Abstract
The in situ stimulation of Fe(III) oxide reduction by Geobacter bacteria leads to the concomitant precipitation of hexavalent uranium [U(VI)] from groundwater. Despite its promise for the bioremediation of uranium contaminants, the biological mechanism behind this reaction remains elusive. Because Fe(III) oxide reduction requires the expression of Geobacter's conductive pili, we evaluated their contribution to uranium reduction in Geobacter sulfurreducens grown under pili-inducing or noninducing conditions. A pilin-deficient mutant and a genetically complemented strain with reduced outer membrane c-cytochrome content were used as controls. Pili expression significantly enhanced the rate and extent of uranium immobilization per cell and prevented periplasmic mineralization. As a result, pili expression also preserved the vital respiratory activities of the cell envelope and the cell's viability. Uranium preferentially precipitated along the pili and, to a lesser extent, on outer membrane redox-active foci. In contrast, the pilus-defective strains had different degrees of periplasmic mineralization matching well with their outer membrane c-cytochrome content. X-ray absorption spectroscopy analyses demonstrated the extracellular reduction of U(VI) by the pili to mononuclear tetravalent uranium U(IV) complexed by carbon-containing ligands, consistent with a biological reduction. In contrast, the U(IV) in the pilin-deficient mutant cells also required an additional phosphorous ligand, in agreement with the predominantly periplasmic mineralization of uranium observed in this strain. These findings demonstrate a previously unrecognized role for Geobacter conductive pili in the extracellular reduction of uranium, and highlight its essential function as a catalytic and protective cellular mechanism that is of interest for the bioremediation of uranium-contaminated groundwater.
Keywords: biological electron transfer, pilus nanowires, Geobacteraceae, metal reduction
Dissimilatory metal-reducing microorganisms gain energy for growth by coupling the oxidation of organic acids or H2 to the reduction of metals. Some can also use uranium (U) as an electron acceptor (1, 2), a process that could be harnessed for the bioremediation of the contaminated aquifers and sediments left by the intensive U mining practices of the Cold War era (3). Interestingly, stimulating the activity of metal-reducing microorganisms in situ resulted in the concomitant removal of soluble hexavalent uranium [U(VI)] from the contaminated groundwater and detection of its sparingly soluble, less mobile form, tetravalent uranium [U(IV)] in sediments (4–8). This suggests that stimulating metal reduction in the subsurface results in the biological reduction of U(VI) to U(IV), thereby preventing plume migration and eliminating the potential for contaminant exposure.
The removal of U(VI) from groundwater following the in situ stimulation of metal reduction is often concomitant with substantial increases in the growth and activity of dissimilarity metal-reducing microorganisms in the family Geobacteraceae (4, 6, 7, 9). Despite extensive efforts to understand the mechanisms and pathways used by these bacteria to reduce U(VI), the nature of its U reductase has remained elusive for almost two decades. Early studies with Geobacter metallireducens GS15 suggested that U was reduced extracellularly to uraninite under conditions conducive to cell growth (1, 10). The development of genetic tools in Geobacter sulfurreducens (11) motivated molecular studies to elucidate the biological mechanism behind this reaction. Because c-cytochromes are abundant in the cell envelope of Geobacter bacteria, studies focused on identifying extracytoplasmic c-cytochromes that could function as dedicated U reductases (12, 13). However, mutations were often pleitrophic (14–16) and showed either no defect or only partial defects in the cell's ability to remove U(VI) (12, 13). Interpretation was also difficult due to inconsistencies in the reported mutant phenotypes, with some mutations reportedly abolishing U(VI) removal activities, yet mutant cells showing extensive mineralization (13). Furthermore, these studies consistently showed that the U precipitated inside the cell envelope. U is not known to be essential for the synthesis of any cell component or for any cellular biological reaction, yet can be reduced and precipitated nonspecifically by the abundant low-potential electron donors of the cell envelope of Gram-negative bacteria (17). This is predicted to compromise the integrity of the cell envelope and its vital functions (18). Because of this, the environmental relevance of these early studies in G. sulfurreducens is questionable.
The energy to support the growth of Geobacter bacteria after in situ stimulation results from the reduction of the abundant Fe(III) oxides, a process that requires the expression of their conductive pili (19). In contrast to the lack of conservation of c-cytochrome sequences (20), the genes encoding the Geobacteraceae pilus subunits or pilins are highly conserved and form an independent line of descent (19). This is consistent with the pili's specialized function as electrical conduits. The pilus apparatus is anchored in the cell envelope of Gram-negative cells (21) and could potentially accept electrons from cell envelope c-cytochromes or the menaquinone pool in the inner membrane. Pili also protrude outside the cell envelope and can reach micrometer lengths, thereby enhancing the cell's reactive surface. Thus, we hypothesized that the pili could catalyze the reduction of U(VI) “at a distance” to maximize the cell's catalytic surface while minimizing exposure to U. Here we show that the conductive pili of G. sulfurreducens catalyze the extracellular reduction of U(VI) to a mononuclear U(IV) phase and prevent its periplasmic mineralization. This mechanism preserves the functioning and integrity of the cell envelope and the cell's viability. These results demonstrate that pili are the elusive U reductase of Geobacter bacteria and that their catalytic function also serves as a protective cellular mechanism. Our findings suggest that pili's expression confers on Geobacter bacteria an adaptive ecological advantage in the contaminated subsurface of potential interest for the optimization of in situ bioremediation.
