Abstract
Inflammation plays a key role in the pathogenesis of obesity. Chronic overfeeding leads to macrophage infiltration in the adipose tissue, resulting in proinflammatory cytokine production. Both microbial and endogenous danger signals trigger assembly of the intracellular innate immune sensor Nlrp3, resulting in caspase-1 activation and production of proinflammatory cytokines IL-1β and IL-18. Here, we showed that mice deficient in Nlrp3, apoptosis-associated speck-like protein, and caspase-1 were resistant to the development of high-fat diet-induced obesity, which correlated with protection from obesity-induced insulin resistance. Furthermore, hepatic triglyceride content, adipocyte size, and macrophage infiltration in adipose tissue were all reduced in mice deficient in inflammasome components. Monocyte chemoattractant protein (MCP)-1 is a key molecule that mediates macrophage infiltration. Indeed, defective inflammasome activation was associated with reduced MCP-1 production in adipose tissue. Furthermore, plasma leptin and resistin that affect energy use and insulin sensitivity were also changed by inflammasome-deficiency. Detailed metabolic and molecular phenotyping demonstrated that the inflammasome controls energy expenditure and adipogenic gene expression during chronic overfeeding. These findings reveal a critical function of the inflammasome in obesity and insulin resistance, and suggest inhibition of the inflammasome as a potential therapeutic strategy.
The discovery of NOD-like receptors (NLRs) as essential components of the immune system triggered significant interest in the study of their contribution to the pathogenesis of inflammatory and autoimmune diseases. NLRs comprise a large family of intracellular proteins that are believed to be primarily involved in the innate immune response to microbial pathogens through the recognition of conserved pathogen-associated molecular patterns (1–3). However, NLRs also contribute to inflammation by sensing “danger signals” (i.e., endogenous molecules that are produced during tissue damage or inflammation) (3–5). A prominent example of an NLR protein implicated in autoinflammatory disease is Nlrp3 (also called Cryopyrin) (6). Nlrp3 activation induces the recruitment and autocatalytic activation of the cystein protease caspase-1 in a large cytosolic protein complex named the “inflammasome” (2–7). The bipartite adaptor protein ASC (apoptosis-associated speck-like protein) bridges the interaction between Nlrp3 and the caspase-1 by means of homotypic interactions involving its pyrin and CARD motifs, making it essential for activation of the inflammasome (7). Activated caspase-1 processes the cytosolic precursors of the related cytokines IL-1β and IL-18, thus allowing secretion of the biologically active cytokines. Hence, mice lacking caspase-1 are defective in the maturation and secretion of IL-1β and IL-18 (8–10). IL-1β participates in the generation of systemic and local responses to infection, injury, and immunological challenges by inducing the “acute phase response” characterized by fever, synthesis of acute phase proteins, and leukocytosis (11). Although IL-18 lacks the pyrogenic activity of IL-1β, it is involved in the induction of several secondary proinflammatory cytokines, chemokines, and cell-adhesion molecules (12, 13).
Obesity is accompanied by the development of a chronic low-grade inflammation that is promoted by expanding adipose tissue (14, 15). Expansion of fat mass characterized by adipocyte enlargement fuels the infiltration of macrophages into the adipose tissue (16, 17). The enhanced inflammatory trait of the adipose tissue instigates the production of cytokines that contribute to the development of insulin resistance (18, 19). IL-1β and IL-18 have also been linked to the development of obesity-induced insulin resistance. IL-1β has been reported to inhibit adipocyte differentiation (20), but the absence of IL-18 induced obesity and insulin resistance (21, 22). Moreover, high-fat diet (HFD) feeding resulted in the activation of caspase-1 in adipose tissue in mice (23). Interestingly, absence of NLRP3 has recently been shown to prevent the development of obesity-induced insulin resitance (24). However, the role of caspase-1 and the inflammasome member ASC in the development of HFD-induced obesity has not been characterized. Chronic activation of inflammasome-mediated caspase-1 activity may underlie the development of obesity-induced insulin resistance. To understand the role of the inflammasome in obesity and insulin resistance, we studied the response of Nlrp3−/−, ASC−/−, and Casp1−/− mice to HFD feeding. Our results indicate a major role for the inflammasome in modulating obesity and reveal its critical function in obesity-induced inflammation and insulin resistance.
Results
Absence of the Inflammasome Protects from HFD-Induced Obesity.
