Abstract
Cells of the Corynebacterium-Nocardia-Mycobacterium group of bacteria are surrounded by an outer membrane (OM) containing mycolic acids that are covalently linked to the underlying arabinogalactan-peptidoglycan complex. This OM presumably acts as a permeability barrier that imparts high levels of intrinsic drug resistance to some members of this group, such as Mycobacterium tuberculosis, and its component lipids have been studied intensively in a qualitative manner over the years. However, the quantitative lipid composition of this membrane has remained obscure, mainly because of difficulties in isolating it without contamination from the inner cytoplasmic membrane. Here we use the extraction, with reverse surfactant micelles, of intact cells of Corynebacterium glutamicum and show that this method extracts the free OM lipids quantitatively with no contamination from lipids of the cytoplasmic membrane, such as phosphatidylglycerol. Although only small amounts of corynomycolate were esterified to arabinogalactan, a large amount of cardiolipin was present in a nonextractable form, tightly associated, possibly covalently, with the peptidoglycan-arabinogalactan complex. Furthermore, we show that the OM contains just enough lipid hydrocarbons to produce a bilayer covering the cell surface, with its inner leaflet composed mainly of the aforementioned nonextractable cardiolipin and its outer leaflet composed of trehalose dimycolates, phosphatidylinositol mannosides, and highly apolar lipids, similar to the Minnikin model of 1982. The reverse micelle extraction method is also useful for extracting proteins associated with the OM, such as porins.
Bacteria of the Corynebacterium-Mycobacterium-Nocardia (CMN) group produce a complex cell envelope containing various lipid species, as well as mycolic acid residues linked covalently to arabinogalactan, which in turn is linked to peptidoglycan. Minnikin originally proposed that the outer part of this envelope, which could be called the outer membrane (OM), consists of a bilayer structure (1). Although the Minnikin model was not universally accepted (see, e.g., refs. 2 and 3), experimental proof of the model was provided in 1993 by X-ray diffraction in our laboratory (4). Because the CMN OM acts as an effective permeability barrier, just like the OM of Gram-negative bacteria (5, 6), it is important to obtain a complete and quantitative accounting of lipid composition in this OM. However, such efforts have not been successful so far, given the difficulty in obtaining an OM preparation that is uncontaminated by components of the cytoplasmic membrane, or inner membrane (IM) (3, 7).
In this study, we took a different approach to achieving this goal. Reverse micellar solutions (RMSs) of some detergents in apolar solvents, such as heptane, have been used for extraction of bacterial enzymes into the intramicellar (aqueous) lumen (8). A remarkable feature of this method is that only periplasmic enzymes are extracted from Gram-negative bacteria, leaving behind cytosolic enzymes, presumably because the reverse micelles cannot traverse the hydrophilic peptidoglycan layer (9, 10). We thought that we might be able to take advantage of this limited access for reverse micelles in cells of the CMN group. We also thought that RMS might extract not only proteins into the micelle lumen, but also lipids into the micellar detergent layer, thereby leading to the specific, complete, and contamination-free extraction of cell wall lipids. We found that this goal can be achieved using the CNM group organism Corynebacterium glutamicum ATCC 13032.
Results
Optimization of Extraction Protocols.
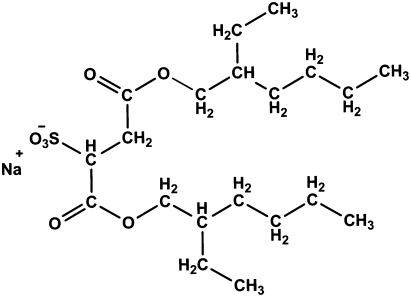
To facilitate the quantitative analysis of all detectable lipid species, we grew the cells in a minimal medium with [1,2-14C]-labeled acetate as the sole carbon source, and quantified various lipid classes obtained by TLC separation with radioactivity detected by phosphorimaging. The extraction processes were optimized for maximum recovery of lipids. RMS extraction was done using 10 mM sulfosuccinic acid 1,4-bis(2-ethylhexyl) ester sodium salt (AOT) (Fig. 1) in heptane. Because AOT contains two bulky, branched hydrocarbon chains connected to one head group, it readily forms reverse micelles. When the solution was mixed with 1% (wt/vol) cells, subsequent centrifugation yielded a single-phase extract. Although the first treatment usually removed 95% of extractable lipids, the extraction was repeated four times (Fig. S1). With chloroform-methanol-water (CMW; 2:1:0.1, vol/vol/v) extraction, again the first two extractions removed >90% of extractable lipids (Fig. S2).
Fig. 1.
Dioctylsulfosuccinate sodium (AOT).
Qualitative and Semiquantitative Analysis of Lipids in Different Extracts.
