Abstract
The increasing sophistication of synthetic biology is creating a demand for robust, broadly accessible methodology for constructing multigene pathways inside of the cell. Due to the difficulty of rationally designing pathways that function as desired in vivo, there is a further need to assemble libraries of pathways in parallel, in order to facilitate the combinatorial optimization of performance. While some in vitro DNA assembly methods can theoretically make libraries of pathways, these techniques are resource intensive and inherently require additional techniques to move the DNA back into cells. All previously reported in vivo assembly techniques have been low yielding, generating only tens to hundreds of constructs at a time. Here, we develop “Reiterative Recombination,” a robust method for building multigene pathways directly in the yeast chromosome. Due to its use of endonuclease-induced homologous recombination in conjunction with recyclable markers, Reiterative Recombination provides a highly efficient, technically simple strategy for sequentially assembling an indefinite number of DNA constructs at a defined locus. In this work, we describe the design and construction of the first Reiterative Recombination system in Saccharomyces cerevisiae, and we show that it can be used to assemble multigene constructs. We further demonstrate that Reiterative Recombination can construct large mock libraries of at least 104 biosynthetic pathways. We anticipate that our system’s simplicity and high efficiency will make it a broadly accessible technology for pathway construction and render it a valuable tool for optimizing pathways in vivo.
Keywords: in vivo DNA assembly, homing endonuclease, cell engineering, metabolic engineering, combinatorial libraries
A key bottleneck to reengineering cells for diverse synthetic biology applications is the technical difficulty of constructing optimized, multigene pathways in vivo. The advent of synthetic biology has raised the tantalizing prospect of reprogramming cells at will for purposes ranging from the biosynthesis of high-value feedstocks and natural product analogs to the development of cell-based sensors and therapeutics (1). Engineering cells for such tasks requires the introduction of numerous exogenous genes into the genome to create customized “pathways.” However, standard molecular biology and genetic techniques, developed for the manipulation of single genes, become unwieldy or ineffective when applied to much larger multigene constructs. A new generation of robust, accessible tools for building pathways inside the cell is needed.
The difficulty of rationally designing complex systems that operate as desired in the cellular milieu (2) further argues that the ability to construct not only individual pathways but also libraries of pathways in vivo will be essential. Precedent has indicated that multicomponent systems introduced into the cell typically require refinement to function optimally (3, 4). By analogy to the directed evolution approaches that empowered the routine discovery of proteins and nucleic acids with prescribed functions, generating large numbers of variant pathways in parallel and screening for those that exhibit the required behavior could streamline optimization efforts (5). Library-based approaches could thus circumvent the gaps in our knowledge, immediately yielding functional systems. One invaluable tool for implementing such strategies will be DNA assembly methods that can reliably generate sizable collections of pathways (> 103) inside of the cell, an especially high standard of efficiency.
While several creative strategies for the in vitro and in vivo assembly of multigene constructs have been developed, building such pathways has not become routine outside of expert laboratories. One class of techniques is comprised of in vitro methods (6–11) such as in vitro recombination (12–14) that stretch the limits of standard molecular biology tools for large-scale DNA assembly. These methods can sometimes meet the standard of being high yielding, but they are resource intensive and do not intrinsically address the issue of moving the DNA into the cell. Though certain of these techniques can theoretically generate large numbers of pathways simultaneously, only a very few examples of constructing libraries that were relatively small (≤ 102 variants) have been reported (15).
The second class of DNA assembly methods exploits in vivo homologous recombination to assemble DNA directly in the cell (16–21). These techniques are attractive for their simplicity, as they require only a straightforward transformation step. In vivo homologous recombination has also been used extensively in the context of library generation for directed evolution applications; the highly efficient recombination machinery of organisms such as Saccharomyces cerevisiae has frequently been employed to build single-gene libraries containing 104–1010 variants (22–26). However, previously reported in vivo assembly techniques for multigene constructs have been low yielding, generating only tens to hundreds of recombinants at a time and making them impractical for the construction of libraries.
