Abstract
Mitochondrial protein import requires cooperation of the machineries called translocators in the outer and inner mitochondrial membranes. Here we analyze the interactions of Tom22, a multifunctional subunit of the outer membrane translocator TOM40 complex, with other translocator subunits such as Tom20, Tom40, and Tim50 and with substrate precursor proteins at a spatial resolution of the amino acid residue by in vivo and in organello site-specific photocross-linking. Changes in cross-linking patterns caused by excess substrate precursor proteins or presequence peptides indicate how the cytosolic receptor domain of Tom22 accepts substrate proteins and how the intermembrane space domain of Tom22 transfers them to Tim50 of the inner-membrane translocator.
Mitochondria, a powerhouse of eukaryotic cells, consist of 1,000–1,500 different proteins that are mainly synthesized in the cytosol and placed in four subcompartments, the outer and inner membranes and the aqueous intermembrane space (IMS) and matrix (1–3). Import and subsequent intramitochondrial sorting of mitochondrial proteins are mediated by the mitochondrial membrane–protein complexes called translocators. The translocator complexes in the outer and inner membranes are not tightly linked with each other, yet dynamically cooperate to achieve protein delivery to each mitochondrial subcompartment. Besides, translocator complexes are dynamic entities on their own, and alter their subunit–subunit interactions to work in demand for imported client proteins with different destinations (4–6). To understand the mechanism of protein transport by the mitochondrial import/sorting systems, it is essential to monitor and analyze dynamic interactions among the constituents of the systems, and in organello and in vivo are techniques providing sufficiently high spatial resolution.
Most matrix proteins and some inner-membrane proteins are synthesized as precursor proteins with an amino-terminal cleavable presequence, which contains a mitochondrial targeting signal. Presequences generally have potentials to form a positively charged amphiphilic helical structure. The outer membrane translocator, the TOM40 complex, functions as an entry gate for most mitochondrial proteins. The yeast TOM40 complex is composed of the core complex consisting of Tom40, Tom22, Tom5, Tom6, and Tom7, and peripheral receptor subunits, Tom20 and Tom70. Tom20 is a general import receptor and anchored to the outer membrane by its N-terminal transmembrane (TM) segment. Tom22 spans the outer membrane by its central TM segment, with its N-terminal and C-terminal domains exposed to the cytosol and IMS, respectively (7, 8). The cytosolic domains of Tom20 and Tom22 cooperate to form a presequence receptor site called the cis site to recognize mitochondrial targeting signals (9–12). After recognition by the receptor subunits of the TOM40 complex, presequence-containing precursor proteins move across the outer membrane through the β-barrel Tom40 channel (13–15), which is stabilized by Tom22 (16). Then the presequence is recognized by another receptor site of the TOM40 complex on the IMS side called the trans site, which consists of the IMS-facing region of Tom40, Tom22, and Tom7 (17–21). The presequence is subsequently forwarded to Tim50, an essential subunit of the inner-membrane translocator, the TIM23 complex (22–24). The TIM23 complex allows presequence transfer to the matrix through the α-helical Tim23 channel utilizing the energy of the membrane potential across the inner membrane (ΔΨ) and ATP hydrolysis by the import motor, mitochondrial Hsp70 in the matrix.
In the present study, we attempt to make protein-interaction mapping of a multifunctional subunit of the TOM40 complex, Tom22, which consists of the three distinct domains that play different roles in mitochondrial protein transport in the cytosol, outer membrane, and IMS. Because no high-resolution structural information is available for Tom22, we adopted in vivo and in organello site-specific photocross-linking to assess protein–protein interactions of Tom22 at its work. Briefly, the gene for Tom22 containing an amber codon for a desired position is introduced into yeast cells that contains an orthogonal pair of amber suppressor tRNA and its cognate aminoacyl–tRNA synthetase specific for DL-2-amino-3-(p-benzoylphenyl)pentanoic acid (BPA) (25). Addition of BPA into culture medium allows incorporation of BPA into Tom22 at the position specified by the amber codon. Subsequent UV irradiation of yeast cells or isolated mitochondria result in photocross-linking of BPA in Tom22 with nearby proteins. Cross-linked partner proteins can be identified by immunoblotting with antibodies against possible candidates or molecular-weight shift of the cross-linked products by epitope tagging of the possible candidates.