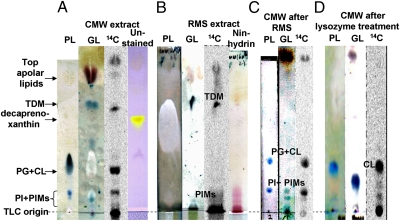
Extracted lipids were separated by TLC (Fig. 2). Each band was identified mainly by specific staining and the use of various solvents and 2D TLC. Clearly, RMS extracts a defined group of lipids both specifically and completely, an important finding because RMS has not been used earlier for lipid extraction to our knowledge. For example, trehalose dimycolate (TDM), a known component of CMN OM (1, 7), was extracted efficiently by RMS (Fig. 2B), and was identified as such because it stained as a glycolipid, yielded corynomycolate but no other fatty acids on hydrolysis (Figs. S3 and S4), and produced the major ion species by electrospray ionization MS (ESI-MS) with a mass of 1,335 (Fig. S5), expected for TDM containing one C33 corynomycolate and one C34 corynomycolate, which are common in Corynebacterium (11, 12). Most importantly, reextraction with CMW of the residue after RMS extraction produced not even a trace of TDM (Fig. 2C), indicating the complete RMS extraction of OM lipids. On the other hand, the RMS extract contained none of the major glycerophospholipids, such as phosphatidylglycerol or phosphatidylinositol (PI), the predominant components of the IM (7). In contrast, these compounds were abundantly present in the CMW extract of the RMS-insoluble fraction (Fig. 2C). Traces of free fatty acids (FFAs) were also found in some extracts, as discussed below. These results suggest that RMS extracts the OM lipids both specifically and completely.
Fig. 2.
TLC profiles of lipids recovered by CMW extraction of whole cells (A), RMS extraction of whole cells (B), CMW extraction of residues after RMS extraction (C), and reextraction with CMW after lysozyme treatment of CMW-extracted cells (D). Thin-layer plates were developed with CMW 30:8:1, followed by visualization of different classes of lipids. Thus, phospholipids (PLs) were detected by spraying with phosphospray and amino-group containing lipids with ninhydrin reagent. Anthrone spray, which produces a characteristic blue color, was used to detect glycolipids (GLs; brown-colored spots are not GLs). Lipids were quantified by radioactivity through phosphorimaging. PIMs, phosphatidylinositol mannosides, PG, phosphatidylglycerol, CL, cardiolipin.
This idea is also supported by the observation that the CMW extracts of RMS-pretreated cells were yellow, whereas the RMS extracts were almost colorless. The absorption spectra of the CMW extract showed maxima at ∼415, 445, and 475 nm, suggesting that the pigment corresponds to a C50 carotenoid decaprenoxanthin reportedly present in C. glutamicum with absorption maxima at 418, 440, and 470 nm (13). This pigment (seen in the unstained lane of Fig. 2A) that comes from IM (14) was found in the CMW extract of whole and RMS-treated cells, but not in the RMS extracts, indicating that the RMS extraction process does not disturb the IM.
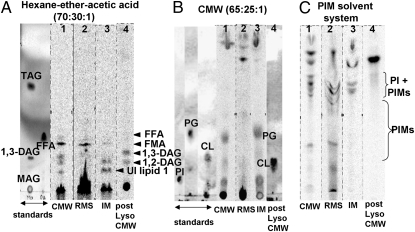
We attempted to identify and quantify the components of various extracts (Table 1). TDM in the RMS extract, mentioned above, comprises ∼0.5% of dry cell mass (dcm). The topmost band of apolar lipids present in all of the extracts showed no positive color reactions with any of the spray reagents. In CMW extracts of RMS-treated cells, this top band contained diacylglycerols and an unidentified apolar component (designated unknown lipid 1) (Fig. 3A and Figs. S6 B–D and S7C). In contrast, RMS extracts contained another unidentified apolar lipid (designated unknown lipid 2) that migrated along with diacylglycerols and was resolved only by 2D TLC (Fig. S7), as well as FFAs and free corynomycolic acids (Fig. 3A and Fig. S7 A and B).
Table 1.
Quantitation of lipids from C. glutamicum (% dry weight)
| CMW extract of RMS-treated cells (all noncovalently bound lipids of the IM) | RMS extract of intact cells (all noncovalently bound lipids of the OM) | CMW extract of lysozyme-treated cells (peptidoglycan-embedded lipids) | |
| PI + PIMs | ∼0.5 | ∼0.18* | 0.2 ± 0.03† |
| Cardiolipin | 0.22 ± 0.01 | — | 2.51 ± 0.1 |
| Phosphatidylglycerol | 0.97 ± 0.2 | — | |
| TDM | — | 0.5 ± 0.1 | — |
| Unknown lipid 1 | ~0.14 | — | — |
| Diacylglycerols | ~0.13 | — | ~0.2 |
| Unknown lipid 2 | — | ~0.15 | — |
| Free mycolic acids with traces of FFAs | — | ~0.46 | — |
| Mycolic acids cleaved from proteins | — | 0.35 ± 0.01 | — |
| Fatty acids cleaved from proteins | — | ~0.01 | — |
Values are averages from at least six experiments.
*This is composed nearly entirely of PIM. The amount is only approximate.
†This is entirely PIM.
Fig. 3.
