Abstract
Type 1 diabetes (T1D) is a complex disease characterized by the loss of insulin-secreting β-cells. Although the disease has a strong genetic component, and several loci are known to increase T1D susceptibility risk, only few causal genes have currently been identified. To identify disease-causing genes in T1D, we performed an in silico “phenome–interactome analysis” on a genome-wide linkage scan dataset. This method prioritizes candidates according to their physical interactions at the protein level with other proteins involved in diabetes. A total of 11 genes were predicted to be likely disease genes in T1D, including the INS gene. An unexpected top-scoring candidate gene was huntingtin-interacting protein (HIP)-14/ZDHHC17. Immunohistochemical analysis of pancreatic sections demonstrated that HIP14 is almost exclusively expressed in insulin-positive cells in islets of Langerhans. RNAi knockdown experiments established that HIP14 is an antiapoptotic protein required for β-cell survival and glucose-stimulated insulin secretion. Proinflammatory cytokines (IL-1β and IFN-γ) that mediate β-cell dysfunction in T1D down-regulated HIP14 expression in insulin-secreting INS-1 cells and in isolated rat and human islets. Overexpression of HIP14 was associated with a decrease in IL-1β–induced NF-κB activity and protection against IL-1β–mediated apoptosis. Our study demonstrates that the current network biology approach is a valid method to identify genes of importance for T1D and may therefore embody the basis for more rational and targeted therapeutic approaches.
Keywords: autoimmunity, huntington's disease, protein network, bioinformatics, programmed cell death
Type 1 diabetes (T1D) is a polygenetic multifactorial autoimmune disease in which the insulin-producing pancreatic β-cells in the islets of Langerhans are destroyed during an inflammatory process known as insulitis (1). Genes in the HLA region are by far the most important T1D susceptibility genes (2, 3). The largest and most recent genome-wide association and linkage studies revealed more than 40 genomic regions with significant association to T1D (4, 5). These insights, however, do not lead to identification of the causative variants and the specific disease genes in these regions. The main challenge is that most of the identified risk loci contain several genes, and prioritization of these is difficult.
An increasing number of integrative bioinformatics tools have been applied to disease susceptibility data with the aim of prioritizing the positional candidate genes. This has mainly been performed by integrating genetic data and other data, such as gene expression or text mining (6–9). These approaches have recently been further developed on the assumption that cellular functions and disease phenotypes are regulated by the coordinated actions of proteins working together in protein complexes and pathways (10–13). Therefore, causal genetic variants in a complex disease such as T1D are likely to translate into changed functionality of a protein network or pathway causing a change in the biological and phenotypic outcome. These kinds of strategies have also been applied on T1D and T2D (14–16) but without biological and functional evaluation of candidate genes.
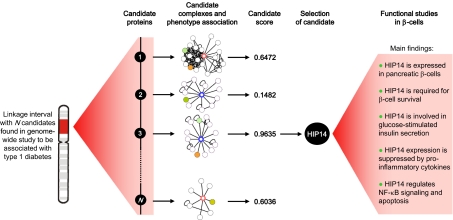
We developed a unique in silico “phenome–interactome network analysis” for predicting proteins involved in disease (17). Starting from high-confidence human protein–protein interaction data, networks for proteins encoded by genes in associated loci are generated. This is followed by scoring and ranking of the proteins according to phenotypic association to disease of the individual proteins in the network, and all candidates are awarded a posterior probability between 0 and 1 on the basis of these data (17). Thus, a candidate is considered likely to be involved in T1D if it interacts physically at the protein level (using only high-confidence interaction data) with proteins already known to be involved in diabetes risk or etiology. Candidates with such a profile will be awarded a high probability by the phenome–interacome prediction method. In the present study, we have applied this method on a T1D genome-wide linkage study (GWLS) dataset (3) to identify T1D candidate proteins. In addition to 10 other proteins, we identified Huntingtin-interacting protein (HIP)-14 as a candidate protein. Functional studies demonstrated that HIP14 is expressed in β-cells, is required for β-cell survival, and is a target of proinflammatory cytokines known to contribute to β-cell destruction in T1D.
Results
Phenome–Interactome Network Analysis Predicts 11 T1D Candidate Proteins.
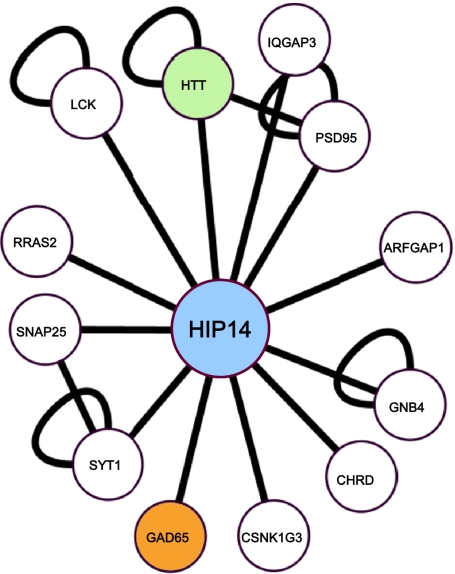
To identify unique disease genes/proteins in T1D, we performed a phenome–interactome network analysis (17) on a published T1D GWLS that revealed 11 genomic regions with linkage to T1D (3). Briefly, genes located within 10 Mb on each side of the most linked markers (Table 1) in the 11 regions were used as input for the in silico generation of protein–protein interaction networks. After the generation of these networks, disease phenotype association for all proteins in all networks was determined, allowing scoring and ranking of individual candidate proteins. A total of 11 proteins were predicted as candidate proteins (Table 1), and their protein complexes are shown in Fig. S1. Notably, insulin (INS), which is an established T1D candidate gene (18, 19), was the highest-scoring candidate protein predicted, thus providing proof of principle for the sensitivity of the method and underlining the feasibility of this approach. Another top-scoring candidate protein predicted by the method was HIP14. The network for HIP14 contained 12 protein interaction partners and is considered highly likely to be involved in T1D, because it interacts physically with the huntingtin (HTT) protein and glutamate decarboxylase 65 (GAD65), two proteins that phenotypically are associated with diabetes (Fig. 1).
Table 1.
T1D candidate proteins predicted by in silico phenome–interactome network analysis
| Candidate protein | Chromosome | Nearest marker | Score |
| INS, insulin | 11p15 | D11S922 | 0.9989 |
| GLS, glutaminase C | 2q-31-q33 | D2S2167 | 0.9969 |
| HIP14, huntingtin-interacting protein 14 | 12q14-q12 | D12S375 | 0.9635 |
| ASS1, arginino succinate synthetase 1 | 9q33-q34 | D9S260 | 0.6472 |
| HLA-DRA | 6q21 | TNFA 2 | 0.6405 |
| SNTB2, syntrophin β-2 | 16q22-q24 | D16S504 | 0.6036 |
| HLA-DRB1 | 6q21 | TNFA 1 | 0.3304 |
| PKCD, protein kinase C-δ | 3p13-p14 | D3S1261 | 0.3237 |
| JNK1, c-jun N-terminal kinase 1 | 10p14-q11 | D10S1426 | 0.1900 |
| AKD1, adenylate kinase domain containing 1 | 6q21 | D6S283 | 0.1577 |
| STX4, syntaxin 4 | 16p12-q11.1 | D16S3131 | 0.1482 |
A total of 11 candidate proteins with scores above 0.1 were predicted. Briefly, all proteins encoded by genes in linkage intervals were identified and used for in silico construction of protein networks according to protein–protein interaction information from several databases. Each protein in each network was then subjected to text-mining for association to diabetes or similar phenotype. Each network was then scored and ranked by a Bayesian predictor. Candidate proteins are listed according to their score.
Fig. 1.
Protein network for HIP14. The network consists of 12 interaction partners, of which 2 (GAD65 and HTT) are phenotypically associated with diabetes.