Results and Discussion
The Cytosol Domain of Tom22 Interacts with Tom20 in a Presequence-Dependent Manner.
Yeast Tom22 is predicted to consist of three domains: the N-terminal cytosolic domain (residues 1–97), central TM domain (residues 98–119), and C-terminal IMS domain (residues 120–152). A modified TOM22 gene with an amber codon in a specific position was placed under the control of the galactose-inducible GAL1 promoter and was introduced into yeast cells that contain an orthogonal pair of amber suppressor tRNA and its cognate aminoacyl–tRNA synthetase specific for BPA. When BPA was added to the culture medium, Tom22 containing BPA at the amber-specified position in addition to the Tom22 derivative truncated at the amber-specified position was produced (Fig. S1A). Because Tom22 is essential for yeast cell growth under most conditions (7, 8, 15), we confirmed that Tom22 with BPA could take over the essential functions of wild-type (WT) Tom22 (Fig. S1B) and be assembled into the TOM40 complex (Fig. S2) when expression of endogenous Tom22 was shut off.
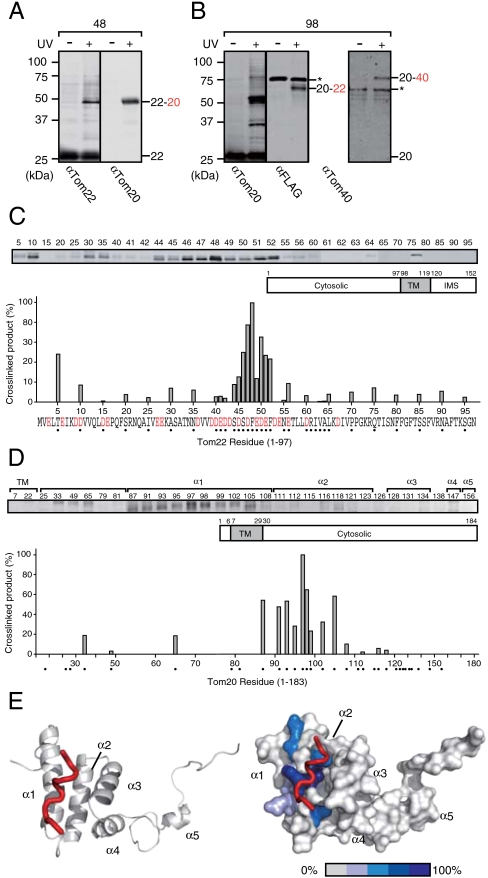
We then overexpressed Tom22 containing BPA in the N-terminal cytosolic domain from the GAL1 promoter in yeast cells in the presence of endogenous WT Tom22, and UV irradiated the cells. Affinity purification of the cross-linked products with the C-terminal His10 tag and subsequent immunoblotting revealed that BPA at residue 48 of Tom22 was cross-linked to Tom20 (Fig. 1A). When the position of BPA was varied, cross-linking to Tom20 was the most prominent around residues 44–52, a region rich in acidic residues (Fig. 1C). Conversely, when we introduced BPA at residue 98 in the cytosolic receptor domain of Tom20, UV irradiation resulted in cross-linking to Tom22 and Tom40 (Fig. 1B). When the position of BPA was varied, cross-linking to Tom22 was the most prominent around helix α1 of Tom20, which constitutes a part of the hydrophobic groove as the presequence binding site in homologous rat Tom20 (9) (Fig. 1 D and E).
Fig. 1.