Apolar and polar lipids resolved by TLC using different solvent systems. (A) Apolar lipids resolved using hexane-diethyl ether-acetic acid (70:30:1). (B) Phospholipids resolved using CMW (65:25:1). (C) PI and PIMs resolved using chloroform–methanol–13 M ammonia–1 M ammonium acetate–water (180:140:9:9:23, vol/vol) (16). Lipids in the extracts were detected with a phosphorimager. Lane 1, CMW extract of whole cells; lane 2, RMS extract of whole cells; lane 3, CMW extract of RMS-treated cells (IM); lane 4, CMW reextract of lysozyme-treated, CMW-extracted cells (post-lyso CMW). Standards: MAG, monoacylglycerol (1-olein); DAG, diacylglycerol; TAG, triacylglycerol (triolein); FFA, oleic acid; PG, phosphatidylglycerol, CL, cardiolipin. UI lipid, unidentified lipid.
A large amount of radioactivity remained at the origin of the RMS and CMW extracts (Fig. 2). Much of this material appeared to contain protein, as suggested by the strong ninhydrin staining (Fig. 2). When the bottom band was subjected to alkaline hydrolysis, TLC after methylation revealed mostly corynomycolyl esters (Figs. S3 and S4). The amount corresponded to ∼0.35% of dcm, consistent with a recent report of corynebacterial porins O-acylated with corynomycolic acids (15).
Large amounts of phospholipids were found in the CMW extract of the cells previously extracted with RMS (Fig. 2). These phospholipids were resolved into phosphatidylglycerol (0.97% of dcm), cardiolipin (0.22% of dcm), and bands containing PI and PI mannosides (PIMs) (Fig. 3 B and C and Table 1). With the specific solvent system used for PIMs (16), two well-resolved bands were found: an upper PI-containing band (which may have overlapping PIMs) and a lower PIM band (Fig. 3C, lane 3). Although the RMS extract did not show the presence of most major phospholipids, it did appear to contain lipids with the retention factor (Rf) values of PIMs (Fig. 3C). It was difficult to get a clear separation between the various PI-containing lipids, a problem compounded in the RMS extracts because of interference by AOT. However, the total amount of extractable PI and PIMs in the entire cell envelope was 0.68% of dcm, and their C12-C20 fatty acid content was approximately one-third of that in phosphatidylglycerol (Fig. S3). This indicates that both the OM and the IM contain PIMs.
Origin of Free Fatty Acids in Stored Cells.
All of the experiments reported in this paper were performed with freshly harvested cells, which produced only trace amounts of FFAs. However, extracts from cells stored frozen for more than 1 mo had large amounts of FFAs (up to 35% of total envelope lipids), although the pattern of other lipids, including phospholipids, was similar to that in fresh cells (Fig. S7 D and E). All of these FFAs were recovered in the RMS extracts and were completely absent in the IM extract, suggesting their location in the OM. Because FFAs are not sufficiently lipophilic to remain stably in the membranes, we suspected that they are degradation products of other lipids. Their likely origin is described below.
Discovery of Cardiolipin Tightly Associated with the Cell Wall.
Expecting to find a large fraction of corynomycolates covalently linked to the cell wall arabinogalactan, we performed alkaline hydrolysis of CMW-extracted (i.e., delipidated) residues of fresh cells. This released the expected corynomycolic acid along with, surprisingly, much larger amounts (5–10 times on a molar scale) of C12-C20 fatty acids (Fig. S8).
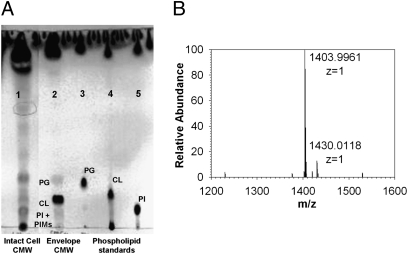
Because the C12-C20 fatty acids could have been released by the hydrolysis of intracellular reserve material, we attempted to rule out this possibility by analyzing the isolated cell envelope fraction (containing IM, OM, and peptidoglycan-arabinogalactan). Cell envelopes were obtained after breaking the cells in a French press followed by ultracentrifugation (Experimental Procedures). The envelopes were extracted with CMW to remove noncovalently linked lipids, and the residual material was then alkali-hydrolyzed. Surprisingly, no significant amounts of C12-C20 fatty acids were found in the hydrolyzed cell envelope (Fig. S9, lane 1). However, the CMW extract of the cell envelopes was found to contain significant amounts of C12-C20 fatty acids (Fig. S9, lane 2), mainly in the form of a phospholipid with an Rf value of cardiolipin (Fig. 4A, lane 2). Given that cardiolipin is only a minor lipid in the IM (Table 1), these results suggest that C. glutamicum contains nonextractable cardiolipin that is embedded in the envelope but becomes extractable during isolation of the cell envelope.
Fig. 4.
Cardiolipin becomes extractable in the isolated cell envelope or after lysozyme treatment of cells. (A) Lipid composition of the CMW extract of intact cells (lane 1) and the CMW extract of the isolated cell envelope (lane 2) as detected by TLC with CMW (65:25:1 vol/vol). Lanes 3–5 contain phospholipid standards. PG, phosphatidylglycerol; CL, cardiolipin. Phosphomolybdotungstate was used for detection. (B) ESI-MS profile of the major lipid band, migrating like CL, isolated from the TLC plate of CMW reextract of lysozyme-treated delipidated cells.
