Abstract
Bacterial cell surfaces are commonly decorated with a layer formed from multiple copies of adhesin proteins whose binding interactions initiate colonization and infection processes. In this study, we investigate the physical deformability of the UspA1 adhesin protein from Moraxella catarrhalis, a causative agent of middle-ear infections in humans. UspA1 binds a range of extracellular proteins including fibronectin, and the epithelial cellular receptor carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1). Electron microscopy indicates that unliganded UspA1 is densely packed at, and extends about 800 Å from, the Moraxella surface. Using a modified atomic force microscope, we show that the adhesive properties and thickness of the UspA1 layer at the cell surface varies on addition of either fibronectin or CEACAM1. This in situ analysis is then correlated with the molecular structure of UspA1. To provide an overall model for UspA1, we have determined crystal structures for two N-terminal fragments which are then combined with a previous structure of the CEACAM1-binding site. We show that the UspA1–fibronectin complex is formed between UspA1 head region and the 13th type-III domain of fibronectin and, using X-ray scattering, that the complex involves an angular association between these two proteins. In combination with a previous study, which showed that the CEACAM1–UspA1 complex is distinctively bent in solution, we correlate these observations on isolated fragments of UspA1 with its in situ response on the cell surface. This study therefore provides a rare direct demonstration of protein conformational change at the cell surface.
Keywords: atomic force microscopy, bacterial adhesins, small-angle X-ray scattering, X-ray crystallography
Bacteria, and all other cells, primarily sense and respond to their environment via proteins extending from their surfaces. For pathogenic bacteria, such interactions form the initial events in infection processes. Whereas there have been extensive studies of the binding activities of cell surface proteins, remarkably little is known of how these molecular associations may translate into physical or conformational changes, in particular in situ directly at the cell surface. Physical changes that accompany receptor binding are widely believed to form an important contribution to biological function, but have rarely been measured directly.
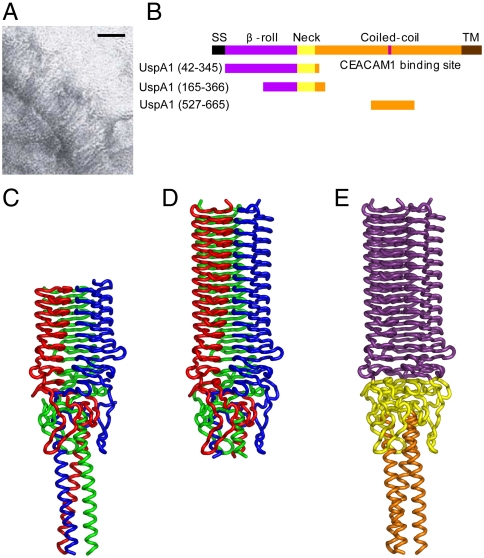
Moraxella catarrhalis (Mx) is a human-specific bacterium associated with respiratory diseases and middle-ear infections (otitis media) (1). Studies of the initial molecular steps by which Mx sense and interact with their environment, leading to infection, focus on surface bacterial adhesin proteins that provide obvious points of mediation for both colonization and infection, and may also assist in the evasion of immune recognition (2). Mx provide an excellent system in which to study the properties of adhesins in situ because electron micrographs (Fig. 1A) indicate their surfaces are dominated by multiple copies of a single, and large, adhesin: the ubiquitous surface protein A1 (UspA1). UspA1 varies in size between different strains of Mx, with each monomer usually comprising between 850–950 amino acids. These monomers form elongated, trimeric structures extending up to 800 Å from the cell surface, where they form a distinctive and densely packed molecular coating (3). Sequence homology studies (3, 4) indicate that UspA1 belongs to a family of bacterial adhesins termed either oligomeric coiled-coil adhesins (Oca) or trimeric autotransporter adhesins (TAAs). These adhesins are believed to share a modular structure consisting of a variable β-propeller amino terminal head group, extended coiled-coil stalk region, and a membrane β-barrel anchor domain at the carboxy terminus (reviewed in ref. 5). This latter domain forms a membrane pore from which the remaining passenger domains are believed to be translocated across the outer membrane (reviewed in ref. 6). Current understanding of TAAs is based on a range of molecular (crystallographic) structures that have been determined for isolated segments of some of these proteins (reviewed in refs. 5 and 6; also see refs. 4, 7, and 8). Because these proteins form large and extended structures, there are no complete structures of a TAA, but electron microscopy studies show these adhesins often form fully extended “lollipop” structures at the cell surface (3, 7) (Fig. 1). The similar overall modular structure of the TAA proteins suggests a common evolutionary precursor, but it is also clear that considerable diversification has taken place.