The cytosol domain of Tom22 is cross-linked to Tom20. The yeast strains TSY1/pTS1-48 (A) and TSY4/pTS4-98 (B) for overexpression of Tom22 with BPA at residue 48 or overexpression of Tom20 with BPA at residue 98 from the GAL1 promoter were cultured in SD or SCD medium, respectively, then shifted to SGal or SCGal medium, respectively, with BPA and grown for 16 h. After UV irradiation, Tom22 or Tom20 with their C-terminally attached His10 tag were affinity purified and analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. In TSY4/pTS4-98 cells, Tom22 possesses a FLAG epitope tag at the C-terminus for detection. The 22, Tom22; 22-20, Tom22 cross-linked with Tom20; 20, Tom20; 20-22, Tom20 cross-linked with Tom22; 20–40, Tom20 cross-linked with Tom40. The asterisks indicate nonspecific bands. (C) BPA was introduced into various positions in the cytosolic domain of Tom22 (indicated with dots) in vivo and subjected to photocross-linking analyses as in A. The amounts of the cross-linked products with Tom20 were quantified and plotted against residue number; the largest amount for residue 48 was set to 100%. The red letters in the sequence show acidic residues. (D) BPA was introduced into various positions in the cytosolic domain of Tom20 (indicated with dots) and subjected to photocross-linking analyses as in A. The amounts of the cross-linked products with Tom22 were quantified and plotted against residue number; the largest amount for residue 97 was set to 100%. (E) The tertiary structure of the receptor domain (residues 86–171) of yeast Tom20 was constructed by homology modeling based on the NMR structure of rat Tom20 [Protein Data Bank (PDB) ID code 1OM2] (10) and shown in ribbon model (Left) and surface model (Right). The bound ALDH peptide is shown by a red tube. The amounts (relative intensities in D) of the cross-linked products with Tom22 were shown in gradation in blue (Right).
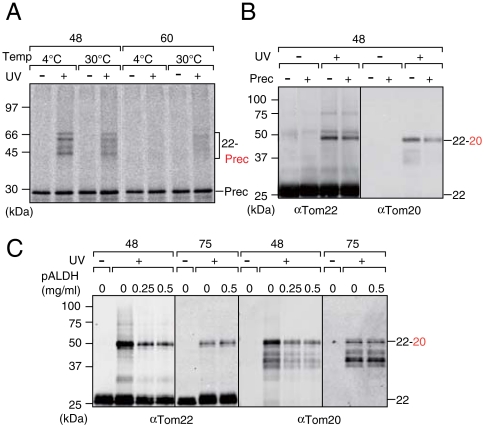
Next, we asked if the Tom20-interacting acidic region of Tom22 is involved in the presequence recognition, as well (12). We isolated mitochondria containing Tom22 with BPA at residue 48 or 60 and incubated them with a radiolabeled model precursor protein pSu9–DHFR, a fusion protein between the 69-residue presequence of the precursor to subunit nine of Neurospora crassa Fo–ATPase and mouse dihydrofolate reductase (DHFR), in the absence of ΔΨ. A previous study showed that, upon incubation with mitochondria lacking ΔΨ, pSu9–DHFR is accumulated at the level of the TOM40 complex, forming two distinct intermediates at stage A (at low temperature) or at stage B (at elevated temperature) (20). At stage A, a positively charged N-terminal segment of the long presequence binds to the trans site through electrostatic interactions without unfolding of the DHFR part whereas at stage B the DHFR part is unfolded and trapped by the inner wall of the Tom40 channel mainly through hydrophobic interactions. Both at stages A and B, BPA at residue 48 (in the acidic Tom20-interacting region), but not at residue 60 (outside the acidic region), of Tom22 was cross-linked to pSu9–DHFR (Fig. 2A). Therefore while the C-terminal half of the pSu9 presequence is in contact with the cytosolic domain of Tom22 as well as Tom20 at stage A (26), the unfolded DHFR part is in contact with the Tom22 cytosol domain at stage B, suggesting the possible chaperoning role of the Tom22 cytosol domain.
Fig. 2.
Cross-linking between Tom20 and Tom22 is affected by substrate precursor proteins/presequence peptides. (A) The yeast strains (TSY2/pTS2-48 and TSY2/pTS2-60) for expression of Tom22 with BPA at residue 48 or 60 from its own promoter were cultured in SCGal medium, then shifted to SCD medium with BPA and grown for 24 h. Then mitochondria were isolated from the cells, subjected to in vitro binding of 35S-labeled pSu9-DHFR for 10 min in the absence of ΔΨ, and UV-irradiated. Tom22 with the His10 tag was affinity purified and subjected to SDS-PAGE and radioimaging. (B) The yeast strains (TSY1/pTS1-48) for overexpression of Tom22 with BPA at residue 48 from the GAL1 promoter with a plasmid (pTS5) for overexpression of pb2(167)Δ19–DHFR from the GAL1 promoter or its control vector (pYO326) were cultured in SD medium, then shifted to SGal medium with BPA and grown for 16 h. After UV-irradiation, proteins were analyzed as in Fig. 1A. (C) Mitochondria with Tom22 containing BPA at residue 48 or 75 were purified and subjected to in vitro binding of indicated amounts of pALDH peptides in the absence of ΔΨ as in A. After UV-irradiation, proteins were analyzed as in Fig. 1A. The 22-Prec, Tom22 cross-linked with 35S-labeled pSu9-DHFR; Prec, pb2(167)Δ19–DHFR; 22, Tom22; 22-20 Tom22 cross-linked with Tom20.