Cardiolipin is believed to be tightly associated with the peptidoglycan-arabinogalactan layer, because it also becomes extractable after lysozyme treatment of CMW-extracted cells. The lysozyme treatment did not release any fatty acid–containing material into aqueous phase; however, reextraction of the lysozyme-treated cells with CMW again produced large amounts of lipids with an Rf value of cardiolipin (Fig. 2D). This phospholipid was purified by preparative TLC; its molecular weight, determined by ESI-MS, was 1,404 (Fig. 4B), corresponding to that of a cardiolipin with two C16:0 acids and two C18:1 acids. There was also a 1,430-Da species, corresponding to a molecule with one C16:0 acid and three C18:1 acids. This agrees with our GC-MS results (presented below), showing that C16:0 and C18:1 are the major fatty acids in the CMW extracts of lysozyme-treated delipidated cells (Fig. S10).
In addition to cardiolipin (2.5% of dcm), smaller amounts of PIMs (0.2% of dcm) and diacylglycerol (0.2% of dcm) were also found in the CMW extract of these lysozyme-treated delipidated cells (Table 1 and Fig. 3) (Note the anthrone-reactive spot in Fig. 2D, which is probably PIM.) Thus, the cell wall of C. glutamicum contains large amount of nonextractable cardiolipin, which is released only when the peptidoglycan layer is disrupted either by enzymatic hydrolysis or mechanically with a French press. During long-term storage, it apparently breaks down, probably by esterases, resulting in the release of extractable FFAs observed in frozen cells. Most published studies show that phosphatidylglycerol is the major component of C. glutamicum cell envelope (17, 18). With many TLC systems, cardiolipin and phosphatidylglycerl have overlapping Rf values. This problem, combined with the fact that cardiolipin is not extractable from intact cells, may be why it has not been detected in C. glutamicum until now.
Quantitative Analysis of Fatty Acid Content in Discrete Layers of the Cell Envelope.
Although the amounts of various lipids were determined on a weight basis (Table 1), we needed to quantitate fatty acids in the OM, IM, and the peptidoglycan-associated fraction on a molar basis, to estimate their contribution to the coverage of cell surface. The results are shown in Table 2. The quantification was based on GC-MS with an internal standard, as described in Experimental Procedures. The GC system that we used did not resolve corynomycolate ester, however, and thus here quantitation was based on the ratio between C12-C20 fatty acid esters and the corynomycolate ester, determined by TLC (Experimental Procedures).
Table 2.
Fatty acid residue contents of C. glutamicum cell envelope (mmol/100 g dcm)
| C12-20 FAs (based on GC-MS) | Corynomycolates (estimated based on radioactivity and GC-MS) | |
| Covalently and noncovalently linked, in the entire cell envelope (whole-cell alkaline hydrolysis) | 9.69 ± 0.2 | 1.79 ± 0.3* |
| OM (RMS extract) | 0.39 ± 0.2 | 1.69 ± 0.4† |
| Peptidoglycan embedded (released in CMW extract after lysozyme treatment) | 4.8 ± 0.7§ | — |
| Covalently bound to the cell wall (released after alkaline hydrolysis of lysozyme-treated delipidated cells) | Negligible | ≥0.22‡ |
| IM (CMW extract − RMS extract) | 4.3 ± 0.6 | 0 |
Values are averages from at least six experiments.
*Calculated based on a C12-20 FA-to-corynomycolate ratio of 5.4 ± 0.9.
†Calculated based on a C12-20 FA-to-corynomycolate ratio of 0.23 ± 0.1 for this fraction.
‡Actual values are expected to be slightly higher because of some loss of cells in preceding CMW extractions and lysozyme treatment steps.
Direct quantitation of all C12-C20 fatty acids (after alkaline hydrolysis) from whole cells resulted in 9.7 mmol/100 g dcm, whereas the sum of IM, OM, and peptidoglycan-associated lipids was 9.5 mmol (Table 2), confirming the validity of this approach to quantitation.
Our analysis showed that C12-C20 fatty acids in the IM (originating from phosphatidylglycerol, PI, PIMs, and diacylglycerols) and the cell wall–embedded lipids (arising mainly from cardiolipin and a few PIMs and diacylglycerols), were mostly composed of nearly equal portions of C16:0 and C18:1 species (Fig. S10). In contrast, the RMS extract, presumably from OM, contained significant amounts of other species, such as C18:0, C14:0, and C12:0 (Fig. S10). The origins of these minor species appear to be mainly PIMs, although this requires further investigation.
Extraction of Cell Envelope Proteins.