HIP14 was first identified in 2002 and is widely expressed but with highest expression in brain, where it colocalizes to Golgi and cytoplasmic vesicle membranes (20). HIP14 possesses palmitoyl transferase activity through a conserved cystein-rich domain (DHHC motif) (21, 22). In neurons, HIP14 has high substrate specificity for several vesicle-associated proteins, including synaptosomal-associated protein 25 (SNAP)-25, GAD65, postsynaptic density protein 95, synaptotagmin 1, and HTT (21, 23). On the basis of these findings, we decided to examine the potential functional role of HIP14 in the target tissue of T1D (i.e., the β-cells in the islets of Langerhans).
HIP14 Is Predominantly Expressed in β-Cells.
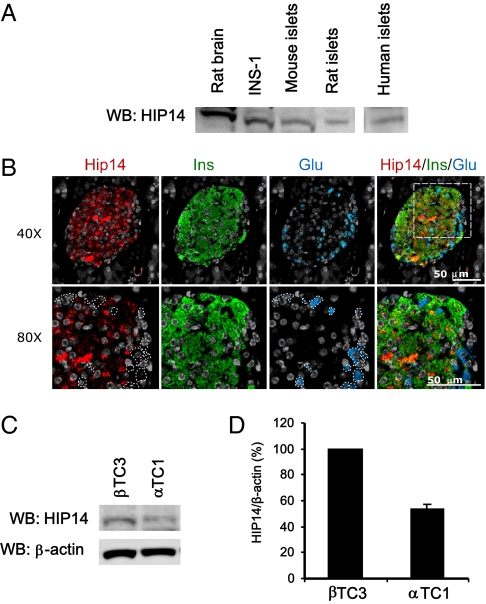
We first wished to demonstrate the presence of HIP14 in islets and β-cells. Western blotting of HIP14 on lysates from insulin-producing rat INS-1 cells and isolated islets from rat, mouse, and human demonstrated immunoreactive bands corresponding to HIP14, indicating that HIP14 is expressed in islets and β-cells (Fig. 2A). We then performed immunohistochemical analyses of mouse pancreatic sections, which confirmed staining of HIP14 in islets (Fig. 2B). Interestingly, HIP14 staining appeared to be mainly restricted to insulin-positive cells. Consistent with this notion, comparison of HIP14 protein levels in an insulin-secreting β-cell line (βTC3) vs. a glucagon-secreting α-cell line (αTC1) suggested that β-cells express more HIP14 than α cells (Fig. 2 C and D). Collectively, these results demonstrate that HIP14 protein is expressed in pancreatic β-cells and provide the basis for investigations of the functional role of HIP14 in β-cells.
Fig. 2.
Expression of HIP14 in pancreatic islets. (A) Expression of HIP14 in rat brain (positive control), INS-1 cells, and pancreatic islets from mouse, rat, and human. Whole-cell lysates were subjected to immunoblotting using an anti-HIP14 antibody. (B) Immunohistochemical stainings of mouse pancreatic sections using antibodies against HIP14, insulin, and glucagon. Nuclei staining was done using DAPI (gray). (C) Immunoblotting of HIP14 in a mouse β-cell line (βTC3) and a mouse α-cell line (αTC1). (D) Quantification of HIP14 protein normalized to β-actin (n = 2–3).
HIP14 Is Required for β-Cell Survival.
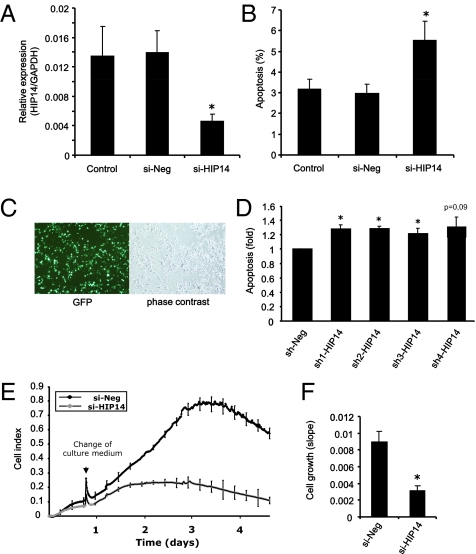
The hallmark of T1D is the induction of β-cell apoptosis by inflammatory mediators. We therefore sought to investigate whether HIP14 may play a functional role in the regulation of β-cell apoptosis. To examine this, we applied RNAi to knockdown the expression of HIP14. We used FACS-purified primary rat β-cells transfected with either negative control siRNA (si-Neg) or siRNA against HIP14 (si-HIP14). These experiments revealed that knockdown of HIP14 (≈70% knockdown efficiency; Fig. 3A) caused an increase in the apoptotic cell death rate by 83% (Fig. 3B). To substantiate this finding, we transfected INS-1 cells with expression vectors encoding four different shRNA molecules, which are processed intracellularly to active siRNAs targeting HIP14 mRNA. Transfection with a vector encoding GFP revealed a transfection efficiency of ≈50% (Fig. 3C). The experiments showed that each vector increased the basal apoptosis rate by ≈30% (Fig. 3D). In line with these observations, we found that INS-1 cells transfected with siRNA against HIP14 had a significantly slower growth rate than cells transfected with control siRNA (Fig. 3 E and F). These findings suggest that HIP14 possesses antiapoptotic properties and is required for survival of β-cells in vitro.
Fig. 3.
HIP14 is required for β-cell survival. (A) Efficiency of siRNA-mediated knockdown of HIP14 expression in FACS-purified primary β-cells was evaluated by real-time PCR. Data are means ± SEM of n = 5. (B) Primary FACS-purified rat β-cells were left untransfected (control) or transfected with negative control siRNA (si-Neg) or siRNA against HIP14 (si-HIP14). Apoptosis was evaluated using Hoechst/PI staining. Data are means ± SEM of n = 5. (C) INS-1 cells were transfected with pGFP. A transfection efficiency of approximately 50% was obtained. (D) INS-1 cells were transfected with negative control shRNA (sh-Neg) or four different shRNAs directed against HIP14 (sh-HIP14 1–4). Apoptosis was determined by Cell Death Detection ELISA measuring the presence of cytosolic histone–DNA complexes (nucleosomes). Data are means ± SEM of n = 4. *P < 0.05 vs. si-Neg. (E) INS-1 cell growth was measured in real time as described in Materials and Methods. Representative growth curves from cells transfected with si-Neg or si-HIP14 are shown. Each experiment was performed in triplicate. (F) Slopes of growth curves. Data are means ± SEM of n = 3. *P = 0.012.
HIP14 Is Required for Glucose-Stimulated Insulin Secretion.
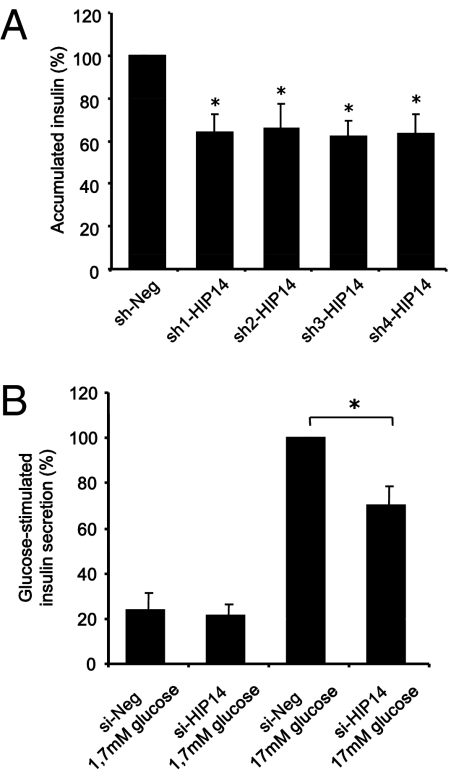
HIP14 plays an important role in the regulation of vesicle exocytosis in neurons (24). We therefore examined whether HIP14 is involved in the exocytosis of insulin granules in β-cells. We first measured accumulated insulin release to the culture medium in cells after HIP14 knockdown. INS-1 cells transfected with shRNAs against HIP14 secreted ≈35% less insulin to the medium compared with cells transfected with negative control shRNA (Fig. 4A). With a transfection rate of ≈50%, these results indicate that insulin release in sh-HIP14–transfected cells is reduced by ≈70%. To get further insight into the mechanisms underlying HIP14-dependent β-cell insulin release, we next transfected INS-1 cells with a mutant form of HIP14 (ΔHIP14), which lacks the DHHC domain necessary for the palmitoyl transferase activity. Similar to knockdown of HIP14, introduction of ΔHIP14 into INS-1 cells caused a reduction (≈25%) in chronic insulin secretion compared with mock-transfected cells (Fig. S2). Transfection of cells with a vector encoding full-length HIP14 caused a minor but significant decrease in insulin secretion (Fig. S2), reflecting that a too-high cellular level of HIP14 also causes some degree of impairment in the secretory activity.