A next question is if the cytosolic domains of Tom20 and Tom22 dissociate from each other to accommodate the presequence. We thus overexpressed Tom22 with BPA at position 48 in vivo with or without simultaneous overexpression of pb2(167)Δ19–DHFR, a fusion protein between the matrix-targeting N-terminal 167 residues of yeast cytochrome b2 precursor with 19-residue deletion of the sorting signal and DHFR, from the GAL1 promoter. We observed accumulation of the precursor form of Mdj1p, a matrix-localized protein, upon overexpression of pb2(167)Δ19–DHFR, suggesting that pb2(167)Δ19–DHFR saturates the protein import machineries in mitochondria (Fig. S3A). The amounts of the cross-linked products of Tom22 with Tom20 now decreased upon overexpression of pb2(167)Δ19–DHFR (Fig. 2B). We isolated mitochondria containing Tom22 with BPA at position 48 or 75, and after dissipation of ΔΨ, we UV-irradiated them in the absence or presence of increasing amounts of residues 1–22 of the rat aldehyde dehydrogenase presequence (pALDH), which is recognized by the import receptor Tom20 (9). The amounts of the cross-linked products of Tom22 with Tom20 decreased for BPA at position 48 (in the acidic region), but not for BPA at position 75 (outside the acidic region), in the presence of pALDH (Fig. 2C), although like Tom22, cross-linking of Tom20 to cis-site bound substrate peptides was hardly observed (21, 26). These results indicate that the acidic region of Tom22 competes with the presequence for the interaction with Tom20, fitting for the model that Tom20 and Tom22 recognize the hydrophobic side and the positively charged side, respectively, of the amphiphilic helix of the presequence simultaneously (12, 27).
The TM Helix of Tom22 Interacts with Two Molecules of Tom40.
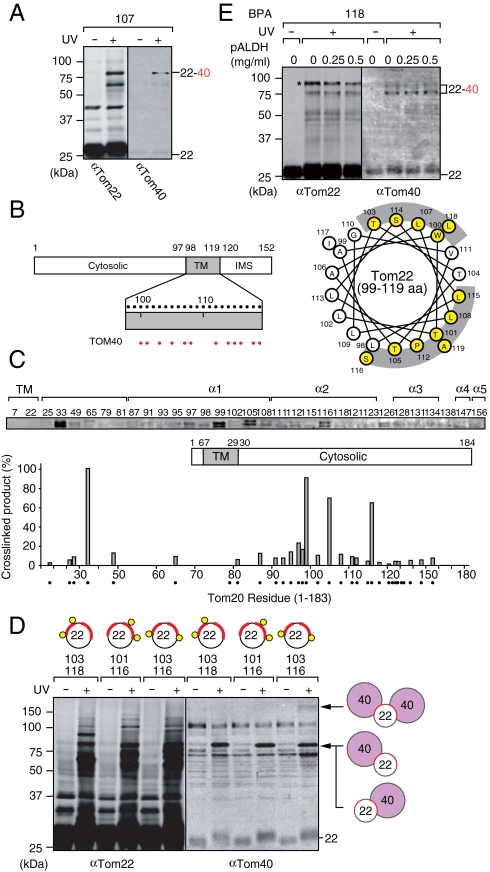
We then probed interactions of the TM segment of Tom22 with other subunits of the TOM40 complex in vivo. BPA at residue 107 in the TM segment of Tom22 was found to be cross-linked to Tom40 (Fig. 3A). When we changed the position of BPA at every residue throughout the TM segment (residues 98–119), Tom40 was cross-linked to BPA at residues 100, 101, 103, 105, 107, 108, 112, 114, 115, 116, 118, and 119 of Tom22 (Fig. 3B, Left) whereas BPA in the cytosol or IMS domain of Tom22 was not cross-linked to Tom40. This is in contrast to Tom20, which was cross-linked to Tom40 mainly through the helices α1 and α2 in the cytosol domain (Fig. 1A and Fig. 3C).