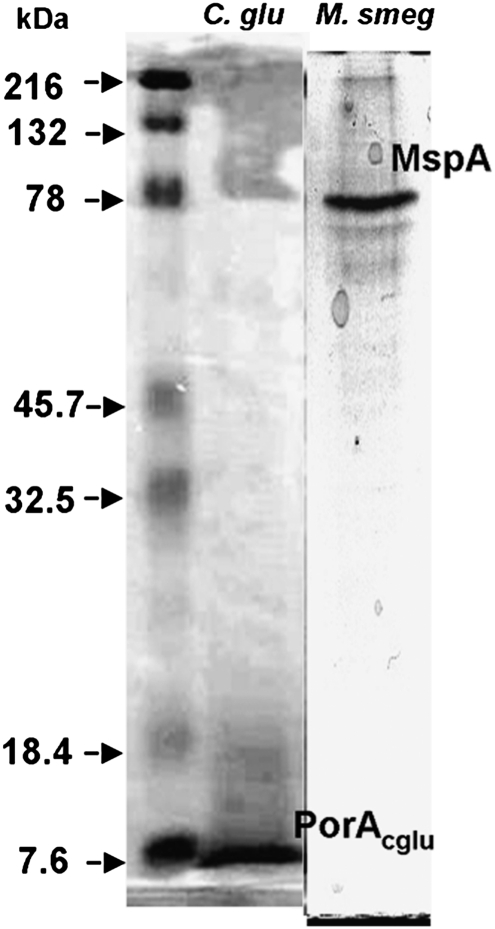
The RMS extract, as well as RMS-extracted cells, were washed with a small amount of water. The pooled aqueous washes and the washed RMS extract were analyzed by SDS/PAGE. As shown in Fig. 5, the major protein found had a molecular weight of ∼5,000, as expected for porin PorA of this organism (19). This sample also contained a diffuse band above PorA, which may correspond to PorH (20, 21). As a control, a similar aqueous extract of RMS-extracted Mycobacterium smegmatis had a major band of ∼80,000 Da (Fig. 5), again corresponding to the stable oligomer of the major MspA porin of this organism (22). The RMS extracts of these two organisms also had major protein bands in the corresponding positions but with lower density, suggesting that most of them were recovered in the aqueous washes. Interestingly, the major protein bands seen after RMS treatment of C. glutamicum were also recovered by CMW extraction. In fact, CMW extraction has been used to recover PorA and PorH from C. glutamicum (23). The CMW extracts of M. smegmatis showed a complete absence of proteins, however.
Fig. 5.
Proteins extracted with RMS analyzed by 12% SDS/PAGE. Lane 1, aqueous wash after RMS treatment of C. glutamicum. Lane 2, aqueous wash after RMS treatment of Mycobacterim smegmatis mc2-155. The leftmost lane shows molecular weight standards (in kDa).
Finally, the RMS extract showed no activity of NADH oxidase, a well-known marker of the IM (24). This finding also is consistent with the notion that RMS extracts lipids and proteins only from the OM. The much more complex patterns of the IM proteins were not detected in these extracts.
Discussion
Our incentive when developing our RMS extraction methodology was the difficulty in preparing contamination-free OM of CMN group bacteria. The methods reported to date involve rate centrifugation (25–27), sucrose density gradient centrifugation (4), and cell surface disruption with glass beads (17). None of these methods has provided a completely clean separation between the OM and IM, however. Although some solvents, such as butanol, have been reported to preferentially extract OM lipids (28), the specificity and exhaustiveness of this method apparently have not yet been investigated. We believe that the results reported herein are important because they show that RMS extracts not only proteins, but also lipids, efficiently and quantitatively only from the OM. The complete absence of glycerophospholipids (except PIMs) in RMS extracts is especially important, because these lipids appeared in every previously reported OM preparation (7, 17, 26, 27).
In the original lipid bilayer model of mycobacterial OM (cell wall) proposed by Minnikin (1), the inner leaflet is composed entirely of the hydrocarbon chains of mycolate residues, covalently linked to arabinogalactan-peptidoglycan. With C. glutamicum, the relative paucity of covalently linked corynomycolate residues did not readily fit into this model; this was an especially serious problem with C. amycolatum, which totally lacks corynomycolic acids (17). A major finding in the present study is the discovery of a large amount of cardiolipin, which is not extractable in intact cells. Because cardiolipin becomes accessible after lysozyme treatment or in the isolated, fragmented cell envelope fraction, it is likely tightly associated with the peptidoglycan(-arabinogalactan) layer. The nature of this association is not known; however, given that cardiolipin has one free OH group available in the central glycerol moiety, the possibility of covalent linkage through this group cannot be excluded.
We can estimate the number of hydrocarbon chains needed for complete coverage of the cell surface. In Salmonella typhimurium, the surface was calculated to be 132 cm2 per mg dry weight, based on cell dimensions of 0.75 × 2.5 μm (29). C. glutamicum cells have reported dimensions of 0.7–1.0 × 1–3 μm (30). Assuming the median value (0.85 × 2 μm), the cell surface area would be expected to be 118 cm2/mg dry weight. Bound corynomycolate and bound C12-C20 fatty acids (Table 2) correspond to 5.2 mmol of hydrocarbon chains in the inner leaflet of the OM per 100 g cells. If we assume that one hydrocarbon chain occupies a 30-Å2 cross-section (31), then we predict that these hydrocarbon chains will cover 94 cm2 of surface in 1 mg of C. glutamicum cells, a value not far from the expected value noted above.