Fig. 4.
HIP14 is required for insulin release. (A) INS-1 cells were transfected with negative control shRNA (sh-Neg) or four different shRNAs directed against HIP14 (sh-HIP14 1–4). Accumulated insulin during 24 h was measured in the culture medium. Data are means ± SEM of n = 5. *P < 0.05. (B) INS-1E cells were transfected with either si-Neg or si-HIP14. Two days after transfection, glucose-stimulated insulin secretion was determined by incubating the cells in either 1.7 mM or 17 mM glucose in Krebs-Ringer buffer or RPMI 1640 for 30 min. Results are expressed as percentage of GSIS in si-Neg. Data are means ± SEM of n = 4. P < 0.05.
We next performed glucose-stimulated insulin secretion (GSIS) experiments to validate the involvement of HIP14 in stimulated insulin release. For these experiments we used INS-1E cells, which are more glucose sensitive than parental INS-1 cells (25). These studies demonstrated that knockdown of HIP14 by siRNA decreased GSIS by ≈40% (Fig. 4B). We further evaluated the requirement of HIP14 for GSIS in RNAi experiments in which we used shRNAs against HIP14, which confirmed involvement of HIP14 in GSIS (Fig. S2). Glucose-induced release of human growth hormone (hGH) has previously been described as a measurement of exocytotic activity (26, 27). We therefore cotransfected INS-1E cells with a vector encoding hGH and with negative control siRNA or siRNA against HIP14 and measured the ability of glucose to stimulate release of hGH. These experiments further revealed that HIP14 is required for glucose-induced secretory activity (Fig. S2). Together, these results demonstrate that HIP14 is involved in the regulation of insulin secretion.
Cytokines Suppress the Expression of HIP14.
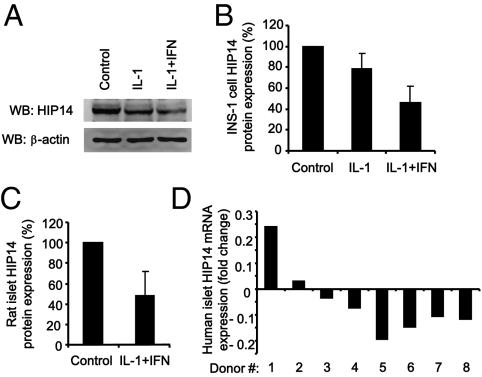
Proinflammatory cytokines, in particular IL-1β, contribute to β-cell dysfunction and apoptosis in T1D (28). We therefore next asked whether cytokines regulate the expression of HIP14 in β-cells. Compared with untreated control cells, exposure of INS-1 cells to IL-1β alone or IL-1β plus IFN-γ for 24 h caused a reduction in the cellular content of HIP14 (Fig. 5 A and B). In particular, the combination of IL-1β and IFN-γ resulted in marked suppression of HIP14 protein expression. Similar findings were obtained using isolated intact rat islets (Fig. 5C). We also examined HIP14 expression in human islets and found that cytokines decreased HIP14 expression in six of eight human islet donor preparations (Fig. 5D). Collectively these results indicate that the expression of HIP14 in β-cells is suppressed by proinflammatory cytokines.
Fig. 5.
Cytokines suppress the expression of HIP14. (A) INS-1 cells were left untreated or exposed to IL-1β (150 pg/mL) alone or IL-1β (150 pg/mL) plus IFN-γ (5 ng/mL) for 24 h. Whole-cell lysates were subjected to immunoblotting using anti-HIP14 and anti–β-actin antibodies. Representative blots of four individual experiments are shown. (B) HIP14 protein in INS-1 cells was quantified and normalized to β-actin. Data are means ± SEM of n = 4. (C) Isolated rat islets were left untreated or exposed to IL-1β (150 pg/mL) plus IFN-γ (5 ng/mL) for 24 h. Islet lysates were subjected to immunoblotting using an anti-HIP14 antibody. HIP14 protein was quantified and data are means ± SEM of n = 3. (D) Expression of HIP14 by real-time PCR in human islet preparations from eight donors left untreated or exposed to cytokines (75 U/mL IL-1β plus 750 U/mL IFN-γ plus 5,000 U/mL TNF-α) for 48 h. HIP14 transcripts were quantified and normalized to a geometric average of three housekeeping genes (GAPDH, 18S-RNA, and PPIA). Data are expressed as fold change after exposure to cytokines.
To address the underlying mechanism of cytokine-mediated down-regulation of HIP14 in β-cells, we examined whether microRNAs are involved in repressing HIP14 expression after cytokine treatment. Using two different microRNA target predictors publicly available (TargetScan 5.1 and Pictar), we searched for microRNAs targeting HIP14 mRNA. MicroRNA 146a (miR146a) was predicted by both predictors to have HIP14 as target. Notably, previous findings demonstrated that cytokines induce miR146a expression in clonal β-cells and isolated islets (29). In line with the prediction that HIP14 is a target of miR146a, transient transfection of INS-1 cells with miR146a resulted in a markedly reduced HIP14 content (Fig. S2), indicating that HIP14 indeed is targeted by miR146a in insulin-secreting cells. These data suggest that miR146a may contribute to cytokine-induced suppression of HIP14 expression in β-cells.
HIP14 Decreases IL-1β–Induced NF-κB Activation and Apoptosis.
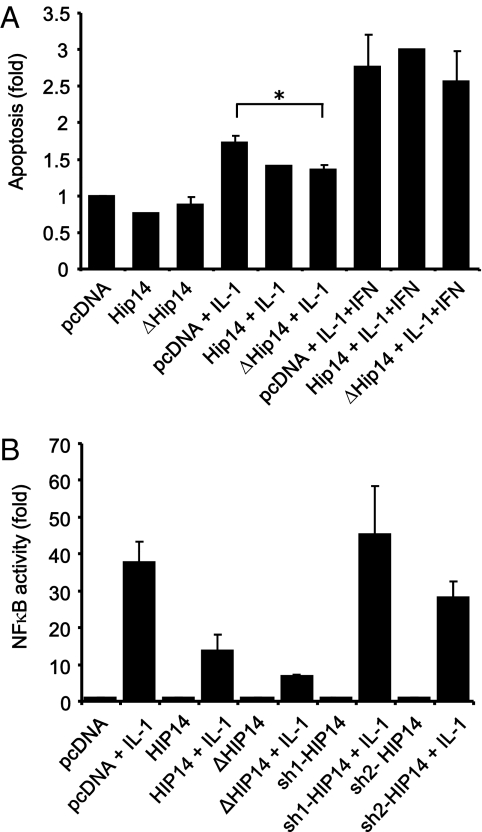
Because HIP14 is required for β-cell survival and because cytokines down-regulate the expression of HIP14, we wanted to investigate whether overexpression of HIP14 could protect against cytokine-induced apoptosis. INS-1 cells were transfected with an empty vector or plasmids encoding either HIP14 or ΔHIP14. After exposure to IL-1β alone or IL-1β plus IFN-γ, apoptotic cell death was measured. Compared with empty vector-transfected control cells, cells transfected with HIP14 or ΔHIP14 were less sensitive to IL-1β–induced apoptosis (Fig. 6A). However, apoptosis induced by IL-1β plus IFN-γ was not modulated by overexpression of HIP14 or ΔHIP14. This probably reflects that the apoptosis rate caused by a mixture of cytokines is too high to be prevented by HIP14, whereas the more moderate apoptosis level induced by IL-1β alone can be suppressed. We also examined the effect of RNAi-mediated HIP14 knockdown on cytokine-induced apoptosis and found that knockdown of HIP14 did not further modulate cytokine-induced apoptosis (3.19 ± 0.40-fold vs. 3.45 ± 0.17-fold, P = 0.54, n = 3–4).