Fig. 3.
The TM segment of Tom22 is cross-linked to two Tom40. (A) The yeast strain (TSY1/pTS1-107) with overexpressed Tom22 containing BPA at residue 107 was UV-irradiated and analyzed as in Fig. 1A. (B) BPA was introduced into various positions in the TM segment of Tom22 (indicated with black dots) in vivo and subjected to photocross-linking. The positions of cross-linking to Tom40 were shown with red dots (Left) and with yellow circle for the helical wheel plot, which were segregated into two regions indicated with gray arcs (Right). (C) BPA was introduced into various positions in Tom20 (indicated with black dots) in vivo and subjected to photocross-linking. The amounts of the cross-linked products with Tom40 were quantified and plotted against residue number; the largest amount for residue 33 was set to 100%. (D) The yeast strains (TSY1/pTS1-103,118, TSY1/pTS1-101,116, and TSY1/pTS1-103,116) with overexpressed Tom22 containing BPA at residues 103/118, residues 101/116, and residues 103/116 were UV-irradiated and analyzed as in Fig. 1A. (E) Mitochondria containing Tom22 with BPA at residue 118 were purified and subjected to in vitro binding of indicated amounts of pALDH peptides in the absence of ΔΨ as in Fig. 2C. After UV-irradiation, proteins were analyzed as in Fig. 1A. The bands with an asterisk is overlapped by the one from the cross-linked product with Tim50 (see Fig. 4C). The 22, Tom22; 22–40, Tom22 cross-linked with Tom40.
Mapping of Tom40-interacting residues on the helical wheel plot of the Tom22 TM segment showed that they are segregated into two sides of the TM helix (Fig. 3B, Right). Does this mean that the TM helix of one Tom22 molecule simultaneously interacts with two molecules of Tom40 or that Tom22 consists of two populations, each of which is in contact with a single Tom40 molecule? To address this question we introduced two amber codons simultaneously into the TOM22 gene. For example, the mutant TOM22 gene with amber codons for residues 103 and 116 allows expression of full length Tom22 with double BPA at both residues 103 and 116 as well as those with a single BPA at either residue 103 or 116. After UV irradiation, those BPA containing Tom22 species generated cross-linked products with not only one Tom40 molecule (85 kDa) but also two Tom40 molecules at the same time (160 kDa), indicating that BPA at residues 103 and 116 are simultaneously in contact with Tom40 (Fig. 3D). On the other hand, BPA at residues 103 and 118 or BPA at residues 101 and 116 generated only single-Tom40 containing cross-linked products, but not double-Tom40 containing cross-linked products (Fig. 3D). Therefore the TM helix of Tom22 interacts with two Tom40 molecules through the two distinct regions containing residues 103 and 118 and residues 101 and 116, which will contribute to stabilization of the Tom40 oligomeric structure. This is consistent with the topological arrangement of three Tom22 and three Tom40 molecules in the TOM40 complex determined by cryo-EM analyses (15).
Does this topological arrangement of Tom40 and Tom22 respond to translocating precursor proteins? We isolated mitochondria containing Tom22 with BPA at position 118 or 105, and UV-irradiated them in the absence of ΔΨ, but in the presence of increasing amounts of the presequence peptide pALDH (Fig. 3E and Fig. S3B). The amounts of the cross-linked products between Tom22 and Tom40 were not affected by pALDH, suggesting that the Tom22–Tom40 interactions do not change upon translocation of substrate presequences.
Interaction of the IMS Domains of Tom22 and Tim50 Responds to the Presequence.