The outer leaflet of the Minnikin model is expected to be composed of “free” or extractable lipids. Here we find 1.69 mmol of corynomycolate in 100 g cells mostly in the form of TDM (Table 2), corresponding to 3.38 mmol of hydrocarbon chains. Together with the 0.39 mmol C12-C20 fatty acids likely coming from PIMs (Table 2) and apolar lipids (not quantified here), these likely are sufficient to cover the outer surface of the OM. Although the amount of C12-C20 fatty acid chains in the IM is somewhat smaller than expected given the cell surface area, possibly a significant area in IM is occupied by intrinsic membrane proteins.
Occupation of the inner leaflet of OM mainly by cardiolipin would produce a striking contrast to that of Mycobacterium spp., where very long, mostly saturated chains of mycolate produce a strong permeability barrier. The susceptibility of some Corynebacterium spp. to erythromycin (32), a large lipophilic drug, in contrast to the generally high resistance of Mycobacterium spp. (33, 34), might be a reflection of this difference.
In conclusion, our analysis allowed us to produce a complete and quantitative inventory of lipids in the OM of a CMN group organism, and our results support the Minnikin model. Our approach also led to the discovery of large amounts of hitherto unsuspected C12-C20 fatty acids linked tightly to the cell wall in the form of cardiolipin. The RMS extraction technique appears to also extract OM proteins efficiently, and it will be useful in the study of OM proteins in this group of organisms.
Experimental Procedures
Bacteria and Growth.
C. glutamicum ATCC 13032 was grown in a minimal medium (35) with 100 mM sodium acetate as the sole carbon source at 30 °C with aeration by shaking. Then [1,2-14C] acetate was added to 0.25 μCi/mL when the OD600 reached 0.5, and cells were harvested at an OD600 of ∼5. Harvested cells were washed with water 10 times to remove contamination of radioactive material in the medium.
Extraction of Lipids.
Noncovalently bound lipids were extracted with either CMW or RMS. An aliquot of fresh or frozen-thawed cells corresponding to 10 mg dry weight was suspended in 3 mL of CMW (2:1:0.1) or 1 mL of RMS (10 mM AOT in heptane). The mixtures were shaken at room temperature, and the monophasic extracts, collected by centrifugation, were cleaned by filtration through a nylon filter (0.2 μm). For the quantitation of IM lipids, RMS-extracted cells were first washed with water and then extracted with CMW.
To recover the peptidoglycan-associated lipids, an aliquot of cells containing ∼10 mg dry mass after CMW extraction was washed with water, followed by treatment with 10 mg of lysozyme in 1 mL of 10 mM sodium phosphate buffer (pH 7.5). The suspension was shaken at 150 rpm for 2 h at 37 °C. The lysozyme-treated cells were then reextracted with CMW in a manner similar to that described above. Lysozyme treatment also was conducted on intact cells.
Lipids were also obtained from isolated cell envelopes, as follows. Harvested cells (∼50 mg dry mass) were suspended in 10 mL of sodium phosphate buffer (50 mM; pH 7.5) and passed six times through a French pressure cell at 15,000 psi. Unbroken cells were removed by centrifugation at 5,000 × g for 15 min, and the supernatant was centrifuged at 200,000 × g for 30 min (rotor TL100, Beckman TL-100 ultracentrifuge). The pellet containing the cell envelope was then extracted twice with 3 mL of CMW (2:1:0.1). The delipidated sample was subjected to alkaline hydrolysis for analysis of the covalently bound fatty acids and mycolic acids, as described below.
Quantitative Analysis of Fatty Acid Content.
Lipids in extracted fractions were converted into fatty acid methyl esters by phase-transfer catalysis (36). Lipids were hydrolyzed with 0.4 mL of 15% tetrabutyl ammonium hydroxide overnight at 100 °C. After the addition of 0.4 mL water, methylation was performed by adding 0.2 mL of dichloromethane and 0.05 mL of iodomethane, followed by agitation for 30 min at room temperature. The lower organic phase was washed with 0.6 mL of dilute HCl and then with 0.6 mL of water. Covalently linked or tightly associated fatty acids were treated similarly except using intact cells, CMW-extracted residues, or the isolated envelopes as the starting material.
Quantitative data on the content of C12-C20 fatty acids were obtained by GC-MS of fatty acid methyl esters on a DB-XLB capillary column (30 m × 0.25 mm, 0.25 μm film thickness), using tridecanoic acid methyl ester as the internal standard. An 8 °C/min temperature gradient from 140 °C to 280 °C was used, and detection was performed using a quadrupole mass spectrometer.
The GC system did not allow the elution of corynomycolate esters. These were quantitated as follows. Fatty acid methyl esters were separated by TLC [developed with petroleum ether/ether (85/15, vol/vol)] into those of corynemycolic acids (Rf = 0.24) and those of the common fatty acids (Rf >0.7), and the radioactivity of each band was determined by phosphorimaging. Based on the known size distributions of the common fatty acids and corynomycolates in this organism, we obtained the molar ratio between the two classes as shown in the footnote to Table 2.