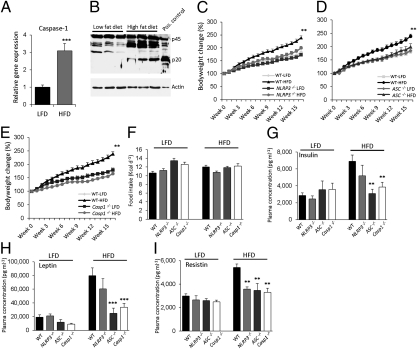
To investigate the role of the inflammasome in the adipose tissue, we first analyzed the expression and activation of the inflammasome downstream molecule caspase-1 in adipose tissue. In line with our previous work (23), caspase-1 gene expression and activation in white adipose tissue (WAT) of HFD-fed wild-type mice were increased (Fig. 1 A and B). We next assessed the functional roles of Nlrp3, ASC, and Caspase-1 in the development and progression of HFD-induced obesity using mice lacking Nlrp3, ASC, or Caspase-1. During 16 wk of HFD-feeding, food intake and bodyweight development were examined weekly. HFD-feeding of wild-type, Nlrp3−/− (Fig. 1C), ASC−/− (Fig. 1D), and Casp1−/− (Fig. 1E) animals demonstrated that the absence of the inflammasome components protects against the development of HFD-induced obesity. The observed lean phenotype in Nlrp3−/−, ASC−/−, and Casp1−/− mice was a result of defective inflammasome activation confirmed by significantly reduced IL-1β production, but not IL-6, in WAT of HFD-fed mice compared with wild-type mice (Fig. S1). The daily caloric food intake of all genotypes fed the low-fat diet (LFD) or HFD was similar (Fig. 1F). Concurrent with the development of HFD-induced obesity, HFD-fed wild-type animals displayed hyperinsulinemia (Fig. 1G). In contrast, HFD-fed ASC−/− and Casp1−/− animals had significantly lower plasma insulin and leptin levels (Fig. 1 G and H). Moreover, plasma concentrations of resistin, which is known to impair glucose tolerance and insulin action (25), were significantly reduced in HFD-fed Nlrp3−/−, ASC−/−, and Casp1−/− mice (Fig. 1I). However, plasma triglycerides and cholesterol levels were not significantly different between all genotypes (Table 1).
Fig. 1.
Absence of the Nlrp3-inflammasome protects against the development of HFD-induced obesity. (A) qPCR analysis of caspase-1 gene expression levels in epididymal WAT of LFD- and HFD-fed wild-type C57/Bl6 animals after 16 wk of diet-intervention. (B) Caspase-1 protein levels in WAT of LFD- vs. HFD-fed wild-type animals. Comparison of bodyweight gain in wild-type, Nlrp3−/−(C), ASC−/− (D), and Casp1−/− (E) animals on LFD or HFD during 16 wk. (F) Daily food intake of wild-type, Nlrp3−/−, ASC−/−, and Casp1−/− animals fed a LFD or HFD. Plasma concentrations of insulin (G), leptin (H), and resistin (I) in LFD- and HFD-fed wild-type, Nlrp3−/−, ASC−/−, and Casp1−/− mice. **P < 0.01; ***P < 0.001; n = 6–8 mice per group. Error bars represent SEM.
Table 1.
Plasma cholesterol and triglyceride concentrations
| Triglycerides (mg/dL) | SEM | Cholesterol (mg/dL) | SEM | |
| Wild-type LFD | 129 | 11 | 163 | 8 |
| Wild-type HFD | 143 | 6 | 185 | 7 |
| Casp1−/− LFD | 146 | 23 | 125 | 5 |
| Casp1−/− HFD | 116 | 14 | 161 | 9 |
| ASC−/− LFD | 131 | 15 | 132 | 10 |
| ASC−/− HFD | 132 | 11 | 174 | 14 |
| NLRP3−/− LFD | 104 | 10 | 178 | 7 |
| NLRP3−/− HFD | 144 | 7 | 157 | 7 |
Total plasma cholesterol and triglyceride concentrations were measured after 16 wk of LFD or HFD feeding.
ASC-Deficient Mice Are Protected from HFD-Induced Insulin Resistance, Liver Steatosis, and Adipocyte Hypertrophy.
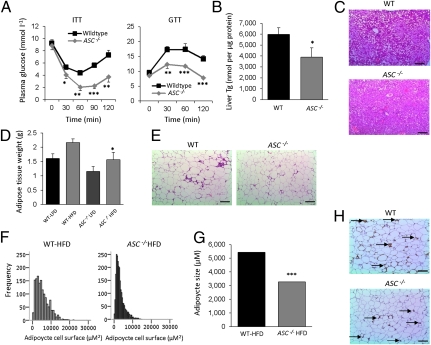
To explore the effects of the key inflammasome adaptor ASC on HFD-induced insulin resistance, we performed insulin and glucose tolerance tests. As shown in Fig. 2A, both insulin sensitivity and glucose tolerance were improved in ASC−/− mice fed the HFD compared with wild-type mice. We next analyzed liver and adipose tissue morphology after 16 wk of HFD intervention in wild-type and ASC−/− mice. The development of liver steatosis was blunted in ASC−/− mice (Fig. 2 B and C). However, such striking effect of reduced liver triglyceride was not observed in Nlrp3−/− mice (Fig. S2A). In line with the lower body weight and plasma leptin levels, epididymal adipose tissue mass was reduced in HFD-fed ASC−/− mice (Fig. 2D). Further analysis of the adipose tissue morphology in HFD-fed ASC−/− mice (Fig. 2E) revealed a significant reduction in adipocyte size (Fig. 2 F and G), suggestive of an improvement in adipose tissue dynamics (26). In contrast, obesity-induced macrophage infiltration into the adipose tissue was not prevented by the absence of ASC as determined by an immunohistochemical localization of macrophages (Fig. 2H). These results were further confirmed by qPCR analysis of the macrophage marker CD68 in total adipose tissue (wild-type LFD: 1 ± 0.29, wild-type HFD: 6.86 ± 1.87; ASC−/− LFD: 1.91 ± 0.31, ASC−/− HFD: 7.17 ± 2.17). Adipose tissue morphology was also examined in Nlrp3−/− mice, which showed a marginal reduction in adipocyte size compared with the wild-type (Fig. S2B).