Fig. 1.
Moraxella cell surface, domain organization, and head region structure of UspA1. (A) Electron micrograph showing extended UspA1 molecules at the Mx surface. (Scale bar: 50 nm.) (B) Schematic showing full length UspA1 comprises five regions, each colored separately and labeled. Constructs for which crystal structures are determined are also shown. TM, transmembrane. (C–E) Ribbon representations of the crystal structures of (C) UspA1(165–366), (D) UspA1(42–345), and (E) composite model of UspA1(42–366). In C and D, each chain is colored separately; E is colored by domain as per B.
Multiple binding partners have been reported for most of these bacterial adhesins. In the case of UspA1, binding to both fibronectin (Fn) and laminin, components of the extracellular matrix, has been reported for constructs that include the head domain along with significant stretches of the stalk (9, 10). UspA1 has additionally been reported to bind with high affinity to the human carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) receptor (CD66a) on epithelial cells (11), which normally forms homophilic cell–cell interactions, and for which the binding site on UspA1 resides within the stalk (7) in a region spatially distant from the head domain. A working model emerging from this spatial separation of a range of binding sites is that the head domain may mediate initial, weak encounters with extracellular components leading to the exposure of more specific binding sites elsewhere that promote tighter binding required to trigger infection processes (7). In the crowded and densely packed layers of UspA1 at the Mx cell surface (Fig. 1), access to the full range of binding sites would be expected to be restricted, and is likely to require substantial physical distortions of the adhesin layer and its component molecules.
In this study, we aimed to explore the physical properties of the UspA1 layer on the Mx surface, how these change in response to binding of Fn or CEACAM1, and then to correlate these in situ effects with molecular details of the modular adhesin structure and its complexes with these proteins.
Results and Discussion
Mechanical Properties of UspA1 at the Bacterial Surface.
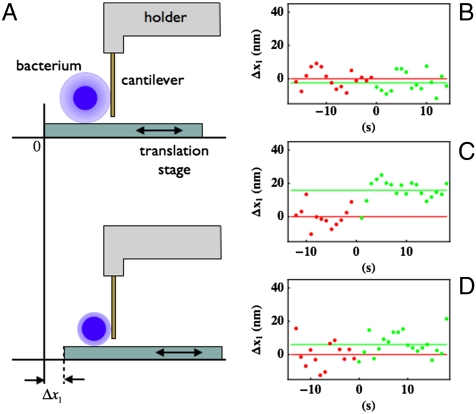
Electron micrographs have previously demonstrated that UspA1 forms the dominant component of, and is densely packed at, the Mx cell surface (3, 7) (Fig. 1) where it forms a layer approximately 80-nm thick, which is significant relative to the average size of the bacterium (ca. 800 nm). We therefore assumed that adhesive and mechanical responses to a cantilever when pressed against a single bacterium would reflect the response of UspA1 to this force sensor. These responses were tested in a force microscope with a vertically mounted Si3N4 microcantilever, the lateral molecular-force microscope (LMFM), as described in Materials and Methods. The LMFM measurements are made by observing the dampening in the thermal fluctuations of the vertical cantilever as it makes initial contact with the bacterium surface. As the cantilever comes into contact with the bacterium surface, the stiffness of the combined system increases and this is used to determine the contact point, without applying any significant force to the bacterium. Relative changes in this initial contact point, which should be independent of any compression of the cell, are used to measure the effective thickness of the adhesin layer.