Fig. 6.
HIP14 decreases IL-1β–induced apoptosis. (A) INS-1 cells were transfected with either an empty control vector (pcDNA) or vectors encoding HIP14 or ΔHIP14. Two days after transfection, cells were left untreated or exposed to IL-1β (150 pg/mL) or IL-1β (150 pg/mL) plus IFN-γ (5 ng/mL) for 24 h. Apoptosis was determined by Cell Death Detection ELISA measuring the presence of cytosolic histone–DNA complexes (nucleosomes). Data are means ± SEM of n = 2–4. *P < 0.01 vs. pcDNA plus IL-1β. (B) INS-1 cells were transfected with an NF-κB–driven Luciferase Firefly reporter gene construct and with pcDNA, HIP14, ΔHIP14, or shRNAs targeting HIP14 (sh-HIP14). Cells were then left untreated or exposed to IL-1β (150 pg/mL) for 4 h. Luciferase activity was measured and results shown are means ± SEM of n = 3.
To gain insight into the putative mechanisms underlying the protective effect of HIP14 against IL-1β–induced apoptosis, we next investigated whether HIP14 affects IL-1β–induced activation of the transcription factor NF-κB, which is well known to contribute to cytokine-induced apoptosis in β-cells (30, 31). Using an NF-κB–driven luciferase reporter gene construct, we found that both HIP14 and ΔHIP14 reduced IL-1β–induced NF-κB activity in INS-1 cells (Fig. 6B), suggesting that a possible mechanism by which HIP14 decreases IL-1β–induced apoptosis is via suppression of NF-κB transactivation.
Association Between SNPs and T1D and Transcription Factor Binding Sites Upstream of HIP14.
Finally, we looked for direct genetic association of HIP14 with T1D. Genetic analyses were performed in a T1D family material comprising DNA from 1,928 Danish T1D families (6,853 individuals). According to information from HapMap (www.hapmap.org), two linkage disequilibrium (LD) blocks were found to cover the HIP14 gene on chromosome 12. We selected one tagSNP in each of these blocks (rs4761444 in intron 1 and rs17813975 in exon 8), as well as one SNP in the promoter region (rs7956544 located 17 kb upstream of the HIP14 start site) to capture variation in the gene. However, we could not demonstrate association to T1D of any of the SNPs analyzed [P (transmission disequilibrium test) = 0.78, 0.42, and 0.49, respectively]. This finding suggests that genetic variation in the HIP14 gene and promoter per se is not associated with T1D. It does not rule out the possibility, however, that there are rare variants or genetic variation in the same chromosomal region but outside the HIP14 gene and promoter that affect HIP14. To test this hypothesis, we looked for genetic association using the full genetic dataset from the recent GWAS and metaanalysis published by the Type 1 Diabetes Genetics Consortium (4). We identified SNPs that were nominally associated (P < 0.01) with T1D in a region located >100 kb upstream of the HIP14/ZDHHC17 locus. Because of a recombination hotspot that separated the associated SNPs from the HIP14 gene, there was no LD between the associated SNPs and SNPs within HIP14 and hence no direct association, supporting the observation made in our genetic analysis of the T1D family material. The most significantly associated SNP (rs2632214, P = 0.0055), however, was located 125 kb upstream of the transcriptional start site of HIP14 and could potentially affect a transcription factor binding site that regulates the expression of HIP14 (Fig. S3). Using ENCODE ChIP-seq data available at the University of California, Santa Cruz genome browser (http://genome.ucsc.edu/), we identified two regions of high-scoring transcription factor binding sites surrounding rs2632214. The first was located 3 kb downstream of rs2632214 and contained an overlapping binding site for c-jun and STAT1. The second one was located 2 kb upstream of rs2632214 and contained an overlapping binding site for multiple transcription factors, of which BRG1 had the highest score. We examined the LD between rs2632214 and SNPs located within these two transcription factor binding sites and found that rs2632214 was highly correlated with rs11114818 (r2 > 0.9) within the binding site for c-jun and STAT1, as well as with rs2632217 (r2 = 1.0) in the binding site for BRG1 (Fig. S3). These findings suggest that SNPs located in the nominally associated T1D region could affect the expression of HIP14 by interrupting with one or more transcription factor binding sequences.
Discussion
The genetic susceptibility to T1D involves a small number of genes with large effects sizes and a large number of genes with smaller contributions. Although GWAS studies have emerged during the last years and enabled identification of a large number of chromosomal risk regions with small odd ratios, linkage studies with the power of detecting larger risk loci still provide valuable information and merit follow-up (32). The 11 candidate genes/proteins identified by our current bioinformatics approach originated from linkage regions for T1D (3). The use of protein interaction information to pinpoint and prioritize positional candidate genes in the linkage loci of T1D has been demonstrated previously by Gao and Wang (16). However, this previous study decreased the number of potential candidate genes without any further functional prioritization or biological evaluation, as presently done.
The top three ranked proteins (ranking score above 0.9) in our study were INS, glutaminase C (GLS), and HIP14. Although INS is already an established candidate gene (19), both HIP14 and GLS are unknown in the context of diabetes. Glutamate, which is produced by GLS, is mainly described as an extracellular messenger in the brain (33). However, evidence that glutamate is also is an intracellular messenger in β-cells regulating glucose-induced insulin secretion has been provided (34). Interestingly, we found GLS to be significantly down-regulated to 79% ± 22% (P = 0.02, n = 8) at the mRNA level by cytokines in human islets. Future studies will be needed to clarify the role of GLS in pancreatic β-cells. Some of the other predicted proteins included c-jun N-terminal kinase 1 (JNK1), protein kinase C-δ (PKCD), syntaxin 4 (STX4), and arginino succinate synthetase (ASS) (Table 1). JNK1 plays a significant role in mediating cytokine-induced β-cell apoptosis (35, 36). Likewise, recent evidence from transgenic mice with β-cell-specific overexpression of a dominant-negative form of PKCD pointed toward that PKCD as an important regulator of β-cell apoptosis (37). Studies from STX4 heterozygous knockout mice and MIN6 β-cells implicated that STX4 is required for glucose-stimulated first-phase insulin secretion (38). ASS regenerates arginine from citrulline, contributing to nitric oxide production and apoptosis in cytokine-treated pancreatic β-cells (39, 40). Thus, several of the predicted T1D candidate proteins in the present study have previously been connected to the regulation of essential β-cell functions.