Fig. 2.
ASC−/− animals are protected against HFD-induced insulin resistance, steatosis, and adipocyte hypertrophy. (A) Insulin tolerance test (ITT) and glucose tolerance tests (GTT) of HFD-fed wild-type and ASC−/− animals. (B) Liver triglyceride (Tg) levels in HFD-fed wild-type and ASC−/− mice. (C) Liver histology as determined by H&E staining. (Scale bars, 100 μm.) (D) Epididymal adipose tissue weight of LFD- and HFD-fed wild-type or ASC−/− mice. (E) Adipose tissue morphology after H&E staining of HFD-fed wild-type and ASC−/− mice. (Scale bars, 100 μm.) (F and G) Quantification of adipocyte size using software analysis. (H) Localization of macrophages in WAT of HFD-fed wild-type and ASC−/− at week 16. Arrow indicates macrophage specific immunoreaction. (Scale bars, 100 μm.)
Hyperinsulinemic Euglycemic Clamp Studies Reveal a Critical Role for Inflammasome in HFD-Induced Insulin Resistance.
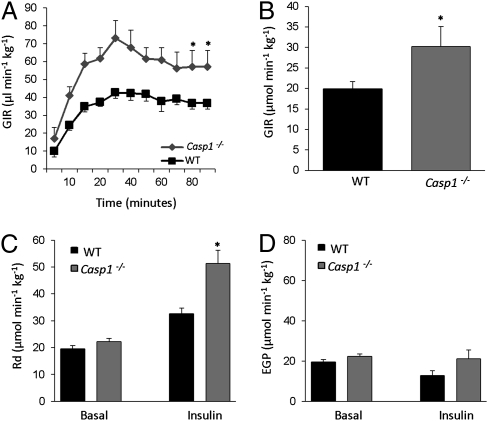
We next investigated the role of inflammasome in HFD-induced insulin resistance. Hyperinsulinemic euglycemic clamp studies revealed that glucose infusion rates were higher in HFD-fed Casp1−/− mice (Fig. 3 A and B), in line with an improvement in insulin sensitivity because of the absence of caspase-1. Blood glucose concentrations during the clamp, as well as basal and hyperinsulinemic plasma, insulin, and free fatty acid (FFA) levels are shown in Fig. S3 and Table S1. The elevated glucose infusion rate during hyperinsulinemia was caused by an increase of peripheral glucose uptake (Fig. 3C), but hepatic glucose production was unchanged (Fig. 3D) compared with HFD-fed wild-type animals. Similarly, inhibition of caspase-1 using pralnacasan in Ob/ob animals improved insulin sensitivity (23), paralleled by an increase in circulating plasma adiponectin levels (Ob/ob + vehicle vs. Ob/ob + pralnacasan: 697.5 ± 40.1 vs. 797.5 ± 24.7, P value< 0.05).
Fig. 3.
Absence of caspase-1 protects against the development of HFD-induced insulin resistance as determined by hyperinsulinemic euglycemic clamp analysis. (A) Glucose infusion rates (GIR) during the euglycemic hyperinsulinemc clamp in HFD-fed wild-type and Casp1−/− animals. (B) Average glucose infusion rate of wild-type and Casp1−/− animals fed a HFD for 16 wk. (C) Rate of disappearance (Rd), a measure of peripheral glucose uptake in wild-type and Casp1−/− animals. (D) Endogenous (hepatic) glucose production (EGP) during the clamp. Mice were maintained on a HFD during 16 wk before the clamp experiment. *P < 0.05; n = 6–8 mice per group. Error bars represent SEM.
Caspase-1 Mediates Macrophage Influx into Adipose Tissue.