Across a range of individual bacterial cells tested, an adhesion force was experienced on contact with the LMFM cantilever (Fig. 2; also see Movies S1–S4). This force was routinely found to be less than 300 pN. Because the Si3N4 cantilevers are hydrophilic and negatively charged under the buffer conditions (12), the adhesion is likely due to a charge effect at the Mx surface. From changes in the thermal fluctuations of the Si3N4 cantilever upon contact with the Mx surface, it was possible to calculate the apparent stiffness (13) of the adhesin layer, which was found to be less than 10 pN/nm. Furthermore, we noted that the apparent stiffness is smaller when the cantilever pushes against the adhesin layer than when it is stretched. In selected cases, the rupture of the adhesion between the cantilever and the bacterium was a single event with a force smaller than 100 pN, suggesting that the observed events were because of a single adhesin molecule. These forces correlate with other measurements of cell adhesins (14) and are substantially less than the forces required to deform a cell (15).
Fig. 2.
Atomic force microscopy analysis of Mx bacteria. (A) A sinusoidal sideways movement is imparted to the sample stage in order to gently push a single adsorbed bacterium (blue sphere; pale-blue outer layer represents surface adhesins) against the cantilever. The change in the sample stage position (Δx1) required to achieve contact is measured upon addition of either ligand (CEACAM1 or Fn) or buffer/control protein. The graphs on the right show representative data measured for the change in the contact point position before (in red) and after (in green) the addition of (B) control, (C) CEACAM1, and (D) FNIII12,13,14. The red and green lines represent the mean value of the red and green points, respectively.
We then proceeded to investigate how these properties of the UspA1 layer might change in response to the presence of known UspA1 binding proteins, specifically CEACAM1 and fibronectin. First, in control experiments, it was observed that addition of buffer or a control protein, a recombinant form of Plasmodium falciparum lactate dehydrogenase, to the experimental chamber did not produce any observable changes in the position of the contact point or in the adhesion force. However, in Mx strain MX2 cells, the addition of a truncated fragment of Fn (FnIII12–14) that binds the UspA1 head region (see below) resulted in an increase in the distance the bacterium needed to be moved in order to establish contact with the cantilever by 7 ± 5 nm (standard deviation of the mean, SDM), and a complete loss of adhesion force in 90% of contacts. These results indicate that Fn-specific binding to UspA1 can be detected by the LMFM. The simplest explanation for the change in the contact point position, relative prior to the addition of Fn, is a reduction in the overall size (diameter) of the MX2 bacteria when Fn binds. Because the adhesive properties also changed, we deduced that the reduction is likely to correlate with a conformational change in the UspA1 layer, potentially of UspA1 from its extended form to a more compact or “bent” form.
These analyses were then extended through the addition of CEACAM1 which binds UspA1 with high affinity. Within 5 s of the addition of CEACAM1 to MX2 strain cells, in which UspA1 includes a CEACAM1-binding site in the stalk region (7, 11), an average reduction of 16 ± 5 (SDM) nm in the overall size of the MX2 bacterium was observed. A significant reduction in adhesion force was also noted (70% of contacts did not produce any adhesion). However, when the same amount of CEACAM1 was added to O35E strain Mx, in which UspA1 lacks the CEACAM1-binding site (7), no change in the size of the bacterium was recorded and the adhesion force was unaffected (see Movie S4). These results act as a further control and again indicate that the LMFM is detecting CEACAM1-specific binding to UspA1.
Finally, we measured the effect of first adding FnIII12–14 and subsequently CEACAM1 (and vice versa) to the MX2. In each case, an initial size reduction was observed on addition of the first protein (as above), which was then followed by a further contraction of the cell by about 10 nm on addition of CEACAM1 after Fn (but not vice versa), giving a maximal contraction equivalent to that obtained with CEACAM1 alone. These results indicate that the mechanism for the observed contraction in each case is independent, consistent with the Fn- and CEACAM1-binding regions on UspA1 being at distal sites, but that the CEACAM1 distortion is dominant.