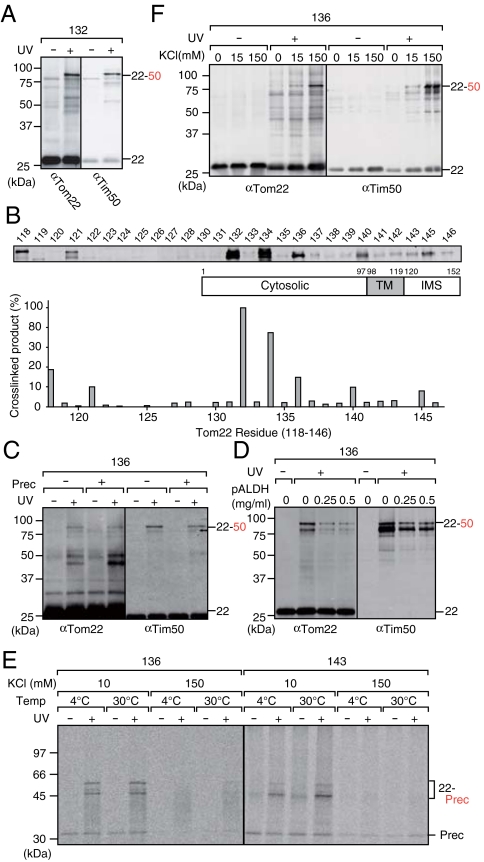
Next we introduced BPA in the IMS domain (residues 120–152) of Tom22 in vivo and UV-irradiated. BPA at residue 132 in the Tom22 IMS domain was found to be cross-linked to Tim50 (Fig. 4A, Left). We detected similar cross-linking between FLAG-tagged Tim50 and nontagged Tom22, ruling out the possibility that the Tim50–Tom22 cross-linking arose from artifacts due to the His10 tag (Fig. S3C). When we changed the position of BPA at every residue throughout the IMS domain except for the very C-terminal 6 residues (residues 120–146), Tim50 was clearly cross-linked to BPA at residues 118, 121, 132, 134, 136, 140, and 145 of Tom22 (Fig. 4B).
Fig. 4.
The IMS domain of Tom22 is cross-linked to Tim50. (A) The yeast strain TSY1/pTS1-132 with overexpressed Tom22 containing BPA at residue 132 was UV-irradiated and subjected to affinity purification of Tom22 with the His10 tag. Proteins were analyzed as in Fig. 1A. (B) BPA was introduced into various positions of Tom22 (indicated with black dots) in vivo and subjected to photocross-linking. The amounts of the cross-linked products with Tim50 were quantified and plotted against residue number; the largest amount for residue 132 was set to 100%. (C) The yeast strains (TSY1/pTS1-136) with overexpressed Tom22 containing BPA at residue 136 and with overexpressed pb2(167)Δ19–DHFR or its vector control were UV-irradiated as in Fig. 2B. Proteins were analyzed as in A. (D) Mitochondria with Tom22 containing BPA at residue 136 were purified and subjected to in vitro import of indicated amounts of pALDH peptides in the absence of ΔΨ as in Fig. 2C. Proteins were analyzed as in A. (E) Mitochondria were isolated from the yeast strains (TSY2/pTS2-136 and TSY2/pTS2-143) with Tom22 containing BPA at the indicated positions, subjected to in vitro binding of 35S-labeled pSu9–DHFR for 10 min at 4 °C or at 30 °C in binding buffer containing 10 mM KCl or 150 mM KCl in the absence of ΔΨ, and UV-irradiated as in Fig. 2A. Tom22 with the His10 tag was affinity purified and subjected to SDS-PAGE and radioimaging. (F) Mitochondria were isolated from the yeast strain (TSY2-136) with overexpressed Tom22 containing BPA at residue 136, suspended in 250 mM sucrose, 10 mM MOPS-KOH (potassium 3-[N-morpholino] propanesulfonate), pH 7.2, for and indicated concentrations of KCl (plus 1 mM EDTA for 0 mM KCl), and UV-irradiated as in Fig. 2A. Proteins were analyzed as in A. The 22, Tom22; Prec, pb2(167)Δ19–DHFR; 22–50 Tom22 cross-linked with Tim50; 22-Prec, Tom22 cross-linked with 35S-labeled pSu9–DHFR.
Tim50 is anchored to the inner membrane by its TM segment near the N terminus, exposing a large C-terminal domain to the IMS, which could interact with mitochondrial presequences (22). We thus asked if Tom22–Tim50 interactions would be affected by the translocating precursor proteins or presequence peptides. When mitochondrial precursor protein pb2(167)Δ19–DHFR was overexpressed, the cross-linking between BPA at residue 136 of Tom22 and Tim50 decreased in vivo (Fig. 4C). When mitochondria containing BPA at residue 136 were isolated and UV-irradiated in the absence of ΔΨ, the cross-linking between Tom22 and Tim50 also decreased with increasing amounts of pALDH (Fig. 4D). Therefore the IMS domains of Tom22 and Tim50 likely dissociate from each other upon precursor protein transfer from the TOM40 complex to the TIM23 complex.