On the basis of our prediction of HIP14 as a candidate protein in T1D, and because there were no previous studies directly describing a role for HIP14 in relation to diabetes, we investigated the functional role of HIP14 in β-cells. In T1D, the β-cells are specifically destroyed by an autoimmune attack involving proinflammatory cytokines, and β-cells have been shown to be particularly prone to apoptosis compared with other cells types in pancreatic islets (28). We therefore investigated whether HIP14 could be involved in the regulation of apoptotis in β-cells. Transient knockdown of HIP14 expression in FACS-purified primary rat β-cells revealed a significant increase in apoptosis, indicating that decreased HIP14 levels in β-cells lead to increased apoptosis. In agreement with these findings in primary rat β-cells, we found that inhibition of HIP14 expression by transient knockdown in INS-1 cells inhibited cell growth and induced apoptosis. Although the level of apoptosis induced by knockdown of HIP14 in both primary and clonal β-cells was modest (≈30–80% increase), it is important to keep in mind that β-cell demise in T1D in man is a slowly progressing process stretching over several years. Thus, even minor increases in β-cell apoptotic cell death rates may be of high importance seen from a pathogenic point of view. Transfection of INS-1 cells with HIP14 or ΔHIP14 decreased IL-1β–induced apoptosis, suggesting that HIP14 irrespective of its palmitoyl transferase activity possesses antiapoptotic properties. Given that cytokines reduced the expression of endogenous HIP14, it also suggests that cytokines cause apoptosis via down-regulation of HIP14 in β-cells. The protective effect of HIP14/ΔHIP14 on IL-1β–induced apoptotic cell death was associated with a decrease in IL-1β–induced NF-κB transactivation, as observed in reporter gene experiments. This finding is in line with previous reports showing that blockade of the NF-κB pathway protects against cytokine-mediated β-cell death in vitro (30, 31).
HIP14 is important for intracellular trafficking and exocytosis in neurons (20, 21, 24, 41). Neurons and pancreatic β-cells have highly overlapping phenotypes and share a similar exocytotic machinery (42). We observed that knockdown of HIP14 expression resulted in a decrease in insulin release, both when measuring long-term accumulated insulin in the culture medium from INS-1 cells and when measuring short-term GSIS in INS-1E cells. Several proteins involved in exocytosis, such as SNAP25 and CSP, are palmitoylated by HIP14 (21). Overexpression of a mutant ΔHIP14 that lacks the palmitoyl transferase domain also decreased accumulated insulin release, albeit this decrease was less prominent than that caused by HIP14 knockdown. This indicates that the palmitoyl transferase activity of HIP14 is important for β-cell insulin release, probably by palmitoylation of proteins involved in the exocytotic machinery. This assumption is in line with results from neurons whereby mutant HIP14 caused decreased exocytotic activity that correlated with severe synaptic mislocation of SNAP25 and CSP in Drosophila melanogaster (24, 41). Of note, SNAP25 is also expressed in pancreatic β-cells, and its inhibition by IL-1β contributes to the loss of first-phase insulin release in response to glucose (43).
HTT, the causative protein for Huntingtons’s disease (HD), is palmitoylated by HIP14 (20, 23), and a transgenic mouse model of HD expressing mutant HTT (140 polyglutamine repeats in the HTT gene) develops diabetes due to β-cell dysfunction (44, 45). Patients with HD have lower acute-phase insulin response after an oral glucose tolerance test and higher prevalence of diabetes (46, 47). The biological function of HTT is not fully established, but several studies indicated its involvement in the intracellular transport of vesicles (48). Thus, a mutant HTT in β-cells decreases both efficiency of intracellular transport and stimulated insulin secretion (49). This may be explained by an increased interaction between mutant HTT and β-tubulin, leading to decreased intracellular transport and consequently decreased insulin release (49). Moreover, palmitoylation of HTT by HIP14 is important for the subcellular localization and prevention of intracellular inclusion of HTT (23). Taking these observations and the present results into consideration, we suggest that HIP14 and HTT are both important for β-cell insulin secretory capacity.
Proinflammatory cytokines such as IL-1β and IFN-γ play an important role in the destruction of β-cells in T1D (28, 50). The intracellular pathways leading to apoptosis are not completely understood, although it is known that pathways such as MAPK, NF-κB, and STAT1 signaling and endoplasmatic reticulum stress play important roles (50, 51). However, the exact genes and proteins that are targeted by cytokine signaling and responsible for promoting β-cell destruction are unclear. Interestingly, HIP14 was down-regulated at the protein level after cytokine treatment in both INS-1 cells and primary rat islets. Furthermore, we found that cytokines suppressed HIP14 mRNA expression in six of eight human islet donor preparations. Collectively, these data indicate that expression of HIP14 is decreased in the β-cells by a T1D-relevant inflammatory stimulus. As a regulatory mechanism of HIP14 gene suppression after cytokine exposure, we suggest that miR146a could be essential. miR146 is induced by cytokines (29, 52) and was predicted to bind to the 3′ UTR of HIP14 mRNA. In transfection experiments, we showed that expression of HIP14 is significantly reduced by miR146a. However, additional experiments are needed to fully establish a potential role of miR146a in the regulation of HIP14 expression in β-cells.
Although we identified HIP14 as a candidate gene in the phenome–interactome analysis, we could not demonstrate T1D association for any of the chosen tagSNPs covering the HIP14 gene and promoter in a large Danish T1D family material. We cannot rule out, however, that there are rare variants and/or other genetic variation in HIP14 that could be associated with T1D. Likewise, the genetic variation could lie outside the HIP14 gene and was therefore not detected in the present study. In line with this, we identified a SNP (rs2632214) 125 kb upstream HIP14 that is nominally associated with T1D when using the detailed information from a recently published GWAS dataset (4). Further analysis revealed that this SNP was in LD (r2 > 0.9) with SNPs located in predicted transcription factor binding sites for c-jun/STAT1 and BRG1. We speculate that these SNPs may have an impact on the transcriptional regulation of HIP14, which would influence β-cell insulin secretion and turnover.
Concluding Remarks.
In the present study, we have used an integrative bioinformatics approach and identified 11 T1D candidate proteins. A top-scoring candidate protein was HIP14, which we selected for functional studies, and we provide evidence that HIP14 regulates apoptosis and insulin secretion in pancreatic β-cells. Thus, our study demonstrates the feasibility and high impact of integrating genetic information and protein–protein interaction data to predict valid candidate proteins that may lead to novel treatment strategies for T1D.
Materials and Methods
Immunohistochemistry.
Pancreata obtained from adult NMRI mice were fixed in 4% paraformaldehyde overnight and transferred to 70% EtOH until paraffin embedding. Four-micrometer-thick sections were cut on a microtome. Subsequently paraffin sections were deparaffinized with 3× Xylene, 3× 99% alcohol, 3× 96% EtOH, 1× 70% EtOH, and finally rinsed in tap water, 5 min each step. Sections were then microwaved in TEG buffer (10 mM Tris, 0.5 mM EGTA) for 4 min at 600 W in 200 mL buffer followed by 15 min at 250 W and finally left to cool for 20 min and incubated with 1% H2O2 in PBS for 30 min and subsequently blocked with 0.5% tris-NaCl blocking reagent (Perkin-Elmer). Sections were incubated with rabbit anti-HIP14 antibody (1:1,000; Sigma H7414) overnight at room temperature. The following day sections were rinsed in PBS, and biotinylated donkey anti-rabbit antibody (1:300; Jackson ImmunoResearch) was added and incubated for 45 min. Sections were then incubated with streptavidin-HRP, washed in PBS, and TSA-cy3 (tyramide signal amplification; Perkin-Elmer) was added, and sections were washed with PBS. On top of this new primary antibodies were added: guinea pig anti-insulin (1:150; Abcam ab7842) and mouse anti-glucagon (1:100; Novo Biolabs Glu001). Guinea pig antibody was visualized with donkey anti-guinea pig-cy5 and mouse antibody with donkey anti-mouse-cy2 (1:300; Jackson ImmunoResearch). DAPI (0.2 μg/mL) was used to stain nuclei. Images were taken on an LSM510 LASER SCANNING confocal microscope (Zeiss). False color images were generated with the LSM software. Final brightness/contrast picture adjustment and layout of the figures were done using Canvas software. Control experiments verifying the specificity of the anti-HIP14 antibody (Sigma H7414) used for staining are shown in Fig. S4.