Overall, the LMFM data indicate that the physical properties of the UspA1 layer at the Mx cell surface change in response to binding of specific proteins. In particular, these in situ data are consistent with the correlated bending previously noted for an isolated UspA1 stalk fragment on binding CEACAM1 (7). The data also show that the extracellular protein fibronectin instigates similar changes. Because direct structural data on UspA1 was limited to the CEACAM1 site of the stalk region, we therefore decided to probe the UspA1–fibronectin interaction in more detail by first extending the structural analysis of UspA1 into the reported site of Fn binding, the head region.
Structure of the UspA1 Head Region.
Because of the high molecular masses and extreme molecular elongation of both UspA1 and Fn, detailed molecular analysis of their interactions first required the generation of truncated forms of each protein (Fig. S1). Crystals of a range of UspA1 constructs were obtained, but only two of these, UspA1(42–345) and UspA1(165–366), proved suitable for structure determination. The two structures obtained, each about 110 Å in length with a diameter up to 40 Å (Fig. 1 C–E), could be readily combined to form a composite structure (Fig. 3C) for UspA1 residues 42–366, which is about 160 Å in length.
Fig. 3.
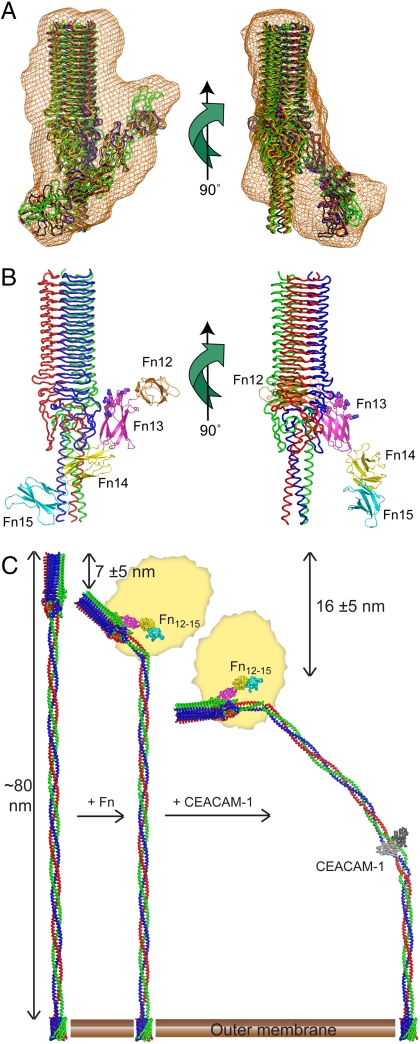
SAXS analysis of UspA1(42–366)-FnIII12–15 complex. (A) Molecular envelope (orange) defined by ab initio bead modeling, with five independent rigid-body reconstructions shown as Cα ribbons, with each reconstruction displayed in a different color. (B) Representative rigid-body reconstruction of UspA1–Fn complex, with UspA1(165–366) colored as in Fig. 1C and FnIII12–15 as a Cα cartoon colored by domain and labeled. Heparin-binding residues are shown as spheres. (C) Overall model for UspA1(154–869) (colored as in Fig. 1C) and deformation associated with sequentially binding Fn (colored as in B) and CEACAM-1 (in gray). The remainder of the Fn molecule is symbolized as a rough yellow ellipsoid (not to scale). The distances and uncertainties shown are from the LMFM analysis and represent the decrease in overall UspA1 length resulting from binding of each ligand. The bending angle on binding CEACAM1 required to achieve this change has been calculated from the linear dimensions of the combined crystal structures and is estimated to be approximately 45 ± 15°, which is within the range of movement observed in a prior molecular dynamics simulation (7).
The UspA1 head domain (residues 42–275) is globally similar to other structures of TAA head domains (Fig. S2) and comprises a trimeric left-handed parallel β-roll, which is formed from 14 repeating 14–16 residue segments that form a ladder of β-strand coils. This head domain differs from YadA (16) in the variability of its repeats (with 9 × 15, then 3 × 14, then 2 × 16 residues per repeat).