Results
Expression of Pili Promotes the Extracellular Reduction of U(VI).
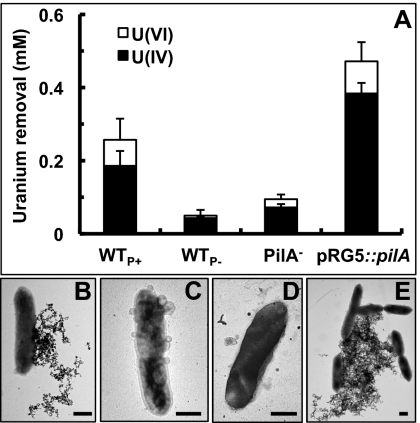
The correspondence between pili expression and U immobilization was examined by monitoring the removal of U(VI) acetate from solution by resting wild-type cells incubated at 25 °C (WTP+) or 30 °C (WTP−) to induce or prevent pili assembly, respectively. Controls with a pilin-deficient mutant (PilA−) and its genetically complemented strain (pRG5::pilA) were also included. The piliated strains WTP+ and pRG5::pilA removed substantially more U(VI) from solution than the nonpiliated strains WTP− and PilA− (Fig. 1A). The activity was biological in nature, inasmuch as heat-killed WTP+ and WTP− controls did not remove U(VI) significantly (0.02 ± 0.04 and 0.05 ± 0.02 mM, respectively). X-ray absorption near-edge structure (XANES) spectroscopy confirmed the reductive nature of the U removal activity and measured an average of 70–85% U(IV) in all samples (Fig. 1A). Furthermore, the expression of the pilA gene relative to the internal control recA did not change during the assay (Fig. S1), thus ruling out any de novo pilin expression. The extent of U(VI) removal corresponded well with the levels of piliation, which were measured as the protein content of purified PilA-containing pili samples (Fig. S2). The pRG5::pilA piliation (3.6 ± 1.7 μg pili/OD600) was 2.5-fold higher than WTP+ (1.5 ± 0.1 μg/OD600), which matched well with its superior capacity to remove U(VI) from solution (1.8 ± 1.0-fold higher than WTP+). By contrast, WTP− and PilA− samples had no detectable pili protein and reduced less U(VI).
Fig. 1.
Reduction of U(VI) to U(IV) (A) and TEM images of unstained whole cells showing the subcellular localization of the U deposits in the WTP+ (B), WTP− (C), PilA− (D), and pRG5::pilA (E) strains. (Scale bar, 0.5 μm.)
The location of the U reductase system was studied by examining the cellular localization of the U deposits in unstained whole cells by transmission electron microscopy (TEM; Fig. 1 B–E). The piliated strains, WTP+ and pRG5::pilA, preferentially deposited the U extracellularly and in a monolateral fashion, consistent with the localization of Geobacter's conductive pili to one side of the cell (19). The pili filaments were interspersed with the dense deposits (Fig. S3). Elemental composition analyses of the pili-associated deposits by TEM energy dispersive spectroscopy (EDS) in the WTP+ confirmed the presence of U (Fig. S3). In contrast, extracellular U mineralization in the nonpiliated strains, WTP− and PilA−, was limited to the cell surface and membrane vesicles. TEM thin sections of the unstained cells confirmed the presence of extracellular, needle-like U deposits in the piliated strains as well as discreet regions of U deposition on the outer membrane (Fig. S4). Only a few cells (8 ± 3% of the WTP+ and <1% of the pRG5::pilA) had periplasmic mineralization. Outer membrane foci of U deposition were also noticeable in the WTP−, but more cells (37 ± 13%) had periplasmic deposition. The increased periplasmic mineralization in the WTP− cannot be attributed to a differential expression of outer membrane c-cytochromes, because outer membrane protein fractions had the same heme profile and content as the WTP+ (Fig. S5). By contrast, the PilA− mutant was partially defective in outer membrane c-cytochrome production (Fig. S5) and had the highest levels of periplasmic mineralization (85 ± 12% of the cells; Fig. S4), which suggests that outer membrane c-cytochromes also contribute to the extracellular reduction of U(VI). It is unlikely that the outer membrane c-cytochrome OmcS, which has been hypothesized to mediate electron transfer between the conductive pili and metals (22), contributed to the pili-mediated reduction of U, because the pRG5::pilA strain expressed OmcS at wild-type levels (Fig. S5) yet reduced more U than the WTP+ (Fig. 1A) and proportionally to the levels of piliation. Furthermore, the pRG5::pilA strain also had a defect in outer membrane, heme-containing proteins (Fig. S5), yet cells had very little U deposition in their cell envelope (Fig. S4). This finding is consistent with the pili functioning as the primary site for U reduction.
X-Ray Absorption Fine Structure (EXAFS) Analyses Demonstrate the Reduction of U(VI) to Mononuclear U(IV).