Cell Culture.
Primary neonatal rat islets were isolated from 3- to 6-d-old Wistar rats (Taconic) and precultured for ≈7 d in RPMI1640 containing 10% newborn calf serum and 100 U/mL penicillin and 100 μg/mL streptomycin (all from Invitrogen). Randomly picked islets were placed in 12-well dishes (Nunc) and left untreated or stimulated with cytokines for 24 h in RPMI 1640 supplemented with 10% FBS and penicillin/streptomycin. Recombinant mouse IL-1β was from BD Pharmingen, and recombinant rat IFN-γ was obtained from R&D Systems. Isolation and culture of primary FACS-sorted rat β-cells (>90% purity) was performed as previously described (53, 54). INS-1 cells were maintained in RPMI 1640 medium (11 mM glucose) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin (all from Invitrogen). In addition, the media contained 50 μM β-mercaptoethanol. Cells were trypsinized and passaged weekly. Human pancreatic islets were obtained from a multicenter European Union-supported program on β-cell transplantation in diabetes directed by Prof. D. Pipeleers. The program has been approved by central and local ethics committees. Human islet preparations were obtained from eight donors (aged 8–57 y); six were male donors, and two were female. Each preparation was stimulated with a mixture of cytokines [TNF-α (5,000 U/mL), IFN-γ (750 U/mL), and IL-1β (75 U/mL)] for 48 h.
Immunoblotting.
For preparation of whole-cell extracts, cells or islets were lysed for 30 min on ice in lysis buffer containing 20 mM Tris-acetate (pH 7.0), 0.27 M sucrose, 1 mM EDTA, 1 mM EGTA, 50 mM NaF, 1% vol/vol Triton X-100, 5 mM sodium pyrophosphate, 10 mM sodium glycerophosphate, 1 mM benzamidine, 1 mM DTT, 1 mM Na3VO4, and 4 μg/mL leupeptin. Detergent-insoluble material was pelleted by centrifugation at 15,000 × g for 5 min at 4 °C. The supernatants (whole-cell extracts) were stored at −80 °C until further use. Protein concentrations in lysates were determined by the Bradford method using the dye reagent concentrate (Bio-Rad). Cell lysates were denatured by addition of 4× lithium dodecyl sulphate sample buffer (Invitrogen) and heating for 5 min at 70 °C. Samples were then subjected to 10% SDS/PAGE followed by electro-transfer onto nitrocellulose membranes. Membranes were blocked in 1× Tris-buffered saline (pH 7.6) containing 0.1% Tween 20 (TBST) and 5% nonfat dry milk for 1 h before primary antibody was added (anti-HIP14; Sigma H7414 diluted 1:250–500 in TBST with 5% dry milk). Filter membranes were incubated with primary antibody at 4 °C for 1–2 d and then washed in TBST and incubated with secondary horseradish peroxidase-conjugated antibodies for 1 h. Immune complexes were detected by chemiluminescence using LumiGLO (Cell Signaling), and images were digitally captured using a FUJI LAS3000. To verify the specificity of the anti-HIP14 antibody (Sigma H7414), HEK293 cells were transfected using Lipofectamine 2000 (Invitrogen) with an expression vector encoding HIP14. The lysates were then examined by immunoblotting (Fig. S4).
Cell Growth.
Cell growth was measured in real time using an xCELLigence apparatus (Roche). INS-1 cells were seeded (3.0 × 104 cells/well) in standard culture conditions in 96-well E-plates. Background impedance was determined according to the manufacturer’s protocol. The plate was incubated for 30 min at room temperature before installation into the Real-Time Cell Analyzer station in a conventional cell incubator at 37 °C and 5% CO2. After 24 h the culture medium was changed to antibiotic-free medium and the cells transfected. The transfection medium was changed to normal culture medium after 24 h. The impedance was measured every 15 min and expressed as cell index (CI), defined as (Rn − Rb)/15, where Rn represents the impedance at a given time point, and Rb is the background impedance. The normalized CI was determined by the CI at a given time point divided with the CI at the normalization time point. Slopes of growth curves were determined between the time points “day 1” and “day 3” [i.e., from the time point when the medium was changed (24 h after transfection) to the time point when si-Neg-transfected cells reached confluence (peak)].
Apoptosis Measurements.
Apoptosis in primary FACS-sorted β-cells was determined by propidium iodine (PI) and Hoechst staining as previously described (54). Apoptotic cell death in INS-1 cells was determined by the detection of DNA-histone complexes present in the cytoplasmic fraction of cells using the Cell Death Detection ELISAPLUS assay (Roche) as described by the manufacturer. Briefly, after exposure of cells to the desired reagents, the culture medium was removed and the cells lysed in 400 μL lysis buffer for 30 min at room temperature. Lysates were centrifuged for 10 min at 200 × g and 20 μL supernatant and 80 μL immunoreagent (anti-DNA-POD antibody and anti-histone-biotin antibody) were added to streptavidin-coated microtiter plates and incubated for 2 h under shaking conditions (300 rpm) at room temperature. The solution was then removed and each well washed three times with 250 μL incubation buffer, after which 100 μL ABTS (2,2-azino-bis-3-ethylbenzthiazoline-6-sufonate) solution was added. Absorbance was measured at 405 nm and 490 nm.
RNAi and Transfection.
Primary FACS-purified β-cells were transfected with TARGETplus SMARTpool siRNA listed below, and transfection was performed as in ref. 54. INS-1 cells were cultured in 24-well (0.2 × 106 cells/well) or 96-well (3.0 × 104 cells/well) plates. Before transfection, the cells were cultured in antibiotic-free medium for at least 4 h. The siRNAs (ON-TARGETplus SMARTpool; Thermo Scientific) directed against rat HIP14 were used with the target sequences: AGGUUAUGACGUACGGCAA, UAUGAACGCUGCCGAGAAU, GUACAUAGCACAUGCGAAU, and GGACAAAGAGAACGUUACA. For the miR-146a transfection experiments we used miR-146a miRIDIAN mimic (#C-300630-03), and as control we used miRIDIAN negative control (#CN-001000-01) (Thermo Scientific). Expression plasmids encoding HIP14 and ΔHIP14 (21) were kindly provided by Dr. Alaa El-Husseini (University of British Columbia, Vancouver, BC, Canada). The vector encoding human growth hormone (pXGH5) was kindly provided by Dr. Romano Regazzi (University of Lausanne, Switzerland). Negative control siRNA (AllStars Negative Control siRNA) was purchased from Qiagen. The shRNA vectors (SureSilencing, catalog no. KR54358N) targeting HIP14 and control vector were purchased from SABiosciences. The siRNA or vectors and the DharmaFECT 1 solution (Thermo Scientific) were diluted separately in OptiMEM and incubated for 5 min at room temperature. Afterward, the two dilutions were mixed and incubated for 20 min at room temperature to allow complex formation. This mixture was subsequently diluted five times in antibiotic-free medium and added to the cells at a final concentration of 30 nmol/L siRNA or 2.5 μg/mL vectors and 1.5 μL/mL DharmaFECT. The cells were then incubated overnight. The next day the medium was changed to standard culture medium, and the cells were incubated for at least 24 h before analysis or further experimentation.
Accumulated Insulin and Glucose-Induced Insulin Secretion.
Accumulated insulin in culture medium or glucose-induced insulin release to Krebs Ringer buffer were determined as previously described (55). For glucose-stimulated insulin release, 0.12 × 106 INS-1E cells were seeded in 24-well dishes. Transfection with siRNAs or vectors was conducted as described above. After a recovery period of 24 h, the cells were starved for 1 h in Krebs Ringer buffer without glucose. Afterward the cells were exposed to Krebs Ringer buffer or culture medium containing either 1.7 mM or 17 mM glucose for 30 min.