The neck region of UspA1 (residues 276–334) features a complex intertwining arrangement that differs from that observed in YadA but is similar to the neck region of BpaA (Fig. S2). The large external loops form a concentration of positive charge in this region that may be indicative of ligand-binding functional activity, and to which sulphate ions are observed to be fortuitously bound in one of the crystal structures. The structure then features the beginning of the stalk (residues 334–372), which extends approximately 60 Å (UspA1(42–366)) as a canonical left-handed coiled coil.
The Fibronectin–UspA1 Complex.
Having determined the structure of the UspA1 head region, we then investigated how this region accommodated a binding site for fibronectin. Following on from earlier reports that extracellular Fn bound to the N-terminal portion of UspA1 (9, 10), we initially undertook binding studies with truncated forms of both proteins (Fig. S1) to better define the Fn–UspA1 complex.
The binding studies used immunoblotting (dot blots), ELISA, and surface plasmon resonance and are fully described in SI Text and Figs. S3 and S4. The consensus of these assays was that FnIII13 plays a key role in binding to the UspA1 region contained in the construct UspA1(42–366) and provides approximately 90% of the binding energy and FnIII12 ∼ 10% (at 25 °C) based on the dissociation constants summarized in Table S1.
This central role for FnIII13 in binding was then validated by heparin competition assays which showed that, with increasing heparin concentration, UspA1 binding to Fn decreased. The heparin-binding site on Fn has previously been characterized in detail and centers on Arg-6, Arg-7, and Arg-23 on FnIII13 (17, 18). Heparin did not bind significantly to UspA1 itself, consistent with a previous observation (19).
The binding data indicated that a moderate affinity (Kd ∼ 5 μM) complex is formed between UspA1(42–366) and the FnIII12–15 fragment. In the absence of crystals of this complex, low-resolution solution X-ray scattering data were collected (Fig. S5) from which a molecular envelope was calculated to which the crystal structures of both components were then fitted (Fig. 3). The distinctive shape of the UspA1 head domain is evident in the outline of the complex. The observed data were poorly fitted by alternative models such as dimers of either component (Fig. S5). Due to the similarity of the FnIII domains, there is potential ambiguity in the polarity of the orientation of the FnIII12–15 fragment, but the orientation shown in the figure is preferred because it maximizes contacts between FnIII13 and UspA1 in accordance with the binding data. UspA1 contacts are predominantly at the base of the β-roll head domain, the site where the extended turn 1 loops of the β-rolls and the loops from the neck region are found.
A notable feature of this low-resolution model of the complex is the distinctive angular (approximately 40°) association between the long axes of each component. Because in the intact Fn protein there is considerable protein mass on both sides of the FnIII13 domain, it is difficult to envisage that this angular contact with UspA1 could be generated without distorting a close array of extended adhesin structures.
An Overall Model for UspA1 Structure, Binding, and Function.
On the bacterial surface, each monomer of UspA1 comprises 821 amino acids for which crystal structures are now available for segments 42–366 (this study) and 527–665 (7). Importantly, these two segments cover two of the main binding sites on UspA1. Although the crystal structures represent only about 60% of the overall structure, the remaining parts of the structure were readily amenable to homology modeling: There is strong coiled-coil propensity for most of the remaining sequence (11) and the transmembrane region is very highly conserved and expected to retain a similar structure to the Hia adhesin (20). An overall composite model for UspA1 was therefore constructed (Fig. 3C).
This overall model for UspA1 bears structural similarity to that previously proposed for YadA (21) although it varies in detail, dimension, and, importantly, can be combined with functional data that indicate responsiveness to ligand binding. From the model, in its fully extended conformation, UspA1 extends 760 Å in length from the membrane, which correlates with earlier electron microscopy estimates (3). These same electron microscopy studies and others (7) indicate that unliganded UspA1 is often both fully extended and densely packed at the bacterial surface such that the stalk and head regions of adjacent UspA1 molecules lie in parallel and in close proximity. This arrangement appears to (partially) preclude access to the Fn-binding site at the base of the head domain, and more fully to the CEACAM1-binding site that is some 400-Å distant from the tip of the molecule. This close packing may provide some physical degree of protection of these conserved regions from immune recognition (2, 7).