Thus, we sought to develop a high-yielding method for constructing multigene pathways that would harness the technical ease of in vivo recombination yet be efficient enough to create large libraries. Given that specific double-strand DNA breaks are known to promote repair by homologous recombination and have formed the basis of robust technologies to seamlessly manipulate genomes (27, 28), we hypothesized that coupling DNA cleavage by a homing endonuclease to DNA assembly could provide the needed boost in efficiency. Here we develop a method that utilizes endonuclease-stimulated homologous recombination for DNA assembly and demonstrate that we can easily and efficiently build large libraries of biosynthetic pathways in vivo.
Results
Design and Construction of a Reiterative Recombination System.
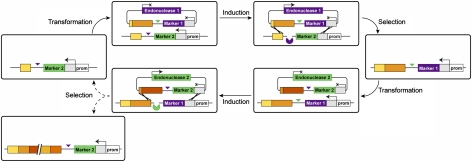
Our DNA assembly system, “Reiterative Recombination,” elongates a construct of interest in a stepwise manner by employing pairs of alternating, orthogonal endonucleases and selectable markers. As shown in Fig. 1, endonuclease cleavage sites are placed between fragments of the construct of interest and selectable markers in “donor” and “acceptor” modules. Following endonuclease cleavage of the acceptor module, the donor module provides homology on either side of the double-strand break through a short region of overlap between the fragments to be assembled on one side and a homology region upstream of the marker on the other side. Repair by homologous recombination adds the donor module’s fragment to the acceptor module’s growing construct while simultaneously replacing the acceptor module’s endonuclease cleavage site and selectable marker. Because only the acceptor module’s marker is actively transcribed, recombinants can be readily identified. During the next round of elongation, the endonuclease cleavage site and selectable marker return to the original configuration, allowing assembly to proceed in a cyclical format.
Fig. 1.
General scheme of Reiterative Recombination, showing two rounds of elongation. Each round of elongation is achieved by recombination between an acceptor module (shown here in the linear chromosome) and a donor module (in the circular plasmid). The two modules contain orthogonal homing endonuclease cleavage sites (triangles) adjacent to different selectable markers (purple and green). Both markers are downstream of a homology region (gray), but only the acceptor module contains a promoter (white) driving marker expression. Endonuclease cleavage of the acceptor module stimulates recombination, joining the fragments being assembled (orange) and replacing the acceptor module’s endonuclease site and expressed selectable marker with those of the donor module. Repeating this procedure with a donor module of the opposite polarity returns the acceptor module to its original state, allowing the assembly to be elongated indefinitely.
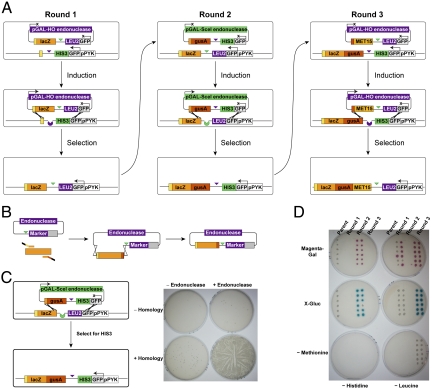
We constructed an initial Reiterative Recombination system in Saccharomyces cerevisiae, which has highly efficient homologous recombination machinery, placing the acceptor module in the chromosome and donor modules in plasmids. For the orthogonal endonucleases, we turned to the two well studied S. cerevisiae enzymes employed throughout the homologous recombination literature, HO (29) and SceI (30), placing them under the control of the inducible GAL promoter on the donor plasmids. For the alternating markers, we used HIS3 and LEU2, which complement the histidine and leucine auxotrophies of many laboratory yeast strains, adding endonuclease cleavage sites downstream of their terminators and fusing GFP genes to their N termini to provide upstream homology regions. The donor plasmids also contain the positive- and negative-selectable URA3 marker, allowing cells to be cured of donor plasmids by growth on 5-fluoroorotic acid (FOA) after each elongation round. Only two modifications were necessary to prepare a standard strain with the appropriate auxotrophies for Reiterative Recombination: elimination of the endogenous HO cleavage site in the MAT locus with a silent mutation (31) via “pop-in/pop-out” gene replacement (32) and integration of the acceptor module. The resulting parental acceptor strain can in theory be used for the assembly of any desired DNA construct. However, if certain applications necessitate the use of a specific background strain, only two established, robust integration steps are required to convert any strain with the appropriate auxotrophies into an acceptor strain.
Proof-of-Principle of Reiterative Recombination.