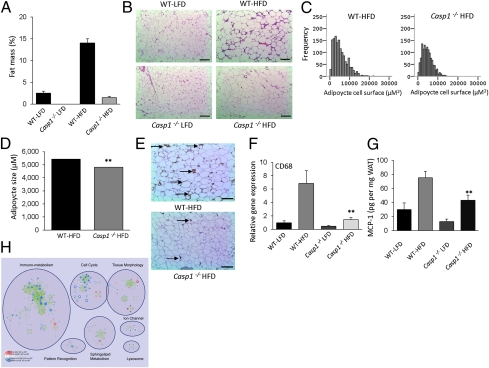
Because caspase-1 is the core molecule of inflammasome complex, we focused on Casp1−/− mice to understand the role of the inflammasome in adipose tissue function after HFD-feeding. The resistance to HFD-induced body weight-gain in Casp1−/− animals (Fig. 1E) was accompanied by a dramatic reduction in WAT mass, as determined by dual-emission X-ray absorptiometry (DEXA)-scan analysis (Fig. 4A). Notably, liver triglyceride storage upon HFD feeding was unchanged in Casp1−/− animals compared with wild-type mice (Fig. S4). We next analyzed morphological changes of the WAT induced by HFD-feeding. As shown in Fig. 4B, histological analysis revealed the presence of smaller adipocytes in HFD-fed Casp1−/− animals (Fig. 4 B–D). Immunohistochemical localization of macrophages in WAT of HFD-fed animals showed a reduction in the number of macrophages in Casp1−/− mice (Fig. 4E). These results were confirmed by qPCR analysis of the macrophage marker CD68 in WAT (Fig. 4F). Additionally, as monocyte chemoattractant protein (MCP)-1 is responsible for macrophage influx (27), we analyzed protein levels of MCP-1 in adipose tissue of HFD-fed animals. As shown in Fig. 4G, MCP-1 protein levels were significantly reduced (P < 0.01) in adipose tissue of Casp1−/− animals compared with wild-type mice. To test the relative contribution of IL-1β and IL-18 to this phenomenon, human adipocytes (SGBS cell line) were differentiated toward adipocytes in the presence of IL-1β or IL-18. As shown in Fig. S5, MCP-1 gene-expression levels were enhanced by treatment of the cells with IL-1β (fold-change vs. control: 3.6, P value < 0.001), whereas IL-18 (fold-change vs. control: 0.57, P value: NS) had no effect. Moreover, blockade of endogenous IL-1 bioactivity by treatment with IL-1 receptor antagonist (IL-1ra) during differentiation of the cells led to a significant reduction in MCP-1 gene-expression levels (fold-change vs. control: 0.25, P value < 0.05), suggesting that the lower levels of MCP-1 in adipose tissue of HFD-fed Casp-1−/− animals (Fig. 4G) can mainly be attributed to the absence of IL-1β. To acquire more information regarding the molecular pathways controlled by caspase-1 in adipose tissue during HFD-feeding, we performed a microarray analysis comparing HFD-fed wild-type and Casp1−/− animals (Fig. 4H and Figs. S6–S8) that revealed a substantial overlap of pathways regulated in both Casp1−/− and ASC−/− animals, including cell cycle and immunometabolism that encompasses functional gene sets involved in both immunity and metabolism. However, several pathways were regulated specifically in one genotype. For example, sphingolipid metabolism that may generate ligands, including ceramide, is specifically regulated in the absence of caspase-1. A complete overview of differentially expressed genes in adipose tissue of HFD-fed animals is available in Dataset S1.
Fig. 4.
HFD-fed Casp1−/− animals are protected against obesity-induced adipocyte hypertrophy and macrophage influx into the adipose. (A) Total percentage of WAT as determined by DEXA-scan analysis of wild-type and Casp1−/− on LFD or HFD for 16 wk. (B) Histology as determined by H&E staining of WAT. (Scale bars, 100 μm.) (C and D) Software analysis of adipocyte size (C) and quantification (D). (E) Macrophage influx into the WAT as determined by immunohistochemistry. Arrow indicates macrophage specific immunoreaction. (Scale bars, 100 μm.) (F) qPCR analysis of the macrophage marker CD68 in white adipose tissue. (G) Concentration of MCP-1 in adipose tissue. (H) Enrichment map for gene expression in WAT of Casp-1−/− (n = 3) (inner node area) and ASC−/− (n = 3) (node borders) compared with WT (n = 4) at 16 wk after HFD intervention. Nodes represent functional gene sets, and edges between nodes their similarity. Color intensity of node area or border is proportional to enrichment significance in Casp-1−/− or ASC−/− mice compared with wild-type; red indicates increased and blue suppressed gene sets in null mice compared with wild-type. Node size represents the gene set size, and edge thickness represent degree of overlap between two connected gene sets. Clusters were manually circled and labeled to highlight the prevalent biological functions among related gene sets. **P < 0.01; n = 6–8 mice per group. Error bars represent SEM.
Defects in Inflammasome Increase Energy Expenditure in HFD-Fed Mice.