Mx targeting epithelial cells would encounter increasingly higher local concentrations of fibronectin as epithelia are approached, because Fn is a major component of extracellular matrix and also adheres to the apical surface of epithelia (22, 23). Because the Fn-binding site is at the base of the head domain, access for a large molecule such as Fn (a 440-kDa dimer) is only possible if the close parallel UspA1 packing is disrupted. Bending of the UspA1 head (Fig. 3C), perhaps initiated by physical contact, makes this site accessible to extracellular proteins. The angular association observed between UspA1 and Fn may reflect the degree of bending required to fully expose the binding site at the surface. The small decrease in overall Mx diameter on addition of Fn measured in this study is consistent with distortion of the UspA1 layer, most easily achieved through bending of the extended UspA1 structure. The binding assays show these initial interactions are of moderate affinity (ca. 5 μM) reflecting the extensive opportunities for multivalent interactions provided by the high numbers of UspA1 exposed at the bacterial cell surface.
This disruption also correlates with previous experimental data suggesting that binding of the high-affinity human CEACAM1 receptor, which takes place in the lower half of the stalk region and closer to the membrane, is associated with distinctive bending of the UspA1 stalk (7). In this previous study, the isolated CEACAM1∶UspA1 stalk complex was found to exhibit pronounced curvature in solution, with molecular dynamics simulations suggesting complex formation destabilizes the extended rod-like conformation of the stalk region. It is unclear if this distortion precedes, accompanies, or results from CEACAM1 binding, although a concerted action seems most likely. The experiments performed with the LMFM in the current study provide indirect evidence that such bending also occurs on the Mx cell surface. These studies show a correlation between the addition of CEACAM1 and a diminution of adhesion and reduction in overall Mx diameter, which implies that the extended UspA1 structure (which determines the thickness of the adhesin layer) is physically distorted by CEACAM1 binding. The measured reduction of 16 ± 5 (SDM) nm in overall diameter (Fig. 2) is similar to the expected change generated (Fig. 3C) by bending of UspA1 by the amounts previously proposed (7) by modeling studies. The broad correlation between these measurements supports the simplest explanation that the extended—and hence compressible—layer of UspA1 molecules becomes more compact on CEACAM1 binding, as would be expected if the stalks were bent. These changes were not observed on the addition of a control protein, implying they are generated by specific interactions of UspA1 with both Fn and CEACAM1.
Contact deformation of the UspA1 structure is perhaps unsurprising in view of its extreme elongation. It is more surprising that bending can be triggered, or at least enhanced, through binding of specific proteins. This direct correlation implies that the induced distortion may arise in or close to the binding regions of UspA1 (Fig. 3C). Previous studies of the UspA1:CEACAM1 interaction have suggested that adherence of CEACAM1 may interrupt the symmetry of the extended coiled-coil stalk leading to destabilization of the extended structure (7). The separate and lower degree of bending observed with Fn binding may simply reflect torsional distortion resulting from the angular association of these molecular components. It is interesting to speculate on the biological significance of these conformational changes. A clear consequence of UspA1 distortion is compaction of the protein layer at the bacterial surface, allowing closer approach of the bacterial and host cell membranes, a requirement for effective infection to proceed. Further, as noted above, this primitive physical deformation switch provides a simple way of locating both the Fn- and, particularly, CEACAM1-binding sites away from the most distal and hence accessible regions of UspA1, which may provide some degree of evasion from immune recognition. Although the data indicate the distortion resulting from Fn and CEACAM1 binding are independent events, a sequential model in which Fn binding initiates bending to facilitate CEACAM1 binding is attractive.
Finally, because binding to cell surface proteins is intimately associated with effective bacterial infection, a direct role for these mechanoselective deformations in the infection process is inferred. Further in situ studies of these dynamic properties of adhesins and related molecules will require the development of different technical approaches.
Materials and Methods
Protein Production, Purification, and Structure Determination.