ESI-MS.
Lipids were isolated by preparative TLC using Analtech silica gel G plates. The isolated lipids were then analyzed in negative-ion mode using either a Waters Q-Tof Premier mass spectrometer or a Thermo Scientific Finnigan LTQ-FT mass spectrometer (for high-resolution data), both of which were equipped with a nanospray ion source.
Supplementary Material
Acknowledgments
We thank the campus QB3 Mass Spectrometer Facility for help with the GC-MS and ES-MS work. This study was supported in part by US Public Health Service Grant AI-09644.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112572108/-/DCSupplemental.
References
- 1.Minnikin D. In: The Biology of Mycobacteria. Ratledge C, Stanford J, editors. Vol 1. London: Academic; 1982. pp. 94–184. [Google Scholar]
- 2.Wheeler PR, Ratledge C. Metabolism in Mycobacterium leprae, M. tuberculosis and other pathogenic mycobacteria. Br Med Bull. 1988;44:547–561. doi: 10.1093/oxfordjournals.bmb.a072267. [DOI] [PubMed] [Google Scholar]
- 3.McNeil MR, Brennan PJ. Structure, function and biogenesis of the cell envelope of mycobacteria in relation to bacterial physiology, pathogenesis and drug resistance: Some thoughts and possibilities arising from recent structural information. Res Microbiol. 1991;142:451–463. doi: 10.1016/0923-2508(91)90120-y. [DOI] [PubMed] [Google Scholar]
- 4.Nikaido H, Kim S-H, Rosenberg EY. Physical organization of lipids in the cell wall of Mycobacterium chelonae. Mol Microbiol. 1993;8:1025–1030. doi: 10.1111/j.1365-2958.1993.tb01647.x. [DOI] [PubMed] [Google Scholar]
- 5.Jarlier V, Nikaido H. Permeability barrier to hydrophilic solutes in Mycobacterium chelonei. J Bacteriol. 1990;172:1418–1423. doi: 10.1128/jb.172.3.1418-1423.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Barry CE, 3rd, Besra GS, Nikaido H. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J Biol Chem. 1996;271:29545–29551. doi: 10.1074/jbc.271.47.29545. [DOI] [PubMed] [Google Scholar]
- 7.Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 8.Bansal-Mutalik R, Gaikar VG. Cell permeabilization for extraction of penicillin acylase from Escherichia coli by reverse micellar solutions. Enzyme Microb Technol. 2003;32:14–26. [Google Scholar]
- 9.Bansal-Mutalik R, Gaikar VG. Purification and concentration of alkaline phosphatase by selective permeabilization of Escherichia coli using reverse micellar solutions. Biotechnol Prog. 2003;19:1713–1720. doi: 10.1021/bp034141r. [DOI] [PubMed] [Google Scholar]
- 10.Bansal-Mutalik R, Gaikar VG. Mass transfer studies of cell permeabilization and recovery of alkaline phosphatase from Escherichia coli by reverse micellar solutions. Biotechnol Prog. 2004;20:1121–1127. doi: 10.1021/bp049911t. [DOI] [PubMed] [Google Scholar]
- 11.Cooper DG, Zajic JE, Gracey DE. Analysis of corynomycolic acids and other fatty acids produced by Corynebacterium lepus grown on kerosene. J Bacteriol. 1979;137:795–801. doi: 10.1128/jb.137.2.795-801.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins MD, Goodfellow M, Minnikin DE. A survey of the structures of mycolic acids in Corynebacterium and related taxa. J Gen Microbiol. 1982;128:129–149. doi: 10.1099/00221287-128-1-129. [DOI] [PubMed] [Google Scholar]
- 13.Krubasik P, et al. Detailed biosynthetic pathway to decaprenoxanthin diglucoside in Corynebacterium glutamicum and identification of novel intermediates. Arch Microbiol. 2001;176:217–223. doi: 10.1007/s002030100315. [DOI] [PubMed] [Google Scholar]
- 14.Sandmann G, Yukawa H. Vitamin synthesis: Carotenoids, biotin and pantothenate. In: Eggeling L, Bott M, editors. Handbook of Corynebacterium glutamicum. Boca Raton, FL: CRC Press; 2005. pp. 397–416. [Google Scholar]
- 15.Huc E, et al. O-mycoloylated proteins from Corynebacterium: an unprecedented post-translational modification in bacteria. J Biol Chem. 2010;285:21908–21912. doi: 10.1074/jbc.C110.133033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haites RE, Morita YS, McConville MJ, Billman-Jacobe H. Function of phosphatidylinositol in mycobacteria. J Biol Chem. 2005;280:10981–10987. doi: 10.1074/jbc.M413443200. [DOI] [PubMed] [Google Scholar]
- 17.Puech V, et al. Structure of the cell envelope of corynebacteria: Importance of the non-covalently bound lipids in the formation of the cell wall permeability barrier and fracture plane. Microbiology. 2001;147:1365–1382. doi: 10.1099/00221287-147-5-1365. [DOI] [PubMed] [Google Scholar]