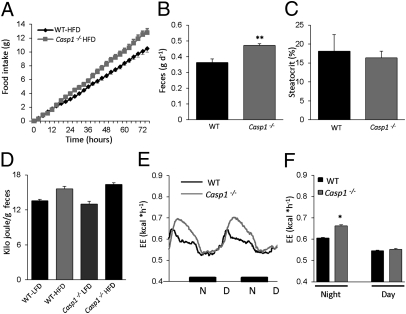
We further analyzed the role of the inflammasome in obesity by examining energy expenditure, fecal output, and caloric content in HFD-fed wild-type and Casp1−/− animals. Detailed analysis of the food intake revealed that, although the absence of caspase-1 protects against the development of obesity, Casp1−/− animals eat more compared with wild-type mice (Fig. 5A). However, as shown in Fig. 5B, fecal output was significantly enhanced in Casp1−/− animals. Analysis of the fat content of the feces, as determined by the percentage of steatocrit, revealed no significant difference between both genotypes (Fig. 5C). Finally, caloric content of the feces did not differ between both genotypes on the HFD (Fig. 5D), suggesting that the net energy intake is similar in HFD-fed wild-type and Casp1−/− mice. However, although total daily caloric intake was highly similar, feeding behavior was strikingly different between wild-type and Casp1−/− mice (Fig. S9A). Whereas wild-type animals displayed a constant intake, the Casp1−/− mice were characterized by the consumption of relatively large amounts of food followed by periods in which the animals did not eat at all. Inasmuch gut hormones are known to control food intake, we measured circulating concentrations of GIP, PYY, and ghrelin before and 2 h after the administration of a lipid bolus. Circulating postprandial levels of the satiety signals GIP and PYY were not different between both phenotypes (Fig. S9B). In contrast, whereas circulating levels of ghrelin were reduced in wild-type animals after receiving the lipid bolus (Fig. S9B), this postprandial reduction was not observed in Casp1−/− mice, suggesting that an orexigenic action of ghrelin may contribute to modulation of the feeding pattern. In contrast, analysis of the energy expenditure in HFD-fed caspase-1–deficient mice unveiled significantly higher energy expenditure (Fig. 5 E and F). These results indicate that although total energy intake does not differ, energy expenditure is enhanced in Casp1−/− animals.
Fig. 5.
Increased energy expenditure without any changes in net energy intake in HFD-fed Casp1−/− mice 16 wk. (A) Cumulative food intake of HFD-fed animals. (B) Fecal output in HFD-fed wild-type and Casp1−/− animals. (C) Fat content of feces from HFD-fed wild-type and Casp1−/− animals as determined by steatocrit measurement. (D) Caloric content of feces from wild-type and Casp1−/− animals determined by bomb calorimetry. (E and F) Energy expenditure of HFD-fed wild-type and Casp1−/− mice. *P < 0.05; **P < 0.01; n = 6–8 mice per group. Error bars represent SEM.
Discussion
The prevalence of obesity has reached epidemic proportions worldwide. During the development of obesity, the morphology and functional properties of adipose tissue change dramatically. In addition to adipocyte hypertrophy, adipose tissue turns into an inflamed tissue characterized by macrophage infiltration and altered secretion of adipokines (14, 17). Whereas the secretion of proinflammatory cytokines is enhanced, the production of insulin-sensitizing adipokines, such as adiponectin, is reduced. To date, several proinflammatory cytokines have been linked to the development of insulin resistance, including IL-1β and IL-18 (14). These cytokines are both produced as inactive precursors in the cytosol and are released following their maturation by the cysteine protease caspase-1. The cytosolic zymogen form of caspase-1 consists of an N-terminal prodomain that is processed and activated within the inflammasomes, which are assembled on NLR protein scaffolds. Because of the metabolic effects described for IL-1β and IL-18, and because of the crucial role of the inflammasome in the activation of these cytokines, we set out to test the role of the inflammasome in obesity. Our studies demonstrate that the mice deficient in inflammasome components are protected from HFD-associated body-weight gain, adipocyte hypertrophy, hyperinsulinemia, and hyperresistinemia. Importantly, HFD-induced production of the protein resistin was significantly reduced in inflammasome-deficient animals, implying its critical role in driving the inflammatory potential of adipocytes. Furthermore, chronic overfeeding resulted in pheno- and genotypical differences in WAT of inflammasome-deficient animals, illustrated by a divergent macrophage influx and gene-expression profile of the adipose tissue.