We first employed Reiterative Recombination to sequentially integrate the reporter genes lacZ (β-galactosidase), gusA (β-glucuronidase), and MET15 (complementation of methionine auxotrophy) using three rounds of assembly, creating an 8.5-kilobase construct (Fig. 2A). Subfragments for integration were PCR amplified as one or two overlapping pieces using primers that incorporated short regions of homology (30–40 bp) (i) to the preceding piece of the growing assembly and (ii) to the donor plasmid. PCR products were cotransformed with a digested, generic donor plasmid into the acceptor strain to generate intact donor plasmids by plasmid gap repair (Fig. 2B) (33). Our procedure thus eliminates any in vitro manipulation (e.g., subcloning) other than basic PCR. Galactose induction of endonuclease expression in the transformants led to a high rate of marker conversion only when both the endonuclease gene and the homology on both sides of the endonuclease cut site were present (Fig. 2C). Phenotypic analysis of recombinants following donor plasmid curing indicated that auxotrophies for histidine and leucine alternated with each round of elongation, as expected (Fig. 2D). Each newly integrated reporter (lacZ, gusA, or MET15) was functional in 75–100% of recombinants when > 40 individual colonies from each round were assayed, and previously integrated reporters were maintained (Fig. 2D, Fig. S1, SI Text). We also confirmed that integration occurred in the expected manner by analyzing the purified genomic DNA of cured recombinants by PCR and restriction digestion (Fig. S2).
Fig. 2.
Reiterative Recombination reporter proof-of-principle system. (A) Details of the assembly process for the proof-of-principle system, in which the three reporter genes lacZ, gusA, and MET15 were sequentially integrated into the chromosome. (B) Construction of donor plasmids by plasmid gap repair, in which a digested universal donor plasmid and PCR fragments with appropriate homology regions were cotransformed into the Reiterative Recombination strain and assembled via homologous recombination. (C) Results of the round 2 induction step are shown as a representative example. As negative controls, cells containing identical donor plasmids lacking the SceI endonuclease gene and/or the gusA fragment with lacZ homology were induced in parallel. A calculated 6 × 106 cells were plated on SC(-Histidine) media to assay for selective marker conversion after a 12-h galactose induction. (D) Phenotypes of 12 unique cured colonies from each round of assembly. In columns, recombinants are assayed for the HIS3 [SC(-Histidine)] and LEU2 [SC(-Leucine)] markers. In rows, recombinants are assayed for lacZ (Magenta-Gal), gusA (X-Gluc), and MET15 [SC(-Methionine)].
Construction of Libraries of Biosynthetic Pathways via Reiterative Recombination.
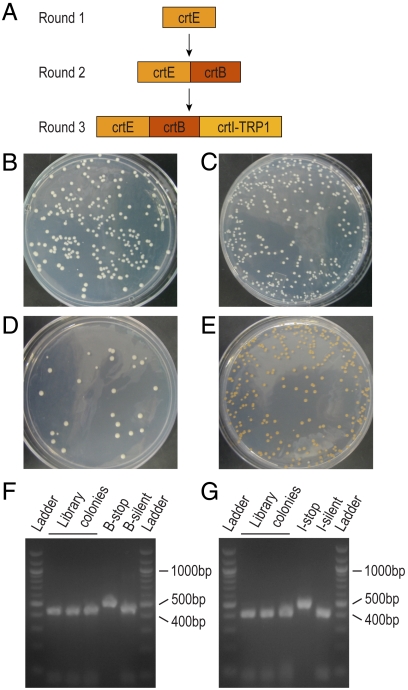
To demonstrate the generality of Reiterative Recombination and its application to a biosynthetic pathway, we integrated codon-optimized versions of Erwinia herbicola crtE (round 1), crtB (round 2), and crtI (round 3) to generate a yeast strain capable of producing the isoprenoid pigment lycopene (Fig. 3A). We also integrated the selectable marker TRP1 during round 3 to provide further verification of correct pathway construction (vide infra). After the third round of assembly, 99% of the resulting recombinants exhibited the expected orange phenotype, indicative of lycopene production (Fig. 3E). In parallel, as negative controls, we built pathways containing nonsense mutations in crtB and/or crtI, and the resulting strains did not produce lycopene (Fig. 3 B–D).
Fig. 3.