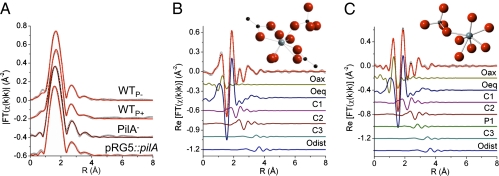
U LIII-edge EXAFS spectra were modeled to determine the atomic coordination about U and characterize the U(IV) product in all of the strains (23). Models for the EXAFS spectra included signals from neighboring P, U, and Fe atoms, but only C neighbors were found to accurately reproduce the measured spectra. The spectra were best described by a mixture of U(IV) and U(VI) coordinated by C-containing ligands. Only the PilA− mutant required an additional P ligand. A U signal corresponding to the U–U distance in uraninite at 3.87 Å was tested but was inconsistent with the measured spectra. Fig. 2A shows the magnitude of the Fourier-transformed spectra and models for each spectrum. Fig. 2 B and C show, as examples, the contribution of each path in the model in the real part of the Fourier transform for the WTP+ and PilA− cells, and Fig. 2 Insets show a molecular moiety of the U(IV) atomic environment that is consistent with the measured EXAFS. The WTP+ model includes one C ligand bound to two O atoms of U in a bidentate fashion and followed by a distant C atom (C3), and another C ligand bonded to one O atom of U(IV) in a monodentate fashion and attached to a distant O atom (Odist). This model was simultaneously refined to all spectra and was insufficient to reproduce the PilA− spectrum, which required an additional monodentate P ligand (Fig. 2C). The distances and σ2 values used to model the spectra are listed in Table S1. The coordination numbers (Table S2) are consistent with 1–2 bidentate C ligands and two monodentate C ligands per U atom. The number of Oax atoms (Noax) was also used to estimate the amount of U(IV) in these samples, as there are two Oax atoms for each U(VI) atom and none for U(IV) (23). An average of 3–4 replicates for each strain gives the values of 72 ± 16% (WTP+), 81 ± 6% (pRG5::pilA), 85 ± 5% (WTP−), and 76 ± 10% (PilA−); this provides additional evidence that although the extent of U(VI) removal depended on the expression of the pili, the ability of the cells to reduce U(VI) to U(IV) did not.
Fig. 2.
U LIII-edge EXAFS spectra (symbols) and models (line). (A) Magnitude of Fourier transform spectra are offset for clarity. (B and C) Real part of Fourier transform of WTP+ (B) and PilA− (C). The components of the model are shown offset beneath the total model. (B and C Insets) U(IV) moiety consistent with the measured EXAFS spectra. U(IV), light gray; O, red; C, black; P, dark gray.
U Reduction via Pili as a Protective Cellular Mechanism.
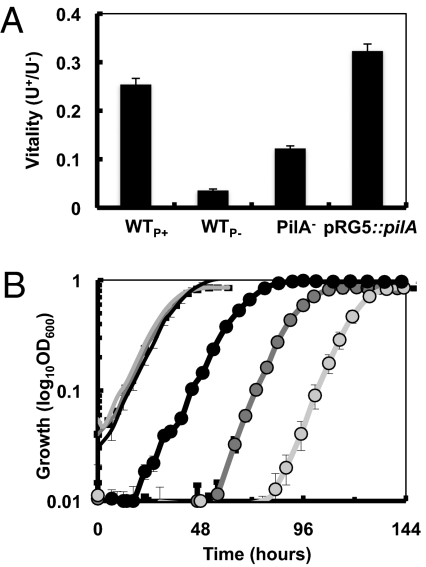
The reverse correlation between piliation and periplasmic mineralization suggested that the pili-mediated reduction prevented U from permeating and being reduced in the periplasm, thus preserving vital functions of the cell envelope. To test this, we used the fluorogenic RedoxSensor Green dye to measure the cell's reductase activity (mostly, respiratory activity) (24) of the strains after U exposure in reference to unexposed controls. The respiratory activity or “vitality” remaining after U exposure was higher in the piliated strains and proportional to the levels of piliation (pRG5::pilA > WTP+; Fig. 3A). Inasmuch as the activity of the electron transport chain is a vital function of the cell, these results suggested that the pili-catalyzed reduction of U also preserved the cell's viability. To test this, we recovered the resting cells in growth medium and studied the cell's survival (defined as the cell's ability to maintain its integrity and undertake division) (25) after exposure to U as a function of the length of the lag phase (Fig. 3B). Though cells that had not been exposed to U recovered rapidly and simultaneously, the strains exposed to U recovered in a step-wise fashion. The lag phase was shortest (∼18 h) in the hyperpiliated pRG5::pilA cells, followed by the WTP+ (∼56 h) and the WTP− (∼81 h), and correlated well with the levels of periplasmic mineralization of the strains (R2 = 0.947). The PilA− mutant recovery was similar to the other nonpiliated strain, WTP−, yet more variable (lag phases ranging from 72 to 82 h). It also grew faster (∼9 h doubling time compared with ∼11 h for the WT and pRG5::pilA strains) than the other strains, which is expected to accelerate recovery. Despite these differences, the survival rates (calculated as the reverse of the length of the lag phase) of all of the strains followed a linear regression (R2 = 0.908) with the levels of pili protein.