Deficiency in caspase-1 or ASC results in the same phenotype, which strongly suggests a role for the inflammasome in obesity. However, there are slight differences between ASC- and caspase-1–deficient mice. Although changes in adipogenic gene expression in caspase-1 and ASC-deficient animals show a considerable overlap, subtle differences exist. Whereas ASC may mediate effects on adipocyte hypertrophy, as observed in Casp-1−/− animals, macrophage influx in the ASC−/− animals may partly be regulated independently of caspase-1. In this regard, recent studies reported that ASC directly mediates gene expression (28) and has a critical role in the development of experimental autoimmune encephalomyelitis and arthritis independently of the inflammasome (29, 30). However, in obesity, several of the parameters tested suggest that ASC and caspase-1 work in the same pathway. Indeed, caspase-1 appears to have a central role in adipose tissue functioning during chronic overfeeding by regulating macrophage influx and systemic insulin sensitivity. Although part of the protective effects in HFD-fed Casp1−/− animals are explained by the absence or lower levels of IL-1β, caspase-1 may also control alternative pathways. The wide range of genes differentially regulated between HFD-fed wild-type and Casp1−/− animals support the hypothesis of novel functions of caspase-1 in adipose tissue. In addition, the increase in energy expenditure observed in Casp1−/− animals identifies caspase-1 as a metabolic regulator. Finally, caspase-1 appears to drive the production of different chemokines in adipose tissue that regulated inflammatory cell influx. We postulate that caspase-1 downstream signaling contributes to the enhanced energy expenditure observed in HFD-fed Casp1−/− animals. Indeed, using the diagonal gel proteomic approach, several proteins that are involved in energy metabolism were identified as caspase-1 substrates (31). Additional substrates of caspase-1 in adipose tissue may be identified by in silico analysis and may help to explain its contribution to the development of adipose tissue dysfunction during HFD-induced obesity.
One of the future challenges is to delineate the exact role of adipose-tissue–specific caspase-1 to the development of HFD-induced obesity and insulin resistance. Moreover, future studies in animals that have adipocyte-specific overexpression of caspase-1 will be helpful to identify the differential contribution of macrophage- or adipocyte-derived caspase-1 to adipose-tissue dysfunction and obesity. It will also be important to identify metabolic signals activating the inflammasome during the development of obesity. Interestingly, ceramide, a lipid molecule, has been described as a potent danger signal for inflammasome activation (24). However, these results were generated in vitro and do not rule out alternative pathways that mediate activation of caspase-1. Indeed, high levels of glucose have also been shown to activate caspase-1 within adipose tissue (32). Possibly, multiple danger signals exist that may use specific inflammasome pathways all leading to caspase-1 activation.
Although we have clearly established the importance of the inflammasome during the development of obesity in animal models, additional studies focused on unraveling the importance of the inflammasome activation in human adipose tissue are needed. In conclusion, our study shows that the inflammasome, in addition to its role in the innate immune response, contributes to the development of obesity-induced insulin resistance. Therefore, the inflammasome represent a useful therapeutic target in the treatment of obesity and insulin resistance.
Materials and Methods
Mice.
Nlrp3−/−, ASC−/−, and Casp1−/− mice backcrossed to C57BL/6 background for at least 10 generations have been described previously (33, 34). Mice were housed in a pathogen-free facility and the animal studies were conducted under protocols approved by St. Jude Children's Research Hospital Committee on Use and Care of Animals. All mice were 8- to 10-wk-old males at the start of the diet intervention, and maintained in a SPF facility. All experiments were conducted under protocols approved by the St. Jude Children's Research Hospital Committee on Use and Care of Animals and the animal experimentation committee of Leiden University Medical Center.
Diet Intervention.
Male mice received a LFD or HFD for 16 wk, providing 10% or 45% energy percent in the form of fat (D12450B or D12451; Research Diets). At the end of the feeding experiment, blood was collected in EDTA-coated tubes and centrifuged to collect plasma. Liver and epididymal WAT were dissected, weighed, and immediately frozen in liquid nitrogen. All studies described below were performed on animals fed the HFD.
Hyperinsulinemic Euglycemic Clamp Study.
The hyperinsulinemic euglycemic clamp study was performed as previously described (21, 35, 36). Plasma glucose, insulin, and FFA levels were determined using commercially available kits (INstruchemie, Crystal Chem Inc., and Wako Pure Chemical Industries).
Calculations.
Turnover rates of glucose (μmol⋅min⋅kg) were calculated during the basal period and in steady-state clamp conditions as the rate of tracer infusion (dpm/min) divided by the plasma-specific activity of 3H-glucose (dpm/μmol). All metabolic parameters were expressed per kilogram of bodyweight. The hepatic glucose production (EGP) is calculated from the rate of disappearance (Rd) and glucose infusion rate (GIR) by the following equation: Rd = EGP + GIR. The Rd is measured from Steele's equation in steady state using the tracer infusion rate (Vin) and plasma-specific activity (SA) of 3H-glucose (dpm/μmol) by the following formula: Rd = Vin/SA.
Indirect Calorimetry.
Groups of eight mice per genotype were subjected to individual indirect calorimetry and metabolic cage measurements for a period of 84 h (Comprehensive Laboratory Animal Monitoring System, Columbus Instruments). A period of 24 h before the start of the experiment allowed the acclimatization of the animals to the cages and single housing. Experimental analysis started at 0700 hours and continued for 60 h.
Statistical analysis indirect calorimetry.
Averages were tested for significant differences between groups using Student t test after the D'Agostino and Pearson test for normality. The statistical significance threshold was set at 0.05.
Histology/Immunohistochemistry.