Assembly of the lycopene biosynthesis pathway using Reiterative Recombination. (A) Order of crt gene insertion. (B–E) Phenotypes of cured round 3 colonies containing wild-type crtE and (B) crtB-stop + crtI-stop, (C) crtB-stop + crtI-silent, (D) crtB-silent + crtI-stop, and (E) crtB-silent + crtI-silent. For (E), 315 out of 317 colonies had an orange phenotype; none of the other plates contained any orange colonies. (F, G) Restriction analysis of the three orange cured recombinants recovered from the 104∶1 (100∶1 crtB stop∶silent + 100∶1 crtI stop∶slient) lycopene library screen. Regions of the crtB (F) and crtI (G) alleles containing the diagnostic mutations were amplified by colony PCR and digested with EcoRV and BsmBI, respectively. Only alleles containing the silent mutations are cut by these enzymes. The plasmids with the B-stop, B-silent, I-stop, and I-silent alleles that served as PCR templates for the subfragments were PCR amplified and digested in parallel as controls. The ladder is a 100 bp DNA ladder from New England Biolabs.
Given that every step of Reiterative Recombination proceeded with high efficiency in these proof-of-principle studies, we expected that we could generate larger libraries of pathways than attainable with other in vivo DNA assembly techniques, which generate only tens to hundreds of variants at a time. Using a basic yeast electroporation protocol, we can obtain as many as 106–108 transformants per transformation (25); the induction, which is readily scalable, typically gives ≫104 recombinants per milliliter of culture. We therefore used the lycopene biosynthesis pathway to explicitly challenge Reiterative Recombination’s ability to generate large libraries. We repeated rounds 2 and 3 of the lycopene pathway assembly, this time transforming various ratios of crtB and crtI alleles that contained either nonsense or silent mutations with diagnostic restriction sites. Initially, we did not recover lycopene-producing colonies from our libraries at the expected frequencies. Further analysis of the pool of cured recombinants obtained from various Reiterative Recombination rounds, both from the reporter proof-of-principle system and the lycopene pathway assembly, revealed that a small percentage of cured recombinants (≤ 0.2%; SI Text, Table S1) acquired both the HIS3 and LEU2 markers. This subpopulation of cells was sufficient to skew the observed ratios of orange colonies after carrying the library forward for multiple rounds. While we are developing a next-generation Reiterative Recombination system that eliminates this problem entirely, we were immediately able to construct large libraries in this first-generation system by simply selecting for the TRP1 marker at the end of the pathway (Fig. 3A) after the last round of assembly. This additional selection served as a stringent final purification step for our libraries and, importantly, is a general solution that could be used for any desired library application. As shown in Table 1, we were readily able to recover lycopene-producing colonies at the expected frequencies from mock libraries of up to 104. These colonies contained the expected silent mutations in crtB and crtI, demonstrating that they arose from the silent alleles rather than from mutation of the genes with nonsense mutations (Fig. 3 F–G, Fig. S3).
Table 1.
Mock screen for lycopene-producing strains via Reiterative Recombination
| Transformed DNA ratios |
Library complexity |
Colonies assayed |
Orange colonies |
Observed percentage of orange colonies |
P* |
|
| crtB stop∶silent | crtI stop∶silent | |||||
| 10∶1 | 0∶1 | 101 | 2,360 | 225 | 10% | 0.5 |
| 100∶1 | 0∶1 | 102 | 587 | 5 | 0.9% | 0.7 |
| 100∶1 | 10∶1 | 103 | 2,079 | 3 | 0.1% | 0.4 |
| 100∶1 | 100∶1 | 104 | 18,450 | 3 | 0.02% | 0.4 |
*Because the plated cells represented a randomly selected aliquot (< 0.1%) of the population, a 1-proportion z-test was used to test if the observed percentages of orange colonies were significantly different than the expected percentages. All P-values were greater than α = 0.1, indicating that none were significantly different.
Discussion
By providing a highly efficient method for the assembly of pathways in vivo, Reiterative Recombination opens the door to the routine construction of gene circuits, pathways, and libraries thereof in the cell. Reiterative Recombination’s high efficiency, together with its technical straightforwardness, makes it a reliable method for building multigene constructs that is accessible to nonexperts without specialized equipment. While a handful of laboratories that are experts in the field have described landmark achievements in the realm of large-scale DNA assembly, these techniques have not yet been widely adopted by the scientific community. Reiterative Recombination distills the construction of individual pathways into a user-friendly process that requires only basic molecular biology tools.