Fig. 3.
Effect of U(VI) exposure on cell vitality (A) and viability (B). (A) Vitality was measured as bacterial reductase (respiratory) activity with the RedoxSensor dye in resting cells of the pili-expressing (WTP+ and pRG5::pilA) and nonexpressing (WTP− and PilA−) strains and expressed as the ratio of relative fluorescence units emitted by cells incubated with (U+) or without (U−) U. (B) Growth recovery of resting cells of the pRG5::pilA (black), WTP+ (dark gray), and WTP− (light gray) after 6 h of U exposure (circles) in comparison with controls without U (lines).
Discussion
Physiological Relevance of the Extracellular Reduction of U by Geobacter's Pili.
Our results show that piliated cells immobilized a greater amount of U and also prevented it from permeating inside the periplasm, where it would have otherwise been reduced nonspecifically by c-cytochromes and other low-potential electron donors (17). As a result, the extracellular reduction of U via pili also preserved the vital functions of the cell envelope and the cell's viability. This mechanism is consistent with field studies showing that the indigenous Geobacter community that is stimulated during in situ bioremediation is metabolically active (9, 26) and gains energy for growth from the reduction of Fe(III) oxides (6, 27), a process that requires the expression of the conductive pili (19). We used a temperature-dependent regulatory switch (SI Materials and Methods) to produce WT controls (WTP−) that did not assemble pili, yet had WT levels and profiles of outer membrane cytochromes. The lack of pili in the WTP− strain significantly diminished the cell's ability to remove U(VI) from solution, increased the degree of periplasmic mineralization, and reduced the respiratory activity of the cell envelope and the cell's viability. WTP− cells also had extensive outer membrane vesiculation, a process linked to the selective detoxification of unwanted periplasmic materials by cells undergoing cell envelope stress (28). Similarly, the inability of a PilA− mutant to produce pili impaired the yields of U reduction. This mutant strain also had a reduced outer membrane cytochrome content and, as a result, more U traversed the outer membrane and precipitated in the periplasm. The fact that the pili of G. sulfurreducens catalyze the extracellular reductive precipitation of U under physiological conditions conducive to growth, is also in agreement with early studies with G. metallireducens suggesting that the reduction of U is extracellular (10) and coupled to cell growth (1). In these studies (1, 10), cells were grown with Fe(III) citrate as an electron acceptor, which are culture conditions that promote pili expression in G. metallireducens (29) but not in G. sulfurreducens (19). Thus, the extracellular precipitation and sustained removal of U reported for G. metallireducens is consistent with pili catalyzing the reaction as well.
Reduction of U to Mononuclear U(IV) Phases.
Despite differences in the mechanism and yields of U reduction, the strains with the lowest levels of periplasmic mineralization (WTP+, pRG5::pilA, and WTP−) produced similar U LIII-edge EXAFS spectra that were modeled as mostly U(IV) coordinated by C-containing ligands in bidentate and monodentate fashion and that lacked any Fe- or P-containing ligands. The bidentate C1–C3 ligand is likely biological in nature, as reported for the carboxyl coordinations involving amino acids and lipolysaccharide sugars (30–32). In contrast, the PilA− mutant, which had the highest degree of periplasmic mineralization, required an additional monodentate P ligand. This signal was small, with a coordination number of 0.5 ± 0.3, indicating that, on average, 50% of the U atoms contained a P ligand, and the other 50% shared the atomic coordination of the other strains. Alternative interpretations such as 25% or 12.5% of U atoms with two or four P ligands, respectively, are unlikely, because the coordination number values for the C1 and C2 signals did not decrease proportionally (50% and 75%, respectively). It is also unlikely that the low levels of U(VI) reduced by the PilA− strain contributed to the distinct spectra, because WTP− cells reduced less U(VI) and did not require a P ligand for U coordination. The P coordination and the generalized periplasmic mineralization observed in the PilA− mutant cells suggest that U(VI) permeated deep and fast into the cell envelope, where it formed carboxyl and phosphoryl-coordinated complexes with periplasmic proteins and the peptidoglycan layer (30, 31) and membrane phospholipids (33), respectively.
The formation of a mononuclear U(IV) phase has also been reported for other bacteria of relevance to U bioremediation (34, 35), yet contrasts with earlier reports of uraninite formation by Geobacter spp. (10, 36). The chemical composition of the medium can influence the nature of the reduced U mineral (34). We used a bicarbonate buffer and conditions used in previous studies with G. sulfurreducens (13), whereas studies reporting uraninite formation used PIPES-buffered solutions (36) or bicarbonate-buffered uncontaminated groundwater (10). Evidence for the microbial reduction of U(VI) to nonuraninite U(IV) products is also emerging from field-scale studies (35, 37), whereas uraninite formation has been linked to conditions of reduced bioreducing activities (38, 39); this suggests that abiotic factors may contribute to the formation of uraninite.
Model for the Reduction of U(VI) by Geobacter Bacteria.