- 18.Hoischen C, Krämer R. Membrane alteration is necessary but not sufficient for effective glutamate secretion in Corynebacterium glutamicum. J Bacteriol. 1990;172:3409–3416. doi: 10.1128/jb.172.6.3409-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa-Riu N, Burkovski A, Krämer R, Benz R. PorA represents the major cell wall channel of the Gram-positive bacterium Corynebacterium glutamicum. J Bacteriol. 2003;185:4779–4786. doi: 10.1128/JB.185.16.4779-4786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hünten P, Costa-Riu N, Palm D, Lottspeich F, Benz R. Identification and characterization of PorH, a new cell wall channel of Corynebacterium glutamicum. Biochim Biophys Acta. 2005;1715:25–36. doi: 10.1016/j.bbamem.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Barth E, Barceló MA, Kläckta C, Benz R. Reconstitution experiments and gene deletions reveal the existence of two-component major cell wall channels in the genus Corynebacterium. J Bacteriol. 2010;192:786–800. doi: 10.1128/JB.01142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stahl C, et al. MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol Microbiol. 2001;40:451–464. doi: 10.1046/j.1365-2958.2001.02394.x. [DOI] [PubMed] [Google Scholar]
- 23.Lichtinger T, Burkovski A, Niederweis M, Krämer R, Benz R. Biochemical and biophysical characterization of the cell wall porin of Corynebacterium glutamicum: The channel is formed by a low molecular mass polypeptide. Biochemistry. 1998;37:15024–15032. doi: 10.1021/bi980961e. [DOI] [PubMed] [Google Scholar]
- 24.Osborn MJ, Gander JE, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium: Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- 25.David HL, Lévy-Frébault V, Thorel MF. Characterization of distinct layers of the Mycobacterium avium envelope in respect of their composition by fatty acids, proteins, oligosaccharides and antigens. Zentralbl Bakteriol Mikrobiol Hyg [A] 1988;268:193–208. doi: 10.1016/s0176-6724(88)80003-8. [DOI] [PubMed] [Google Scholar]
- 26.Puech V, Bayan N, Salim K, Leblon G, Daffé M. Characterization of the in vivo acceptors of the mycoloyl residues transferred by the corynebacterial PS1 and the related mycobacterial antigens 85. Mol Microbiol. 2000;35:1026–1041. doi: 10.1046/j.1365-2958.2000.01738.x. [DOI] [PubMed] [Google Scholar]
- 27.Ortalo-Magné A, et al. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J Bacteriol. 1996;178:456–461. doi: 10.1128/jb.178.2.456-461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morita YS, et al. Compartmentalization of lipid biosynthesis in mycobacteria. J Biol Chem. 2005;280:21645–21652. doi: 10.1074/jbc.M414181200. [DOI] [PubMed] [Google Scholar]
- 29.Smit J, Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studies with lipopolysaccharide mutants. J Bacteriol. 1975;124:942–958. doi: 10.1128/jb.124.2.942-958.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe S, Takayama K, Kinoshita S. Taxonomic studies on glutamic acid-producing bacteria. J Gen Appl Microbiol. 1967;13:279–301. [Google Scholar]
- 31.Stoffel W, Pruss H-D. Monolayer studies with synthetic saturated, mono- and polyunsaturated mixed 1,2-diglycerides, 1,2-diacylphosphatidylethanolamines and phosphatidylcholines at the air–water interface. Hoppe Seylers Z Physiol Chem. 1969;350:1385–1393. doi: 10.1515/bchm2.1969.350.2.1385. [DOI] [PubMed] [Google Scholar]
- 32.Gómez-Garcés J-L, Alos J-I, Tamayo J. In vitro activity of linezolid and 12 other antimicrobials against coryneform bacteria. Int J Antimicrob Agents. 2007;29:688–692. doi: 10.1016/j.ijantimicag.2006.11.032. [DOI] [PubMed] [Google Scholar]
- 33.Naik S, Ruck R. In vitro activities of several new macrolide antibiotics against Mycobacterium avium complex. Antimicrob Agents Chemother. 1989;33:1614–1616. doi: 10.1128/aac.33.9.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown BA, Wallace RJ, Jr, Onyi GO, De Rosas V, Wallace RJ., 3rd Activities of four macrolides, including clarithromycin, against Mycobacterium fortuitum, Mycobacterium chelonae, and M. chelonae–like organisms. Antimicrob Agents Chemother. 1992;36:180–184. doi: 10.1128/aac.36.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eikmanns BJ, Metzger M, Reinscheid D, Kircher M, Sahm H. Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl Microbiol Biotechnol. 1991;34:617–622. doi: 10.1007/BF00167910. [DOI] [PubMed] [Google Scholar]
- 36.Besra GS. Preparation of cell wall fractions from mycobacteria. In: Parish T, Stoker NG, editors. Methods in Molecular Biology: Mycobacteria Protocols. Vol 1. New York: Humana Press; 1998. pp. 91–107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.