The use of recyclable markers and endonucleases in Reiterative Recombination should also make it useful for the assembly and integration of very large DNA constructs. In this work, we have built constructs of only < 10 kilobases in three rounds of assembly. However, we anticipate no difficulty in continuing the procedure for more rounds, and we are currently working to construct significantly longer biosynthetic pathways. Recycling markers in Reiterative Recombination eliminates the perennial problem of running out of selectable markers during complex strain constructions, which greatly complicates or limits the scope of more traditional methods for introducing multiple genes and libraries thereof into the same yeast strain (e.g., repeated integration steps and/or mating strategies) (34, 35). Furthermore, our assembly system’s modular protocol minimizes the effort needed to design and execute each integration step. Finally, an important aspect of Reiterative Recombination is the generality of its design; Reiterative Recombination systems could be developed in other organisms that have efficient endogenous or engineered recombination systems (36). Alternatively, standard molecular biology techniques could be used to shuttle pathways constructed in yeast into other organisms that are preferred hosts for specific applications (21).
Reiterative Recombination’s robustness makes it capable of generating sizable libraries of multigene pathways (at least 104) containing diversity at multiple loci. Though methods for constructing multicomponent DNA constructs are proliferating, there are surprisingly few examples of using them to build libraries. The in vivo assembly methods described to date have proceeded with efficiencies that are simply too low (≤ 102 colonies) to be useful for the one-pot assembly of collections of pathways. Even after modification and optimization, most in vitro assembly techniques based on restriction digestion and ligation or on enzymatic recombination typically display profound losses in cloning efficiency for ≥3 fragments (≤ 103 colonies per reaction) (37, 38). Several particularly efficient in vitro approaches (8), notably in vitro recombination (12–14), could theoretically generate libraries of ∼104. However, none of these in vitro assembly technologies directly address the problem of moving the resulting pathways into the cell efficiently enough to maintain library complexity, an especial challenge if constructs must be stably integrated in the chromosome. Tellingly, even the high-yielding isothermal assembly method has only once been used to construct a library that consisted of ∼102 two-gene constructs and was incompletely characterized (15). Our mock library experiment is key because it explicitly tests the library sizes Reiterative Recombination can generate and shows that members of the library are present in the expected proportions. To our knowledge, no other DNA assembly method's ability to create such libraries in vivo has been rigorously characterized in this way. In addition, though we only attempted to build libraries of up to 104—due to the limits of our ability to readily screen large numbers of colonies for lycopene production visually—the high efficiency and straightforward scalability of the recombination step suggests that it is only the transformation efficiency of yeast (∼106–108 transformants per transformation) that will limit library size in Reiterative Recombination.
The development of highly efficient DNA assembly methods is an essential first step towards the combinatorial optimization of pathways in vivo. In spite of enormous advances in our understanding of systems-level biology in the past decade, our ability to rationally predict the effects of changes to cell circuitry remains limited (2, 5). Library approaches can provide a direct route for obtaining and refining functional in vivo systems (39–41). In the field of metabolic engineering, for example, researchers have repeatedly improved natural product yields and synthesized analogs by searching collections of isozymes (42, 43), mutant biosynthetic enzymes (3, 44), or promoters and regulatory regions that modulate the expression levels of genes that alter pathway flux (45, 46). Testing these multiple variables in the context of a pathway causes library sizes to rapidly swell (e.g., testing 100 mutants of enzyme A against 100 mutants of enzyme B is already 104 combinations). Pioneering efforts to combinatorially mutagenize the chromosome of Escherichia coli have driven home the message that it is essential to comprehensively explore this potential diversity to identify unexpected synergistic effects (40, 41). An assortment of methods for efficient combinatorial mutagenesis will therefore be needed to fully bring the power of directed evolution and other library optimization approaches to bear on metabolic engineering and synthetic biology problems.