The direct correspondence observed between piliation, extent of U(VI) reduction, cell envelope respiratory activities, and cell viability support a model in which the conductive pili function as the primary mechanism for U reduction and cellular protection (Fig. S6). Pili can reach several micrometers in length, thereby increasing the redox-active surface area available for binding and reducing U(VI) outside the cell. Although most of the U reduced by the piliated cells was extracellular and associated with the pili, discreet regions of the outer membrane also participated in the reduction of U. In G. sulfurreducens, most of the redox activity of the outer membrane is provided by abundant c-cytochromes that decorate the cell surface as defined foci (40). Thus, they could provide a mechanism for reducing U in localized regions of the membrane and prevent it from permeating into the periplasm. In support of this, the PilA− mutant cells, which had reduced outer membrane cytochrome content, preferentially reduced U in the periplasm. Some of the most abundant metalloproteins on the outer membrane of G. sulfurreducens also are loosely bound to and easily detach from the membrane (40–42), providing a natural mechanism for releasing the U deposits. Some of these cytochromes also may be anchored to a recently identified exopolysaccharide matrix (43), which may promote the extracellular reduction of U. Furthermore, although some areas of the outer membrane were voided of U reductase activity, U(VI) was effectively prevented from permeating into the periplasm. The outer leaflet of the outer membrane of Gram-negative bacteria is mostly composed of LPS, which acts as an efficient permeability barrier against soluble toxic compounds (44). G. sulfurreducens produces a rough LPS, i.e., it is composed of lipid A and a core oligosaccharide but lacks the O antigen (45). The core oligosaccharide is the most highly charged region of the LPS and is stabilized by metallic cations (46). Models suggest that rough LPS preferentially chelates and immobilizes uranyl ions over other ions (32) and produces carboxyl and hydroxyl coordinations (32), which is consistent with the C and O coordinations modeled from the EXAFS spectra. These findings suggest that the rough LPS of G. sulfurreducens also functions as a protective barrier to prevent U(VI) from penetrating in the cell envelope.
Implications for the in Situ Bioremediation of U.
Insufficient knowledge of the biological mechanisms of contaminant transformation often limits the performance of in situ subsurface bioremediation and long-term stewardship strategies. The identification of Geobacter's pili as their primary U reductase provides a much-needed, fundamental mechanistic understanding of U reduction by Geobacter spp. required to design effective in situ bioremediation strategies. Analyses of transcript abundance for key Geobacteraceae genes are useful tools to predict the metabolic and physiological state of Geobacter bacteria during in situ bioremediation (26, 47–49), yet provide no information about the mechanism of U bioreduction. However, similar tools could be applied to monitor the activity of conserved components of Geobacter's pilus apparatus to assess the effectiveness of in situ bioremediation schemes. The possibility that conductive appendages such as the pili of Geobacter are a widespread mechanism for U reduction also warrants special attention. The production of conductive appendages has been demonstrated in another U-reducing bacterium, Shewanella oneidensis (50). Furthermore, nanowire-mediated electrical currents have been proposed to couple spatially separated geochemical processes in sediments (51). The extracellular needle-like U deposits observed in TEM thin sections of the piliated strains of G. sulfurreducens (Fig. S4) also resembled the uraninite structures of Desulfovibrio desulfuricans biofilms (52), which some authors have suggested represent mineralized nanowire-like appendages (17). Thus, the contribution of microbial nanowires to U reduction may be significant and, therefore, relevant for the optimization of in situ bioremediation strategies.
Materials and Methods
Strains and Culture Conditions.
WT G. sulfurreducens PCA (ATCC 51573), a pilin-deficient mutant (PilA−) (19), and its genetically complemented strain (pRG5::pilA) (19) were grown in fresh water (FW) medium supplemented with 15 mM acetate and 40 mM fumarate. All cultures were incubated at pili-inducing temperatures (25 °C), except for the nonpiliated WTP− controls, which were grown at 30 °C (SI Materials and Methods).
U(VI) Resting-Cell Suspension Assays.
Resting-cell suspensions were prepared as described elsewhere (13), except that cells were harvested from mid–log-phase cultures (OD600, 0.3–0.5) and resuspended in 100 mL reaction buffer with 20 mM sodium acetate to a final OD600 of 0.1. Heat-killed controls were prepared by autoclaving the cultures for 30 min. Suspensions were incubated for 6 h at 30 °C with 1 mM uranyl acetate (Electron Microscopy Sciences), as previously described (13). After incubation, 500-μL samples were withdrawn, filtered (0.22-μm Millex-GS filter; Millipore) to separate the cells, acidified in 2% nitric acid (500 μL), and stored at −20 °C. All procedures were performed anaerobically inside a vinyl glove bag (Coy Labs) containing a H2:CO2:N2 (7:10:83) atmosphere. The amount of U(VI) removed from solution was estimated from the initial and final concentration of U(VI) measured in the acidified samples using a platform inductively coupled plasma mass spectrometer (Micromass, Thermo Scientific).
X-Ray Adsorption Spectroscopy (XAS) Analyses.
