–1864. doi: 10.1016/j.cub.2006.07.052. [DOI] [PubMed] [Google Scholar]
- 20.Cavalier-Smith T, Chao EE. Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista) J Mol Evol. 2006;62:388–420. doi: 10.1007/s00239-004-0353-8. [DOI] [PubMed] [Google Scholar]
- 21.Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 22.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 23.Baxter L, et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010;330:1549–1551. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Brien EA, et al. TBestDB: A taxonomically broad database of expressed sequence tags (ESTs) Nucleic Acids Res. 2007;35(Database issue):D445–D451. doi: 10.1093/nar/gkl770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Auwera G, et al. The phylogeny of the Hyphochytriomycota as deduced from ribosomal RNA sequences of Hyphochytrium catenoides. Mol Biol Evol. 1995;12:671–678. doi: 10.1093/oxfordjournals.molbev.a040245. [DOI] [PubMed] [Google Scholar]
- 29.Kemen E, et al. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 2011;9:e1001094. doi: 10.1371/journal.pbio.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cultrone A, et al. Convergent evolution of hydroxylation mechanisms in the fungal kingdom: Molybdenum cofactor-independent hydroxylation of xanthine via alpha-ketoglutarate-dependent dioxygenases. Mol Microbiol. 2005;57:276–290. doi: 10.1111/j.1365-2958.2005.04686.x. [DOI] [PubMed] [Google Scholar]
- 31.Sealy-Lewis HM, Scazzocchio C, Lee S. A mutation defective in the xanthine alternative pathway of Aspergillus nidulans: Its use to investigate the specificity of uaY mediated induction. Mol Gen Genet. 1978;164:303–308. doi: 10.1007/BF00333161. [DOI] [PubMed] [Google Scholar]
- 32.Sexton AC, Howlett BJ. Characterisation of a cyanide hydratase gene in the phytopathogenic fungus Leptosphaeria maculans. Mol Gen Genet. 2000;263:463–470. doi: 10.1007/s004380051190. [DOI] [PubMed] [Google Scholar]
- 33.Ullah AH, Gibson DM. Extracellular phytase (E.C. 3.1.3.8) from Aspergillus ficuum NRRL 3135: purification and characterization. Prep Biochem. 1987;17:63–91. doi: 10.1080/00327488708062477. [DOI] [PubMed] [Google Scholar]
- 34.Reddy NR, Sathe SK, Salunkhe DK. Phytates in legumes and cereals. Adv Food Res. 1982;28:1–92. doi: 10.1016/s0065-2628(08)60110-x. [DOI] [PubMed] [Google Scholar]
- 35.de Jonge R, et al. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science. 2010;329:953–955. doi: 10.1126/science.1190859. [DOI] [PubMed] [Google Scholar]
- 36.Qutob D, et al. Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell. 2006;18:3721–3744. doi: 10.1105/tpc.106.044180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.