We view Reiterative Recombination as part of an advancing wave of pioneering technologies for effecting large-scale modifications to the genome. One strategy is de novo genome synthesis, which allows complete customization of the genome. However, in spite of recent heroic feats in this area (16, 47) and the falling price of chemical DNA synthesis (48), such ambitious undertakings are neither technically nor economically feasible for most researchers. Alternatively, a second strategy is reprogramming well characterized host organisms, such as E. coli and S. cerevisiae, for designer functions through genetic engineering. This approach will require tools for incorporating two basic classes of genetic modifications: (i) the alteration of genes already in the genome and (ii) the introduction of multiple exogenous genes into the chromosome. To meet the first of these needs, altering the genetic background, classic mutagenesis techniques such as mutator strains can be useful for phenotypic optimization, but they do not provide control over the extent and location of mutations (49), unlike several notable, recently reported techniques. Exploiting sexual reproduction in S. cerevisiae, Suzuki et al. have used iterative cycles of mating and sporulation along with a quantitative GFP marker to gather up to 16 deletion mutations in an individual strain (50). Using an E. coli strain engineered with the λ Red recombination system (36), multiplex automated genome engineering automates the transformation of mutagenic single-stranded oligonucleotides ∼90 bases in length to create a powerful method for introducing specified deletions, point mutations, and very short insertions of ≤ 30 base pairs anywhere in the chromosome, but it has not been shown to be capable of efficient whole-gene insertion in its current form (see Note.) (40). Reiterative Recombination is one of several techniques that tackle the second issue, integrating exogenous pathways of genes into the chromosome (18–20). However, Reiterative Recombination is uniquely able to integrate pathways in a highly efficient manner to access large numbers of variant strains.
In conclusion, we foresee Reiterative Recombination becoming a powerful addition to the 21st-century molecular biology toolkit. Our method's simplicity and robustness will make it a user-friendly option for assembling seamless multigene constructs by any lab equipped for basic molecular biology. Reiterative Recombination's cyclical format means that it can be used to build pathways of indefinite length. Because our system is highly efficient, in contrast to other in vivo DNA assembly technologies, it can be used to assemble libraries of at least 104 pathways directly in the chromosome. Reiterative Recombination, as part of the expanding arsenal of cutting-edge cell engineering tools, will ensure the continued rapid development of synthetic biology as the scale of our ambitions increases and our applications move into the cell.
Materials and Methods
General Methods.
Standard methods for molecular biology in S. cerevisiae and E. coli were used (51, 52). Complete methods and additional details are provided in SI Text.
Fragment Design.
For clarity, a “fragment” refers to the total pathway-specific region of each donor plasmid, shown in orange in the figures. When convenient, fragments were divided into “subfragments” that were PCR amplified from different templates and assembled into the full fragment by plasmid gap repair upon transformation into yeast. Fragments contain 30 bp of homology to the donor plasmid and 20 bp of homology to the adjacent fragments, providing a total of 40 bp of homology for each integration event (Fig. S4). Overlapping ends of subfragments (within fragments) contained a total of 40 bp of homology. All regions of homology were incorporated with PCR primers. Illustrative examples are provided in the Table S2.
Reiterative Recombination Protocol.
The protocol for an even-numbered round (e.g., round 2) of Reiterative Recombination is described. Descriptions of strains and plasmids are provided in SI Text, and plasmid maps are provided in Fig. S5.
Preparation of subfragments: Subfragments were amplified with primers that added appropriate homology to adjacent fragments and to the donor plasmid (see above and SI Text). All PCR products were gel purified.
Transformation: The PCR products were cotransformed with the digested donor plasmid (pLW2593) in a 100∶1 molar ratio into the cured round 1 strain. Transformants were selected on synthetic complete media lacking leucine and uracil [SC(-Leucine, -Uracil)].
Induction: After 2 d of growth, transformants were lifted from the transformation plates, washed once with sterile water, resuspended in preinduction media [SC(Lactate, -Leucine, -Uracil)] to an OD600 of 1, and shaken at 30 °C for 3 h. Cells were then harvested, washed once with sterile water, and resuspended in induction media [SC(-Uracil, 2% galactose, 2% raffinose)] to an OD600 of 0.1. Cells were shaken at 30 °C for 12 h.
Selection: For control experiments, aliquots of the induction cultures were immediately plated on selective media [SC(-Histidine)] to determine the efficiency of marker switching. Colonies were counted after 2 d of growth. The remaining cells were inoculated into SC(-Histidine) liquid media, shaken at 30 °C for 1 d, and plated on SC(-Histidine, 0.1% FOA) to cure recombinants of the donor plasmid.