Resting cells incubated with U for 6 h were harvested by centrifugation (13,000 × g, 10 min, 4 °C) and loaded into custom-made plastic holders, triply packaged in Kapton film and sealed with Kapton tape (DuPont) under anaerobic conditions. Samples were stored at −80 °C and kept frozen during XAS measurements, which were performed with a multielement germanium detector in fluorescence mode, using the Pacific Northwest Consortium Collaborative Access Team beamline (sector 20-BM) at the Advanced Photon Source (Argonne National Laboratory) and standard beamline parameters, as described elsewhere (37). XANES measurements were used to calculate the relative amount of U(VI) to U(IV) by linear combination fitting of the spectrum with U(VI) and U(IV) standards. The spectra were energy aligned by simultaneously measured uranyl nitrate standards. EXAFS analyses are described in SI Materials and Methods.
Transmission Electron Microscopy.
After U exposure, resting cells were adsorbed onto 300-mesh carbon-coated copper grids (Ted Pella), fixed with 1% glutaraldehyde for 5 min, and washed three times with ddH2O for 2 min. Unstained cells were directly imaged with a JEOL100CX operated at a 100-kV accelerating voltage. Thin sections for TEM were prepared as described in SI Materials and Methods.
Vitality and Viability Assays.
The RedoxSensor Green vitality assay (Invitrogen) was used to measure the cell's vitality (broadly defined as the levels of activity of a cell's vital reactions) remaining after U exposure and in reference to controls not exposed to U. This reagent yields green fluorescence when modified by the bacterial reductases, which are mostly located in the electron transport system of the cell envelope (24, 53). Resting cells were harvested in a microcentrifuge (12,000 × g), washed, and resuspended in 100 μL Tris-buffered saline (TBS) before mixing it with an equal volume of a working concentration of the dye. Fluorescence was measured in two biological replicates, with two technical replicates each, using a SpectraMax M5 plate reader (Molecular Devices) with an excitation of 490 nm and emission of 520 nm. Cell viability after U exposure in comparison with controls without U was assayed by recovering the resting cells in NB medium with acetate and fumarate (NBAF) and measuring the length of the lag phase, as described previously (54). Before inoculation, resting-cell suspensions were gassed for 15 min with filter-sterilized air to reoxidize the U deposits (13) without compromising G. sulfurreducens viability (54). Cells were harvested by centrifugation (1,200 × g, 5 min), resuspended in 1 mL of wash buffer (final OD600 of 0.4), and mixed with 10 mL of NBAF in pressure tubes. The cultures were incubated at 25 °C, and growth was periodically monitored as OD600.
Supplementary Material
Acknowledgments
We thank Matthew Marshall, Evgenya Shelobolina, Kazem Kashefi, Alicia Pastor, XouDong Fan, Blair Bullard, and the Advanced Photon Source (APS) and Sector 20 staff at Argonne National Laboratory for assistance during different phases of this work. This research was supported by National Institute of Environmental Health Science's Superfund Grant R01 ES017052-03 and US Department of Energy (DOE) Office of Science Office of Biological and Environmental Research Grant DE-SC0000795 (to G.R.). Support was also provided by a Michigan State University College of Natural Science Hensley Fellowship and a Biogeochemistry Environmental Research Initiative Fellowship (to D.L.C.). Pacific Northwest Consortium X-ray Sciences Division facilities and research at the APS are supported by the US DOE Basic Energy Sciences, a Major Resources Support grant from Natural Sciences and Engineering Research Council, the University of Washington, Simon Fraser University, and the APS. The APS is an Office of Science User Facility operated for the US DOE's Office of Science by Argonne National Laboratory and supported by US DOE Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108616108/-/DCSupplemental.