We then isolated mitochondria with Tom22 containing BPA at residue 136 or 143 in the IMS domain and incubated them with radiolabeled pSu9–DHFR in the absence of ΔΨ. At both stages A (4 °C) and stage B (30 °C), the interactions between the presequence of the translocation intermediate of pSu9–DHFR and the IMS domain (residues 136 and 143) of Tom22 were abolished by high concentration of KCl (Fig. 4E). A previous study showed that cross-linking of the presequence of the stage B intermediate of pSu9–DHFR to Tim50 was observed only after wash of mitochondria with 150 mM KCl, but not with 15 mM KCl (26). Therefore, whereas the presequence interacts with Tom22 in the IMS primarily through electrostatic interactions, the interactions between the presequence and Tim50 are hydrophobic rather than electrostatic. We now examined if the interactions between the IMS domains of Tom22 and Tim50 are sensitive to KCl concentrations. When we increased the salt concentration from 15 to 150 mM, the cross-link between the Tom22 IMS domain and Tim50 increased (Fig. 4F), suggesting that the interactions between Tom22 and Tim50 in the IMS are, like those between the presequence and Tim50, mainly hydrophobic rather than electrostatic. Therefore the presequence and the IMS domain of Tom22 likely compete with each other to bind to Tim50 by hydrophobic interactions.
Conclusion
In the present study, we employed in vivo site-specific incorporation of BPA into Tom22, a multifunctional subunit of the TOM40 complex, to analyze its interactions with the subunits of the TOM40 and TIM23 complexes and substrate precursor proteins by photocross-linking. We assessed possible changes in interactions of Tom22 with other translocator subunits in response to substrate translocating proteins by overexpressing mitochondrial precursor proteins in vivo or adding chemical amounts of mitochondrial presequence peptides in vitro to partly saturate the import machineries of the TOM40 complex. By doing so, we found the followings. First, the clamp of the cytosolic receptor domains of Tom22 and Tom20 opens to accept presequences for targeting signal recognition. Second, whereas the cytosolic domain of Tom20 interacts with Tom40, the TM helix of Tom22 tethers two Tom40 molecules, yet those interactions are not affected by substrate presequences. Third, dynamic interactions between the IMS domains of Tom22 and Tim50 promote transfer of the mitochondrial presequence from Tom22 to Tim50, suggesting the following scenario. Whereas the transient interaction between Tim50 and Tom22 brings the TIM23 complex closer to the TOM40 complex, the presequence of the incoming precursor protein binds to the IMS domain of Tom22 when the Tom22 IMS domain is not occupied with Tim50. Then binding of Tim50 to Tom22 clears the presequence from Tom22, and Tim50 receives the presequence, thereby facilitating efficient presequence transfer from the TOM40 complex to the downstream TIM23 complex. The present study demonstrates that in vivo site-specific photocross-linking can serve as a useful tool to provide interaction-map snapshots for membrane–protein complexes at work.
Materials and Methods
Plasmids and Strains.
Constructions of yeast strains and plasmids (Tables S1–S4) are described in SI Materials and Methods.
Growth conditions.
Cells were grown in YPD (1% yeast extract, 2% polypeptone, and 2% glucose), SD (0.67% yeast nitrogen base without amino acids and 2% glucose), SCD (0.67% yeast nitrogen base without amino acids, 0.5% casamino acid, and 2% glucose), SGal (0.67% yeast nitrogen base without amino acids and 2% galactose), or SCGal (0.67% yeast nitrogen base without amino acids, 0.5% casamino acid, and 2% galactose) media with appropriate supplements.
In Vivo and in Organello Photocross-Linking.
The plasmids for in vivo BPA cross-linking were provided by P.G. Schultz (The Genomics Institute of the Novartis Research Foundation). Site-specific photocross-linking in vivo and in organello was performed as described previously (6, 20, 26) and in SI Materials and Methods. Preparation of pALDH is described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Peter G. Schultz (The Genomics Institute of the Novartis Research Foundation) for the system for in vivo BPA incorporation and the members of the Endo laboratory for discussions and comments. T.S is and K.Y was a Research Fellow of the Japan Society for the Promotion of Science (JSPS). We acknowledge support of this work by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) and a grant from the Japan Science and Technology Corporation (JST).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.N. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105921108/-/DCSupplemental.