Morphometry of individual fat cells was assessed using digital image analysis. Microscopic images were digitized in 24-bit RGB (specimen level pixel size 1.28 × 1.28 μm2). Recognition of fat cells was initially performed by applying a region growing algorithm on manually indicated seed points, and minimum Feret diameter was calculated. For detection of macrophages/monocytes, an F4/80+ antibody (Serotec) was used. Visualization of the complex was done using 3,3′-diaminobenzidene for 5 min. Negative controls were used by omitting the primary antibody. H&E staining of sections was done using standard protocols.
Liver Triglycerides.
Liver triglycerides were determined in 10% liver homogenates prepared in buffer containing 250 mM sucrose, 1 mM EDTA, and 10 mM Tris-HCl at pH 7.5.
Cell Culture.
Human SGBS cells were differentiated toward adipocytes using a standard protocol. In short, differentiation of cells was induced by treatment with transferrin, insulin, cortisol, T3, dexamethasone, and IBMX. IL-1β (5 ng/mL), IL-18 (25 ng/mL), or IL-1ra (5 μg/mL) were added during differentiation. After 10 d of differentiation, RNA was isolated, cDNA was prepared, and qPCR analysis was performed as described below.
RNA Isolation and qPCR Analysis.
RNA from animal tissues or cultured cells was isolated using TRIzol Reagent (Invitrogen) following the manufacturer's instructions. RNA was reverse-transcribed (iScript cDNA Synthesis Kit; Bio-Rad Laboratories) and real-time PCR was done with a Power Sybr Green PCR master mix (Applied Biosystems) using a 7300 Real-Time PCR System (Applied Biosystems). Melt curve analysis was included to assure a single PCR product was formed. Values were corrected using the housekeeping gene 36B4 or β2-microglobulin.
Plasma Adipokines/Adipose Tissue-Chemokines.
Plasma concentrations of insulin, leptin and resistin are determined using Luminex techniques following the manufacturer's instructions. Concentration of different chemokines in adipose tissue was determined by ELISA following the manufacturer's instructions (R&D Systems) or radio immune assays. Values were expressed as total amount per milligram WAT.
Microarray Gene-Expression Analysis.
RNA quality was determined by analysis on the Agilent 2100 Bio-analyzer, and all samples had a RIN > 8. Total RNA (100 ng) was labeled, and processed automatically on an HT MG-430 PM array plate using the Affymetrix GeneTitan system in the St. Jude microarray core, according to the manufacturer's instructions. The probes on the HT MG-430 PM array were redefined according to Dai et al. (37). Normalized expression estimates were generated from the raw intensity values using the RMA algorithm in the Bioconductor library AffyPLM (38). Differentially expressed probesets were identified using linear models, applying moderated t-statistics that implement empirical Bayes regularization of SEs (39). To adjust for both the degree of independence of variances relative to the degree of identity and the relationship between variance and signal intensity, the moderated t-statistic was extended by a Bayesian hierarchical model to define a intensity-based moderated t-statistic (IBMT) (40). P values were corrected for multiple testing using a false-discovery rate method (41). Probesets with a false-discovery rate < 10% (q value < 0.1) were considered to be significantly regulated. Changes in gene expression were related to functional changes using gene set enrichment analysis (GSEA) (42). Gene sets were derived from Gene Ontology, KEGG, National Cancer Institute, PFAM, and Biocarta pathway databases. Enrichment Map was used for interpretation of the GSEA results (43). Only gene sets consisting of more than 10 and fewer than 500 genes were taken into account. The enrichment map was generated with gene sets that passed the significance threshold of P value < 0.005 and similarity cutoff value of 0.6, resulting in a network of 226 nodes (gene sets) and 670 edges (interactions). All singletons were removed to create the final gene set interaction network.
Statistical Analysis.
Statistical significant differences were calculated using a Student's t-test. The cut-off for statistical significance was set at a P value of 0.05 or below.
Supplementary Material
Acknowledgments
We thank Anthony Coyle, John Bertin, Ethan Grant, Gabriel Nunez, Richard Flavell, and Shizuo Akira for generous supply of mutant mice, and Tim Koenen for technical assistance. This work was supported by National Institutes of Health Grants AR056296 and AI088177; the American Lebanese and Syrian Associated Charities (to T.-D.K.); the Dutch Diabetes Research Foundation (R.S., M.G.N., C.J.T.); the European Association for the Study of Diabetes; a Vici grant of the Netherlands Organization for Scientific Research (to M.G.N.); the Netherlands Organization for Scientific Research Grant 917.76.301 (to J.A.v.D.); and the seventh framework program of the European Union-funded “LipidomicNet” 202272 (to I.V.). P.C.N.R. is an Established Investigator of the Netherlands Heart Foundation (Grant 2009T038).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE25205).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100255108/-/DCSupplemental.