References
- 1.Lovley DR, Phillips EJP, Gorby YA, Landa ER. Microbial reduction of uranium. Nature. 1991;350:413–416. [Google Scholar]
- 2.Sanford RA, et al. Hexavalent uranium supports growth of Anaeromyxobacter dehalogenans and Geobacter spp. with lower than predicted biomass yields. Environ Microbiol. 2007;9:2885–2893. doi: 10.1111/j.1462-2920.2007.01405.x. [DOI] [PubMed] [Google Scholar]
- 3.Lovley DR. Bioremediation. Anaerobes to the rescue. Science. 2001;293:1444–1446. doi: 10.1126/science.1063294. [DOI] [PubMed] [Google Scholar]
- 4.Anderson RT, et al. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl Environ Microbiol. 2003;69:5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Istok JD, et al. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ Sci Technol. 2004;38:468–475. doi: 10.1021/es034639p. [DOI] [PubMed] [Google Scholar]
- 6.Chang YJ, et al. Microbial incorporation of 13C-labeled acetate at the field scale: Detection of microbes responsible for reduction of U(VI) Environ Sci Technol. 2005;39:9039–9048. doi: 10.1021/es051218u. [DOI] [PubMed] [Google Scholar]
- 7.North NN, et al. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl Environ Microbiol. 2004;70:4911–4920. doi: 10.1128/AEM.70.8.4911-4920.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu WM, et al. Pilot-scale in situ bioremedation of uranium in a highly contaminated aquifer. 2. Reduction of u(VI) and geochemical control of u(VI) bioavailability. Environ Sci Technol. 2006;40:3986–3995. doi: 10.1021/es051960u. [DOI] [PubMed] [Google Scholar]
- 9.Wilkins MJ, et al. Proteogenomic monitoring of Geobacter physiology during stimulated uranium bioremediation. Appl Environ Microbiol. 2009;75:6591–6599. doi: 10.1128/AEM.01064-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorby YA, Lovley DR. Enzymatic uranium precipitation. Environ Sci Technol. 1992;26:205–207. [Google Scholar]
- 11.Coppi MV, Leang C, Sandler SJ, Lovley DR. Development of a genetic system for Geobacter sulfurreducens. Appl Environ Microbiol. 2001;67:3180–3187. doi: 10.1128/AEM.67.7.3180-3187.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd JR, et al. Reduction of actinides and fission products by Fe(III)-reducing bacteria. Geomicrobiol J. 2002;19:103–120. [Google Scholar]
- 13.Shelobolina ES, et al. Importance of c-Type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol. 2007;7:16. doi: 10.1186/1471-2180-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim BC, et al. OmcF, a putative c-type monoheme outer membrane cytochrome required for the expression of other outer membrane cytochromes in Geobacter sulfurreducens. J Bacteriol. 2005;187:4505–4513. doi: 10.1128/JB.187.13.4505-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim BC, Lovley DR. Investigation of direct vs. indirect involvement of the c-type cytochrome MacA in Fe(III) reduction by Geobacter sulfurreducens. FEMS Microbiol Lett. 2008;286:39–44. doi: 10.1111/j.1574-6968.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- 16.Kim BC, Qian X, Leang C, Coppi MV, Lovley DR. Two putative c-type multiheme cytochromes required for the expression of OmcB, an outer membrane protein essential for optimal Fe(III) reduction in Geobacter sulfurreducens. J Bacteriol. 2006;188:3138–3142. doi: 10.1128/JB.188.8.3138-3142.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wall JD, Krumholz LR. Uranium reduction. Annu Rev Microbiol. 2006;60:149–166. doi: 10.1146/annurev.micro.59.030804.121357. [DOI] [PubMed] [Google Scholar]
- 18.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reguera G, et al. Extracellular electron transfer via microbial nanowires. Nature. 2005;435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 20.Butler JE, Young ND, Lovley DR. Evolution of electron transfer out of the cell: Comparative genomics of six Geobacter genomes. BMC Genomics. 2010;11:40. doi: 10.1186/1471-2164-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strom MS, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 22.Leang C, Qian X, Mester T, Lovley DR. Alignment of the c-type cytochrome OmcS along pili of Geobacter sulfurreducens. Appl Environ Microbiol. 2010;76:4080–4084. doi: 10.1128/AEM.00023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly SD, et al. Advanced Photon Source Activity Report 2003. Argonne, IL: Argonne Natl Lab; 2005. Comparison of U valence state ratio determined from U L3-Edge XANES to EXAFS measurements. [Google Scholar]
- 24.Gray D, Yue RS, Chueng CY, Godfrey W. Abstracts of the American Society of Microbiology Meeting. Washington, DC: Am Soc Microbiol; 2005. Bacterial vitality detected by a novel fluorogenic redox dye using flow cytometry. [Google Scholar]
- 25.Barer MR, Harwood CR. Bacterial viability and culturability. Adv Microb Physiol. 1999;41:93–137. doi: 10.1016/s0065-2911(08)60166-6. [DOI] [PubMed] [Google Scholar]
- 26.Holmes DE, et al. Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl Environ Microbiol. 2005;71:6870–6877. doi: 10.1128/AEM.71.11.6870-6877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finneran KT, Anderson RT, Nevin KP, Lovley DR. Potential for Bioremediation of uranium-contaminated aquifers with microbial U(VI) reduction. Soil Sediment Contam. 2002;11:339–357. [Google Scholar]
- 28.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Childers SE, Ciufo S, Lovley DR. Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature. 2002;416:767–769. doi: 10.1038/416767a. [DOI] [PubMed] [Google Scholar]