References
- 1.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Endo T, Yamano K. Multiple pathways for mitochondrial protein traffic. Biol Chem. 2009;390:723–730. doi: 10.1515/BC.2009.087. [DOI] [PubMed] [Google Scholar]
- 3.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 4.Chacinska A, et al. Mitochondrial presequence translocase: Switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Popov-Čeleketić D, Mapa K, Neupert W, Mokranjac D. Active remodeling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 2008;27:1469–1480. doi: 10.1038/emboj.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamura Y, et al. Tim23–Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J Cell Biol. 2009;184:129–141. doi: 10.1083/jcb.200808068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lithgow T, Junne T, Suda T, Gratzer S, Schatz G. The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc Natl Acad Sci USA. 1994;91:11973–11977. doi: 10.1073/pnas.91.25.11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakai M, Endo T. Identification of yeast MAS17 encoding the functional counterpart of the mitochondrial receptor complex protein MOM22 of Neurospora crassa. FEBS Lett. 1995;357:202–206. doi: 10.1016/0014-5793(94)01362-5. [DOI] [PubMed] [Google Scholar]
- 9.Abe Y, et al. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- 10.Saitoh T, et al. Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J. 2007;26:4777–4787. doi: 10.1038/sj.emboj.7601888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolliger L, Junne T, Schatz G, Lithgow T. Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 1995;14:6318–6326. doi: 10.1002/j.1460-2075.1995.tb00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamano K, et al. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J Biol Chem. 2008;283:3799–3807. doi: 10.1074/jbc.M708339200. [DOI] [PubMed] [Google Scholar]
- 13.Künkele KP, et al. The preprotein translocation channel of the outer membrane of mitochondria. Cell. 1998;93:1009–1019. doi: 10.1016/s0092-8674(00)81206-4. [DOI] [PubMed] [Google Scholar]
- 14.Model K, et al. Protein translocase of the outer mitochondrial membrane: role of import receptors in the structural organization of the TOM complex. J Mol Biol. 2002;316:657–666. doi: 10.1006/jmbi.2001.5365. [DOI] [PubMed] [Google Scholar]
- 15.Model K, Meisinger C, Kühlbrandt W. Cryo-electron microscopy structure of a yeast mitochondrial preprotein translocase. J Mol Biol. 2008;383:1049–1057. doi: 10.1016/j.jmb.2008.07.087. [DOI] [PubMed] [Google Scholar]
- 16.van Wilpe S, et al. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- 17.Mayer A, Neupert W, Lill R. Mitochondrial protein import: Reversible binding of the presequence at the trans side of the outer membrane drives partial translocation and unfolding. Cell. 1995;80:127–137. doi: 10.1016/0092-8674(95)90457-3. [DOI] [PubMed] [Google Scholar]
- 18.Rapaport D, Neupert W, Lill R. Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J Biol Chem. 1997;272:18725–18731. doi: 10.1074/jbc.272.30.18725. [DOI] [PubMed] [Google Scholar]
- 19.Moczko M, et al. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol Cell Biol. 1997;17:6574–6584. doi: 10.1128/mcb.17.11.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanamori T, et al. Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc Natl Acad Sci USA. 1999;96:3634–3639. doi: 10.1073/pnas.96.7.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esaki M, et al. Mitochondrial protein import: Requirement of the presequence elements and TOM components for precursor binding to the TOM complex. J Biol Chem. 2004;279:45701–45707. doi: 10.1074/jbc.M404591200. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto H, et al. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell. 2002;111:519–528. doi: 10.1016/s0092-8674(02)01053-x. [DOI] [PubMed] [Google Scholar]
- 23.Geissler A, et al. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell. 2002;111:507–518. doi: 10.1016/s0092-8674(02)01073-5. [DOI] [PubMed] [Google Scholar]
- 24.Mokranjac D, et al. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 2003;22:816–825. doi: 10.1093/emboj/cdg090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin JW, et al. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto H, et al. Dual role of the receptor Tom20 in specificity and efficiency of protein import into mitochondria. Proc Natl Acad Sci USA. 2011;108:91–96. doi: 10.1073/pnas.1014918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Likić VA, et al. Patterns that define the four domains conserved in known and novel isoforms of the protein import receptor Tom20. J Mol Biol. 2005;347:81–93. doi: 10.1016/j.jmb.2004.12.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.