Reiteration: To begin the next round of Reiterative Recombination, after 2 d of growth, a single cured colony (in the reporter proof-of-principle experiment) or the pool of recombinants (in the lycopene pathway experiments) was lifted from the SC(-Histidine, 0.1% FOA) plates and inoculated into SC(-Histidine) liquid media to begin an overnight culture for the next transformation.
For odd rounds of Reiterative Recombination, pLW2592 was used as the donor plasmid, and the use of histidine and leucine in dropout media was reversed. All other aspects of the protocol remained the same. For library experiments, sufficient cells were carried through each step to ensure at least threefold coverage of the library.
Supplementary Material
Acknowledgments.
We thank Dr. D. Romanini and N. Ostrov for comments on the manuscript. We are grateful to Prof. Gregory Stephanopoulos and Dr. P. Ajikumar (Massachusetts Institute of Technology) for helpful discussions and the kind gift of plasmids containing codon-optimized crt genes. We thank Prof. K. Nasmyth (U. Oxford) for a strain with the functional HO endonuclease gene, Prof. J. Lindsley (U. Utah) for the gift of a plasmid containing the SceI gene, and Prof. D. Stillman (U. Utah) for an integration plasmid targeting the HO locus. L.M.W. was supported by a National Science Foundation Graduate Research Fellowship. This research was partially supported by National Institutes of Health (NIH) Grant R01 GM062867.
Note.
While this paper was under review, Isaacs, et al. reported that multiplex automated genome engineering could be used to insert double-stranded DNA encoding a selectable marker gene into the E. coli genome, but with efficiencies ranging from 10-5 to 10-7, vs. efficiencies of > 10-1 for almost all loci tested for point mutations introduced by single-stranded oligonucleotides (ref. 53).
Footnotes
Conflict of interest statement: L.M.W. and V.W.C. are inventors on a patent filed regarding this method.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100507108/-/DCSupplemental.
References
- 1.Baker D, et al. Engineering life: building a fab for biology. Sci Am. 2006;294:44–51. doi: 10.1038/scientificamerican0606-44. [DOI] [PubMed] [Google Scholar]
- 2.Purnick PEM, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 3.Leonard E, et al. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci USA. 2010;107:13654–13659. doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci USA. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haseltine EL, Arnold FH. Synthetic gene circuits: design with directed evolution. Annu Rev Biophys Biomol Struct. 2007;36:1–19. doi: 10.1146/annurev.biophys.36.040306.132600. [DOI] [PubMed] [Google Scholar]
- 6.Kodumal SJ, Santi DV. DNA ligation by selection. BioTechniques. 2004;37:34–40. doi: 10.2144/04371BM02. [DOI] [PubMed] [Google Scholar]
- 7.Reisinger SJ, Patel KG, Santi DV. Total synthesis of multi-kilobase DNA sequences from oligonucleotides. Nat Protoc. 2006;1:2596–2603. doi: 10.1038/nprot.2006.426. [DOI] [PubMed] [Google Scholar]
- 8.Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieniossek C, et al. Automated unrestricted multigene recombineering for multiprotein complex production. Nat Methods. 2009;6:447–450. doi: 10.1038/nmeth.1326. [DOI] [PubMed] [Google Scholar]
- 10.Blake WJ, et al. Pairwise selection assembly for sequence-independent construction of long-length DNA. Nucleic Acids Res. 2010;38:2594–2602. doi: 10.1093/nar/gkq123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011;6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 13.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 14.Gibson DG, Smith HO, Hutchison CA, 3rd, Venter JC, Merryman C. Chemical synthesis of the mouse mitochondrial genome. Nat Methods. 2010;7:901–903. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- 15.Ramon A, Smith HO. Single-step linker-based combinatorial assembly of promoter and gene cassettes for pathway engineering. Biotechnol Lett. 2011;33:549–555. doi: 10.1007/s10529-010-0455-x. [DOI] [PubMed] [Google Scholar]
- 16.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 17.Gibson DG, et al. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci USA. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itaya M, Tsuge K, Koizumi M, Fujita K. Combining two genomes in one cell: stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome. Proc Natl Acad Sci USA. 2005;102:15971–15976. doi: 10.1073/pnas.0503868102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itaya M, Fujita K, Kuroki A, Tsuge K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat Methods. 2008;5:41–43. doi: 10.1038/nmeth1143. [DOI] [PubMed] [Google Scholar]