- 30.Barkleit A, Moll H, Bernhard G. Complexation of uranium(VI) with peptidoglycan. Dalton Trans. 2009;(27):5379–5385. doi: 10.1039/b818702a. [DOI] [PubMed] [Google Scholar]
- 31.Benavides-Garcia MG, Balasubramanian K. Structural insights into the binding of uranyl with human serum protein apotransferrin structure and spectra of protein-uranyl interactions. Chem Res Toxicol. 2009;22:1613–1621. doi: 10.1021/tx900184r. [DOI] [PubMed] [Google Scholar]
- 32.Lins RD, Vorpagel ER, Guglielmi M, Straatsma TP. Computer simulation of uranyl uptake by the rough lipopolysaccharide membrane of Pseudomonas aeruginosa. Biomacromolecules. 2008;9:29–35. doi: 10.1021/bm700609r. [DOI] [PubMed] [Google Scholar]
- 33.Koban A, Bernhard G. Uranium(VI) complexes with phospholipid model compounds—a laser spectroscopic study. J Inorg Biochem. 2007;101:750–757. doi: 10.1016/j.jinorgbio.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Fletcher KE, et al. U(VI) reduction to mononuclear U(IV) by Desulfitobacterium species. Environ Sci Technol. 2010;44:4705–4709. doi: 10.1021/es903636c. [DOI] [PubMed] [Google Scholar]
- 35.Bernier-Latmani R, et al. Non-uraninite products of microbial U(VI) reduction. Environ Sci Technol. 2010;44:9456–9462. doi: 10.1021/es101675a. [DOI] [PubMed] [Google Scholar]
- 36.Sharp JO, et al. Structural similarities between biogenic uraninites produced by phylogenetically and metabolically diverse bacteria. Environ Sci Technol. 2009;43:8295–8301. doi: 10.1021/es901281e. [DOI] [PubMed] [Google Scholar]
- 37.Kelly SD, et al. Speciation of uranium in sediments before and after in situ biostimulation. Environ Sci Technol. 2008;42:1558–1564. doi: 10.1021/es071764i. [DOI] [PubMed] [Google Scholar]
- 38.Kelly SD, Hesterberg D, Ravel B. Analysis of soils and minerals using X-ray absorption spectroscopy. In: Ulery AL, Drees LR, editors. Methods of Soil Analysis. Part 5—Mineralogical Methods. Madison, WI: Soil Sci Soc Am; 2008. pp. 367–463. [Google Scholar]
- 39.Kelly SD, et al. Uranium transformations in static microcosms. Environ Sci Technol. 2010;44:236–242. doi: 10.1021/es902191s. [DOI] [PubMed] [Google Scholar]
- 40.Qian X, Reguera G, Mester T, Lovley DR. Evidence that OmcB and OmpB of Geobacter sulfurreducens are outer membrane surface proteins. FEMS Microbiol Lett. 2007;277:21–27. doi: 10.1111/j.1574-6968.2007.00915.x. [DOI] [PubMed] [Google Scholar]
- 41.Mehta T, Coppi MV, Childers SE, Lovley DR. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl Environ Microbiol. 2005;71:8634–8641. doi: 10.1128/AEM.71.12.8634-8641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehta T, Childers SE, Glaven R, Lovley DR, Mester T. A putative multicopper protein secreted by an atypical type II secretion system involved in the reduction of insoluble electron acceptors in Geobacter sulfurreducens. Microbiology. 2006;152:2257–2264. doi: 10.1099/mic.0.28864-0. [DOI] [PubMed] [Google Scholar]
- 43.Rollefson JB, Stephen CS, Tien M, Bond DR. Identification of an extracellular polysaccharide network essential for cytochrome anchoring and biofilm formation in Geobacter sulfurreducens. J Bacteriol. 2011;193:1023–1033. doi: 10.1128/JB.01092-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vinogradov E, Korenevsky A, Lovley DR, Beveridge TJ. The structure of the core region of the lipopolysaccharide from Geobacter sulfurreducens. Carbohydr Res. 2004;339:2901–2904. doi: 10.1016/j.carres.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 46.Schindler M, Osborn MJ. Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry. 1979;18:4425–4430. doi: 10.1021/bi00587a024. [DOI] [PubMed] [Google Scholar]
- 47.Holmes DE, et al. Genes for two multicopper proteins required for Fe(III) oxide reduction in Geobacter sulfurreducens have different expression patterns both in the subsurface and on energy-harvesting electrodes. Microbiology. 2008;154:1422–1435. doi: 10.1099/mic.0.2007/014365-0. [DOI] [PubMed] [Google Scholar]
- 48.Holmes DE, Nevin KP, Lovley DR. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl Environ Microbiol. 2004;70:7251–7259. doi: 10.1128/AEM.70.12.7251-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes DE, et al. Transcriptome of Geobacter uraniireducens growing in uranium-contaminated subsurface sediments. ISME J. 2009;3:216–230. doi: 10.1038/ismej.2008.89. [DOI] [PubMed] [Google Scholar]
- 50.Gorby YA, et al. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc Natl Acad Sci USA. 2006;103:11358–11363. doi: 10.1073/pnas.0604517103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen LP, Risgaard-Petersen N, Fossing H, Christensen PB, Sayama M. Electric currents couple spatially separated biogeochemical processes in marine sediment. Nature. 2010;463:1071–1074. doi: 10.1038/nature08790. [DOI] [PubMed] [Google Scholar]
- 52.Marsili E, et al. Uranium removal by sulfate reducing biofilms in the presence of carbonates. Water Sci Technol. 2005;52:49–55. [Google Scholar]
- 53.Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Real-time detection of actively metabolizing microbes by redox sensing as applied to methylotroph populations in Lake Washington. ISME J. 2008;2:696–706. doi: 10.1038/ismej.2008.32. [DOI] [PubMed] [Google Scholar]
- 54.Lin WC, Coppi MV, Lovley DR. Geobacter sulfurreducens can grow with oxygen as a terminal electron acceptor. Appl Environ Microbiol. 2004;70:2525–2528. doi: 10.1128/AEM.70.4.2525-2528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.