Gene Expression Analysis.
mRNA was extracted from human islet preparations by TRIzol according to the manufacturer’s protocol (Invitrogen). cDNA was prepared from total RNA by oligo-dT–primed reverse transcription, as described by the manufacturer (TaqMan RT reagents; Applied Biosystems). Relative expression levels of HIP14 (TaqMan primer Hs00392305_m1) and the house-keeping genes (PPIA, GAPDH, and 18S) were evaluated by the use of TaqMan assays. The Low Density Array system (Applied Biosystems) containing assays for the individual genes as well as housekeeping genes was used on TaqMan 7900HT (Applied Biosystems). For evaluation, expression levels of genes were normalized against the geometric mean of three housekeeping genes and evaluated using the ΔΔCT method (56). For primary FACS-sorted rat β-cells, RNA extraction, cDNA synthesis, and real-time PCR were conducted as previously described (54).
Luciferase Reporter Gene Assay.
INS-1 cells (0.15 × 106 cells) in duplicates were transfected with an NF-κB–driven luciferase reporter construct containing five repeats of NF-κB consensus binding sites (0.4 μg/well) (Stratagene) and cotransfected with 0.5 μg/well of either empty vector (pcDNA) or vectors encoding HIP14 or ΔHIP14 using the transfection reagent Superfect (Qiagen). Two days after transfection, cells were stimulated with IL-1β for 4 h. Luciferase activity was measured by the dual luciferase reporter system according to the specifications of the manufacturer (Promega).
Genetic Analyses.
T1D families were ascertained through the Danish Society of Childhood Diabetes, a population-based cohort, and were all of Danish Caucasoid origin. Probands in these families were diagnosed with T1D before 15 y of age and had continued insulin treatment since diagnosis. For genotyping, commercially available predesigned TaqMan SNP genotyping assays were used (Applied Biosystems) with TaqMan 7900 HT equipment (Applied Biosystems). SNPs genotyped were rs4761444, rs17813975, and rs7956544. All three SNPs were previously evaluated in the HapMap project and were common in Caucasian populations (minor allele frequencies: 0.4, 0.19, and 0.45, respectively). Association to T1D was evaluated by means of the transmission disequilibrium test in PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/summary.shtml).
The summary statistics were retrieved from the full GWAS and metaanalysis dataset generated by the Type 1 Diabetes Genetics Consortium (4). The genetic association P values across the HIP14/ZDHHC17 locus were visualized using the web-based software LocusZoom (57). The SNP rs2632214 had the lowest P value in a region located 125 kb upstream of HIP14. The chromosomal region surrounding rs2632214 was examined for potential transcription factor binding sites using the ENCODE ChIP-seq data available at the University of California, Santa Cruz genome browser (http://genome.ucsc.edu/). SNPs located within sequences occupied by transcription factors were investigated for LD with rs2632214 using phase II genotypes for the HapMap CEU samples (www.hapmap.org) in Haploview (58).
Statistical Analysis.
In bar graphs, data are means ± SEM. Student’s paired t test was used for statistical analysis. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Fie Hillesø, Tine Wille, Hanne Foght, and Rikke Bonne for technical assistance, and Jens Høiriis Nielsen for providing human islet preparations for gene expression analyses. The work conducted in this study was supported in part by grants from the P. Carl Petersen Foundation, the Danish Agency for Science Technology and Innovation, Novo Nordisk A/S, Juvenile Diabetes Research Foundation Grant 33-2008-391, Fonds National de la Recherche Scientifique Belgium, and the European Union (project Naimit, in the Framework Programme 7 of the European Community).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 15023.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104384108/-/DCSupplemental.
References
- 1.Donath MY, Størling J, Maedler K, Mandrup-Poulsen T. Inflammatory mediators and islet beta-cell failure: A link between type 1 and type 2 diabetes. J Mol Med. 2003;81:455–470. doi: 10.1007/s00109-003-0450-y. [DOI] [PubMed] [Google Scholar]
- 2.Rich SS. Mapping genes in diabetes. Genetic epidemiological perspective. Diabetes. 1990;39:1315–1319. doi: 10.2337/diab.39.11.1315. [DOI] [PubMed] [Google Scholar]
- 3.Concannon P, et al. Type 1 Diabetes Genetics Consortium Type 1 diabetes: Evidence for susceptibility loci from four genome-wide linkage scans in 1,435 multiplex families. Diabetes. 2005;54:2995–3001. doi: 10.2337/diabetes.54.10.2995. [DOI] [PubMed] [Google Scholar]
- 4.Barrett JC, et al. Type 1 Diabetes Genetics Consortium. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Concannon P, et al. Type 1 Diabetes Genetics Consortium. Genome-wide scan for linkage to type 1 diabetes in 2,496 multiplex families from the Type 1 Diabetes Genetics Consortium. Diabetes. 2009;58:1018–1022. doi: 10.2337/db08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eaves IA, et al. Combining mouse congenic strains and microarray gene expression analyses to study a complex trait: The NOD model of type 1 diabetes. Genome Res. 2002;12:232–243. [PubMed] [Google Scholar]
- 7.Zhu J, et al. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet Genome Res. 2004;105:363–374. doi: 10.1159/000078209. [DOI] [PubMed] [Google Scholar]
- 8.Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiffin N, et al. Integration of text- and data-mining using ontologies successfully selects disease gene candidates. Nucleic Acids Res. 2005;33:1544–1552. doi: 10.1093/nar/gki296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J, Li Y. Discovering disease-genes by topological features in human protein-protein interaction network. Bioinformatics. 2006;22:2800–2805. doi: 10.1093/bioinformatics/btl467. [DOI] [PubMed] [Google Scholar]
- 11.Köhler S, Bauer S, Horn D, Robinson PN. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franke L, et al. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet. 2006;78:1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet. 2010;86:581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elbers CC, et al. Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genet Epidemiol. 2009;33:419–431. doi: 10.1002/gepi.20395. [DOI] [PubMed] [Google Scholar]
- 15.Sharma A, Chavali S, Tabassum R, Tandon N, Bharadwaj D. Gene prioritization in Type 2 Diabetes using domain interactions and network analysis. BMC Genomics. 2010;11:84. doi: 10.1186/1471-2164-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao S, Wang X. Predicting type 1 diabetes candidate genes using human protein-protein interaction networks. J Comput Sci Syst Biol. 2009;2:133. doi: 10.4172/jcsb.1000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 18.Anjos S, Polychronakos C. Mechanisms of genetic susceptibility to type I diabetes: Beyond HLA. Mol Genet Metab. 2004;81:187–195. doi: 10.1016/j.ymgme.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Pociot F, et al. Genetics of type 1 diabetes: What’s next? Diabetes. 2010;59:1561–1571. doi: 10.2337/db10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singaraja RR, et al. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Hum Mol Genet. 2002;11:2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- 21.Huang K, et al. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 22.Ducker CE, Stettler EM, French KJ, Upson JJ, Smith CD. Huntingtin interacting protein 14 is an oncogenic human protein: Palmitoyl acyltransferase. Oncogene. 2004;23:9230–9237. doi: 10.1038/sj.onc.1208171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanai A, et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat Neurosci. 2006;9:824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohyama T, et al. Huntingtin-interacting protein 14, a palmitoyl transferase required for exocytosis and targeting of CSP to synaptic vesicles. J Cell Biol. 2007;179:1481–1496. doi: 10.1083/jcb.200710061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merglen A, et al. Glucose sensitivity and metabolism-secretion coupling studied during two-year continuous culture in INS-1E insulinoma cells. Endocrinology. 2004;145:667–678. doi: 10.1210/en.2003-1099. [DOI] [PubMed] [Google Scholar]