References
- 1.Ye Z, Ting JP. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr Opin Immunol. 2008;20(1):3–9. doi: 10.1016/j.coi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 3.Sutterwala FS, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 5.Duewell P, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aganna E, et al. Allelic variants in genes associated with hereditary periodic fever syndromes as susceptibility factors for reactive systemic AA amyloidosis. Genes Immun. 2004;5:289–293. doi: 10.1038/sj.gene.6364070. [DOI] [PubMed] [Google Scholar]
- 7.Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 8.Ghayur T, et al. Caspase-1 processes IFN-gamma-inducing factor and regulates LPS-induced IFN-gamma production. Nature. 1997;386:619–623. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 9.Kuida K, et al. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 10.Li P, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 11.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- 12.Horwood NJ, et al. Interleukin 18 inhibits osteoclast formation via T cell production of granulocyte macrophage colony-stimulating factor. J Clin Invest. 1998;101:595–603. doi: 10.1172/JCI1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olee T, Hashimoto S, Quach J, Lotz M. IL-18 is produced by articular chondrocytes and induces proinflammatory and catabolic responses. J Immunol. 1999;162:1096–1100. [PubMed] [Google Scholar]
- 14.Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol. 2008;8:923–934. doi: 10.1038/nri2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odegaard JI, Chawla A. Mechanisms of macrophage activation in obesity-induced insulin resistance. Nat Clin Pract Endocrinol Metab. 2008;4:619–626. doi: 10.1038/ncpendmet0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisberg SP, et al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 19.Shoelson SE, Herrero L, Naaz A. Obesity, inflammation, and insulin resistance. Gastroenterology. 2007;132:2169–2180. doi: 10.1053/j.gastro.2007.03.059. [DOI] [PubMed] [Google Scholar]
- 20.Jager J, Grémeaux T, Cormont M, Le Marchand-Brustel Y, Tanti JF. Interleukin-1beta-induced insulin resistance in adipocytes through down-regulation of insulin receptor substrate-1 expression. Endocrinology. 2007;148:241–251. doi: 10.1210/en.2006-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Netea MG, et al. Deficiency of interleukin-18 in mice leads to hyperphagia, obesity and insulin resistance. Nat Med. 2006;12:650–656. doi: 10.1038/nm1415. [DOI] [PubMed] [Google Scholar]
- 22.Zorrilla EP, et al. Interleukin-18 controls energy homeostasis by suppressing appetite and feed efficiency. Proc Natl Acad Sci USA. 2007;104:11097–11102. doi: 10.1073/pnas.0611523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stienstra R, et al. The inflammasome-mediated caspase-1 activation controls adipocyte differentiation and insulin sensitivity. Cell Metab. 2010;12:593–605. doi: 10.1016/j.cmet.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandanmagsar B, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. 2011;17(2):179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steppan CM, et al. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 26.Goossens GH. The role of adipose tissue dysfunction in the pathogenesis of obesity-related insulin resistance. Physiol Behav. 2008;94:206–218. doi: 10.1016/j.physbeh.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Kanda H, et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest. 2006;116:1494–1505. doi: 10.1172/JCI26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasegawa M, et al. Mechanism and repertoire of ASC-mediated gene expression. J Immunol. 2009;182:7655–7662. doi: 10.4049/jimmunol.0800448. [DOI] [PubMed] [Google Scholar]
- 29.Shaw PJ, et al. Cutting edge: Critical role for PYCARD/ASC in the development of experimental autoimmune encephalomyelitis. J Immunol. 2010;184:4610–4614. doi: 10.4049/jimmunol.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ippagunta SK, et al. Inflammasome-independent role of apoptosis-associated speck-like protein containing a CARD (ASC) in T cell priming is critical for collagen-induced arthritis. J Biol Chem. 2010;285:12454–12462. doi: 10.1074/jbc.M109.093252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao W, Yeretssian G, Doiron K, Hussain SN, Saleh M. The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J Biol Chem. 2007;282:36321–36329. doi: 10.1074/jbc.M708182200. [DOI] [PubMed] [Google Scholar]
- 32.Koenen TB, et al. Hyperglycemia activates caspase-1 and TXNIP-mediated IL-1beta transcription in human adipose tissue. Diabetes. 2011;60:517–524. doi: 10.2337/db10-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaki MH, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas PG, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voshol PJ, et al. In muscle-specific lipoprotein lipase-overexpressing mice, muscle triglyceride content is increased without inhibition of insulin-stimulated whole-body and muscle-specific glucose uptake. Diabetes. 2001;50:2585–2590. doi: 10.2337/diabetes.50.11.2585. [DOI] [PubMed] [Google Scholar]
- 36.Voshol PJ, et al. Increased hepatic insulin sensitivity together with decreased hepatic triglyceride stores in hormone-sensitive lipase-deficient mice. Endocrinology. 2003;144:3456–3462. doi: 10.1210/en.2002-0036. [DOI] [PubMed] [Google Scholar]
- 37.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolstad BM. Berkeley: University of California; 2004. Low level analysis of high-density oligonucleotide array data: Background, normalization and summarization. PhD dissertation. [Google Scholar]
- 39.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 40.Sartor MA, et al. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 2006;7:538. doi: 10.1186/1471-2105-7-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.