A series of truncated forms of Mx UspA1 from strain MX2 ATCC 25238 and recombinant human fibronectin constructs (Fig. S1) were expressed in Escherichia coli and purified as described in SI Text. Crystals of UspA1(42–366) and UspA1(42–345) were obtained as described in SI Text. Diffraction data were collected at the Diamond Light Source (beamlines IO2 and IO4), and structures were solved using a combination of molecular replacement and experimental phasing from bromide soaks as described in SI Text. Data collection and final refinement statistics for both structures are summarized in Table S2. Homology models were constructed for the regions of UspA1 not covered by the available crystal structures, as described in SI Text.
Binding Studies.
The interactions of UspA1 and Fn fragments were studied by standard methods using plasmon resonance, dot Immunoblot, and enzyme-linked immunosorbent assays as described in SI Text.
Small-Angle X-Ray Scattering (SAXS) Analysis.
SAXS data of the FnIII12–15/UspA1(42–366) complex were collected at the Diamond Light Source (beamline I22) and analyzed as described in SI Text.
Lateral Molecular-Force Microscope.
The mechanical properties of UspA1 at the outer Mx surface were tested using a newly developed LMFM, described in ref. 13. The spring constant of the cantilevers used in this study was 2.4 pN/nm ± 15% and the cross-section was 1 μm ± 10%. To determine UspA1 stiffness, softer cantilevers were used with the same cross-section but of double length, resulting in a stiffness of 2.7 pN/nm ± 15%.
Single bacteria were detected optically using total internal reflection illumination. A sinusoidal sideways movement was imparted to the sample stage in order to gently push a single bacterium against the cantilever. The position of the bacterium was adjusted to produce a maximum deflection of the cantilever of between 50 and 300 nm. The position of the stage when contact was established between the bacterium and the cantilever was monitored on the addition of control protein/buffer, CEACAM1, and FnIII12–14. Changes in these positions can be directly correlated to changes in the size of the bacterium.
Coordinate Deposition.
Coordinates and structure factors of the UspA1(165–366) and UspA1(42–345) crystal structures have been deposited with the Research Collaboratory for Structural Bioinformatics Protein Data Bank (ID codes 3NTN and 3PR7), and coordinates of the models for the fibronectin–UspA1 complex and of the entire UspA1 structure are available from the authors.
Supplementary Material
Acknowledgments.
We thank the staff at the Diamond Light Source for providing support for X-ray diffraction and SAXS data collection, and Prof. Martin Humphries (University of Manchester, United Kingdom) for the constructs FnIII12–15, FnIII12-IIICS-15, and FnIII6–10. These studies were supported by grants from the Wellcome Trust (07746), UK Biological and Biotechnology Research Council (BBSRC) (BB/F007256), and were partly conducted in the Spencer Dayman Meningitis Laboratories. C.A. is supported by a studentship provided by the BBSRC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3NTN and 3PR7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106341108/-/DCSupplemental.
References
- 1.Karalus R, Campagnari A. Moraxella catarrhalis: A review of an important human mucosal pathogen. Microbes Infect. 2000;2:547–559. doi: 10.1016/s1286-4579(00)00314-2. [DOI] [PubMed] [Google Scholar]
- 2.Hultgren SJ, et al. Pilus and nonpilus bacterial adhesins: Assembly and function in cell recognition. Cell. 1993;73:887–901. doi: 10.1016/0092-8674(93)90269-v. [DOI] [PubMed] [Google Scholar]
- 3.Hoiczyk E, Roggenkamp A, Reichenbecher M, Lupas A, Heesemann J. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 2000;19:5989–5999. doi: 10.1093/emboj/19.22.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szczesny P, et al. Structure of the head of the Bartonella adhesin BadA. PLoS Pathog. 2008;4:e1000119. doi: 10.1371/journal.ppat.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wells TJ, Tree JJ, Ulett GC, Schembri MA. Autotransporter proteins: Novel targets at the bacterial cell surface. FEMS Microbiol Lett. 2007;274:163–172. doi: 10.1111/j.1574-6968.2007.00833.x. [DOI] [PubMed] [Google Scholar]