- 26.Wick PF, Senter RA, Parsels LA, Uhler MD, Holz RW. Transient transfection studies of secretion in bovine chromaffin cells and PC12 cells. Generation of kainate-sensitive chromaffin cells. J Biol Chem. 1993;268:10983–10989. [PubMed] [Google Scholar]
- 27.Antinozzi PA, Garcia-Diaz A, Hu C, Rothman JE. Functional mapping of disease susceptibility loci using cell biology. Proc Natl Acad Sci USA. 2006;103:3698–3703. doi: 10.1073/pnas.0510521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donath MY, Størling J, Berchtold LA, Billestrup N, Mandrup-Poulsen T. Cytokines and beta-cell biology: From concept to clinical translation. Endocr Rev. 2008;29:334–350. doi: 10.1210/er.2007-0033. [DOI] [PubMed] [Google Scholar]
- 29.Roggli E, et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes. 2010;59:978–986. doi: 10.2337/db09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heimberg H, et al. Inhibition of cytokine-induced NF-kappaB activation by adenovirus-mediated expression of a NF-kappaB super-repressor prevents beta-cell apoptosis. Diabetes. 2001;50:2219–2224. doi: 10.2337/diabetes.50.10.2219. [DOI] [PubMed] [Google Scholar]
- 31.Giannoukakis N, Rudert WA, Trucco M, Robbins PD. Protection of human islets from the effects of interleukin-1beta by adenoviral gene transfer of an Ikappa B repressor. J Biol Chem. 2000;275:36509–36513. doi: 10.1074/jbc.M005943200. [DOI] [PubMed] [Google Scholar]
- 32.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med. 2009;360:1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 33.Ozkan ED, Ueda T. Glutamate transport and storage in synaptic vesicles. Jpn J Pharmacol. 1998;77:1–10. doi: 10.1254/jjp.77.1. [DOI] [PubMed] [Google Scholar]
- 34.Maechler P, Wollheim CB. Mitochondrial glutamate acts as a messenger in glucose-induced insulin exocytosis. Nature. 1999;402:685–689. doi: 10.1038/45280. [DOI] [PubMed] [Google Scholar]
- 35.Ammendrup A, et al. The c-Jun amino-terminal kinase pathway is preferentially activated by interleukin-1 and controls apoptosis in differentiating pancreatic beta-cells. Diabetes. 2000;49:1468–1476. doi: 10.2337/diabetes.49.9.1468. [DOI] [PubMed] [Google Scholar]
- 36.Bonny C, Oberson A, Negri S, Sauser C, Schorderet DF. Cell-permeable peptide inhibitors of JNK: novel blockers of beta-cell death. Diabetes. 2001;50:77–82. doi: 10.2337/diabetes.50.1.77. [DOI] [PubMed] [Google Scholar]
- 37.Hennige AM, et al. Overexpression of kinase-negative protein kinase Cdelta in pancreatic beta-cells protects mice from diet-induced glucose intolerance and beta-cell dysfunction. Diabetes. 2010;59:119–127. doi: 10.2337/db09-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spurlin BA, Thurmond DC. Syntaxin 4 facilitates biphasic glucose-stimulated insulin secretion from pancreatic beta-cells. Mol Endocrinol. 2006;20:183–193. doi: 10.1210/me.2005-0157. [DOI] [PubMed] [Google Scholar]
- 39.Flodström M, Niemann A, Bedoya FJ, Morris SM, Jr, Eizirik DL. Expression of the citrulline-nitric oxide cycle in rodent and human pancreatic beta-cells: Induction of argininosuccinate synthetase by cytokines. Endocrinology. 1995;136:3200–3206. doi: 10.1210/endo.136.8.7628352. [DOI] [PubMed] [Google Scholar]
- 40.Ortis F, et al. Cytokines interleukin-1beta and tumor necrosis factor-alpha regulate different transcriptional and alternative splicing networks in primary beta-cells. Diabetes. 2010;59:358–374. doi: 10.2337/db09-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stowers RS, Isacoff EY. Drosophila huntingtin-interacting protein 14 is a presynaptic protein required for photoreceptor synaptic transmission and expression of the palmitoylated proteins synaptosome-associated protein 25 and cysteine string protein. J Neurosci. 2007;27:12874–12883. doi: 10.1523/JNEUROSCI.2464-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas-Reetz AC, De Camilli P. A role for synaptic vesicles in non-neuronal cells: Clues from pancreatic beta cells and from chromaffin cells. FASEB J. 1994;8:209–216. doi: 10.1096/fasebj.8.2.7907072. [DOI] [PubMed] [Google Scholar]
- 43.Ohara-Imaizumi M, Cardozo AK, Kikuta T, Eizirik DL, Nagamatsu S. The cytokine interleukin-1beta reduces the docking and fusion of insulin granules in pancreatic beta-cells, preferentially decreasing the first phase of exocytosis. J Biol Chem. 2004;279:41271–41274. doi: 10.1074/jbc.C400360200. [DOI] [PubMed] [Google Scholar]
- 44.Hurlbert MS, et al. Mice transgenic for an expanded CAG repeat in the Huntington’s disease gene develop diabetes. Diabetes. 1999;48:649–651. doi: 10.2337/diabetes.48.3.649. [DOI] [PubMed] [Google Scholar]
- 45.Björkqvist M, et al. The R6/2 transgenic mouse model of Huntington’s disease develops diabetes due to deficient beta-cell mass and exocytosis. Hum Mol Genet. 2005;14:565–574. doi: 10.1093/hmg/ddi053. [DOI] [PubMed] [Google Scholar]
- 46.Podolsky S, Leopold NA, Sax DS. Increased frequency of diabetes mellitus in patients with Huntington’s chorea. Lancet. 1972;1:1356–1358. doi: 10.1016/s0140-6736(72)91092-6. [DOI] [PubMed] [Google Scholar]
- 47.Lalić NM, et al. Glucose homeostasis in Huntington disease: Abnormalities in insulin sensitivity and early-phase insulin secretion. Arch Neurol. 2008;65:476–480. doi: 10.1001/archneur.65.4.476. [DOI] [PubMed] [Google Scholar]
- 48.Caviston JP, Holzbaur EL. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith R, et al. Mutant huntingtin interacts with beta-tubulin and disrupts vesicular transport and insulin secretion. Hum Mol Genet. 2009;18:3942–3954. doi: 10.1093/hmg/ddp336. [DOI] [PubMed] [Google Scholar]
- 50.Eizirik DL, Mandrup-Poulsen T. A choice of death—the signal-transduction of immune-mediated beta-cell apoptosis. Diabetologia. 2001;44:2115–2133. doi: 10.1007/s001250100021. [DOI] [PubMed] [Google Scholar]
- 51.Cnop M, et al. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: Many differences, few similarities. Diabetes. 2005;54(Suppl 2):S97–S107. doi: 10.2337/diabetes.54.suppl_2.s97. [DOI] [PubMed] [Google Scholar]
- 52.Fred RG, Bang-Berthelsen CH, Mandrup-Poulsen T, Grunnet LG, Welsh N. High glucose suppresses human islet insulin biosynthesis by inducing miR-133a leading to decreased polypyrimidine tract binding protein-expression. PLoS ONE. 2010;5:e10843. doi: 10.1371/journal.pone.0010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rasschaert J, et al. Toll-like receptor 3 and STAT-1 contribute to double-stranded RNA+ interferon-gamma-induced apoptosis in primary pancreatic beta-cells. J Biol Chem. 2005;280:33984–33991. doi: 10.1074/jbc.M502213200. [DOI] [PubMed] [Google Scholar]
- 54.Moore F, et al. PTPN2, a candidate gene for type 1 diabetes, modulates interferon-gamma-induced pancreatic beta-cell apoptosis. Diabetes. 2009;58:1283–1291. doi: 10.2337/db08-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nielsen K, et al. Beta-cell maturation leads to in vitro sensitivity to cytotoxins. Diabetes. 1999;48:2324–2332. doi: 10.2337/diabetes.48.12.2324. [DOI] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.Pruim RJ, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]