- 20.Shao ZY, Zhao H, Zhao HM. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Z, Luo Y, Zhao H. Rapid characterization and engineering of natural product biosynthetic pathways via DNA assembler. Mol Biosyst. 2011;7:1056–1059. doi: 10.1039/c0mb00338g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abecassis V, Pompon D, Truan G. High efficiency family shuffling based on multi-step PCR and in vivo DNA recombination in yeast: statistical and functional analysis of a combinatorial library between human cytochrome P450 1A1 and 1A2. Nucleic Acids Res. 2000;28:e88. doi: 10.1093/nar/28.20.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benatuil L, Perez JM, Belk J, Hsieh CM. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng Des Sel. 2010;23:155–159. doi: 10.1093/protein/gzq002. [DOI] [PubMed] [Google Scholar]
- 24.Cherry JR, et al. Directed evolution of a fungal peroxidase. Nat Biotechnol. 1999;17:379–384. doi: 10.1038/7939. [DOI] [PubMed] [Google Scholar]
- 25.Peralta-Yahya P, Carter BT, Lin H, Tao H, Cornish VW. High-throughput selection for cellulase catalysts using chemical complementation. J Am Chem Soc. 2008;130:17446–17452. doi: 10.1021/ja8055744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swers JS, Kellogg BA, Wittrup KD. Shuffled antibody libraries created by in vivo homologous recombination and yeast surface display. Nucleic Acids Res. 2004;32:e36. doi: 10.1093/nar/gnh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lartigue C, et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science. 2009;325:1693–1696. doi: 10.1126/science.1173759. [DOI] [PubMed] [Google Scholar]
- 28.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc Natl Acad Sci USA. 2003;100:14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 30.Colleaux L, Dauriol L, Galibert F, Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci USA. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiffenbach B, et al. Deletions and single base pair changes in the yeast mating type locus that prevent homothallic mating type conversions. Proc Natl Acad Sci USA. 1983;80:3401–3405. doi: 10.1073/pnas.80.11.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scherer S, Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 34.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of ura3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bendixen C, Gangloff S, Rothstein R. A yeast mating-selection scheme for detection of protein-protein interactions. Nucleic Acids Res. 1994;22:1778–1779. doi: 10.1093/nar/22.9.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costantino N, Court DL. Enhanced levels of λ Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci USA. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sone T, et al. Multi-gene gateway clone design for expression of multiple heterologous genes in living cells: modular construction of multiple cDNA expression elements using recombinant cloning. J Biotechnol. 2008;136:113–121. doi: 10.1016/j.jbiotec.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 38.Roure A, et al. A multicassette Gateway vector set for high throughput and comparative analyses in Ciona and vertebrate embryos. PLoS One. 2007;2:e916. doi: 10.1371/journal.pone.0000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bikard D, Julie-Galau S, Cambray G, Mazel D. The synthetic integron: an in vivo genetic shuffling device. Nucleic Acids Res. 2010;38:e153. doi: 10.1093/nar/gkq511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol. 2005;23:612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 42.Steen EJ, et al. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb Cell Fact. 2008;7:36. doi: 10.1186/1475-2859-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menzella HG, et al. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat Biotechnol. 2005;23:1171–1176. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- 44.Fischbach MA, Lai JR, Roche ED, Walsh CT, Liu DR. Directed evolution can rapidly improve the activity of chimeric assembly-line enzymes. Proc Natl Acad Sci USA. 2007;104:11951–11956. doi: 10.1073/pnas.0705348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 47.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 48.Carr PA, Church GM. Genome engineering. Nat Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 49.Sauer U. Evolutionary engineering of industrially important microbial phenotypes. Adv Biochem Eng Biot. 2001;73:129–169. doi: 10.1007/3-540-45300-8_7. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki Y, et al. Knocking out multigene redundancies via cycles of sexual assortment and fluorescence selection. Nat Methods. 2011;8:159–164. doi: 10.1038/nmeth.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams A, Gottschling D, Kaiser C, Stearns T, editors. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 52.Ausubel F, et al. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 53.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.