- 6.Cotter SE, Surana NK, St Geme JW., 3rd Trimeric autotransporters: A distinct subfamily of autotransporter proteins. Trends Microbiol. 2005;13:199–205. doi: 10.1016/j.tim.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Conners R, et al. The Moraxella adhesin UspA1 binds to its human CEACAM1 receptor by a deformable trimeric coiled-coil. EMBO J. 2008;27:1779–1789. doi: 10.1038/emboj.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards TE, et al. Structure of a Burkholderia pseudomallei trimeric autotransporter adhesin head. PLoS One. 2010;5:e12803. doi: 10.1371/journal.pone.0012803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan TT, Nordstrom T, Forsgren A, Riesbeck K. The respiratory pathogen Moraxella catarrhalis adheres to epithelial cells by interacting with fibronectin through ubiquitous surface proteins A1 and A2. J Infect Dis. 2005;192:1029–1038. doi: 10.1086/432759. [DOI] [PubMed] [Google Scholar]
- 10.Tan TT, Forsgren A, Riesbeck K. The respiratory pathogen Moraxella catarrhalis binds to laminin via ubiquitous surface proteins A1 and A2. J Infect Dis. 2006;194:493–497. doi: 10.1086/505581. [DOI] [PubMed] [Google Scholar]
- 11.Hill DJ, Edwards AM, Rowe HA, Virji M. Carcinoembryonic antigen-related cell adhesion molecule (CEACAM)-binding recombinant polypeptide confers protection against infection by respiratory and urogenital pathogens. Mol Microbiol. 2005;55:1515–1527. doi: 10.1111/j.1365-2958.2005.04487.x. [DOI] [PubMed] [Google Scholar]
- 12.Senden TJ, Drummond KJ. Surface chemistry and tip-sample interactions in atomic force microscopy. Colloids Surfaces A Physicochem Eng Asp. 1995;94:29–51. [Google Scholar]
- 13.Scholz T, et al. Processive behaviour of kinesin observed using micro-fabricated cantilevers. Nanotechnology. 2011;22:095707. doi: 10.1088/0957-4484/22/9/095707. [DOI] [PubMed] [Google Scholar]
- 14.Dupres V, et al. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat Methods. 2005;2:515–520. doi: 10.1038/nmeth769. [DOI] [PubMed] [Google Scholar]
- 15.Lulevich V, Zimmer CC, Hong HS, Jin LW, Liu GY. Single-cell mechanics provides a sensitive and quantitative means for probing amyloid-beta peptide and neuronal cell interactions. Proc Natl Acad Sci USA. 2010;107:13872–13877. doi: 10.1073/pnas.1008341107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nummelin H, et al. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 2004;23:701–711. doi: 10.1038/sj.emboj.7600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Busby TF, et al. Heparin binding by fibronectin module III-13 involves six discontinuous basic residues brought together to form a cationic cradle. J Biol Chem. 1995;270:18558–18562. doi: 10.1074/jbc.270.31.18558. [DOI] [PubMed] [Google Scholar]
- 18.Bloom L, Ingham KC, Hynes RO. Fibronectin regulates assembly of actin filaments and focal contacts in cultured cells via the heparin-binding site in repeat III13. Mol Biol Cell. 1999;10:1521–1536. doi: 10.1091/mbc.10.5.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMichael JC, et al. Isolation and characterization of two proteins from Moraxella catarrhalis that bear a common epitope. Infect Immun. 1998;66:4374–4381. doi: 10.1128/iai.66.9.4374-4381.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng G, Surana NK, St Geme JW, 3rd, Waksman G. Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 2006;25:2297–2304. doi: 10.1038/sj.emboj.7601132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koretke KK, Szczesny P, Gruber M, Lupas AN. Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica. J Struct Biol. 2006;155:154–161. doi: 10.1016/j.jsb.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Abraham SN, Beachey EH, Simpson WA. Adherence of Streptococcus pyogenes, Escherichia coli, and Pseudomonas aeruginosa to fibronectin-coated and uncoated epithelial cells. Infect Immun. 1983;41:1261–1268. doi: 10.1128/iai.41.3.1261-1268.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simpson WAaB, Edwin H. Adherence of group A streptococci to fibronectin on oral epithelial cells. Infect Immun. 1983;39:275–279. doi: 10.1128/iai.39.1.275-279.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.