Abstract
Circadian rhythms are a fundamental property of most organisms, from cyanobacteria to humans. In the unicellular obligately photoautotrophic cyanobacterium Synechococcus elongatus PCC 7942, essentially all promoter activities are controlled by the KaiABC-based clock under continuous light conditions. When Synechococcus cells are transferred from the light to continuous dark (DD) conditions, the expression of most genes, including the clock genes kaiA and kaiBC, is rapidly down-regulated, whereas the KaiC phosphorylation cycle persists. Therefore, we speculated that the posttranslational oscillator might not drive the transcriptional circadian output without de novo expression of the kai genes. Here we show that the cyanobacterial clock regulates the transcriptional output even in the dark. The expression of a subset of genes in the genomes of cells grown in the dark was dramatically affected by kaiABC nullification, and the magnitude of dark induction was dependent on the time at which the cells were transferred from the light to the dark. Moreover, under DD conditions, the expression of some dark-induced gene transcripts exhibited temperature-compensated damped oscillations, which were nullified in kaiABC-null strains and were affected by a kaiC period mutation. These results indicate that the Kai protein-based posttranslational oscillator can drive the circadian transcriptional output even without the de novo expression of the clock genes.
Keywords: dark-induced genes, kaiABC genes, digA, hspA, pilH/rre7
Most organisms exhibit circadian oscillations in their physiological activities, with a period of ≈24 h, as part of their adaptation to external environmental changes. The unicellular cyanobacterium Synechococcus elongatus PCC 7942 is an obligate photoautotroph and the simplest model organism in circadian biology. In S. elongatus, most genes display circadian expression rhythms that are regulated by three clock genes, kaiA, kaiB, and kaiC, under continuous light (LL) conditions (1, 2). When the cells are transferred to continuous dark (DD) conditions after 12 h in the light, expression of the kaiA and kaiBC genes is rapidly down-regulated to zero, whereas the KaiC phosphorylation cycle persists in the dark, even in the presence of excess transcription/translation inhibitors (3). Therefore, the basic oscillation is generated via a posttranslational process and does not need a translation/transcription feedback loop in the kai genes. The reconstitution of the temperature-compensated KaiC phosphorylation rhythm in vitro when KaiC is incubated with KaiA, KaiB, and ATP (4) supports this conclusion. Kitayama et al. (5) demonstrated rhythmic kaiBC expression and KaiC accumulation with a lengthened period of ≈60 h, even after two phosphorylation sites in KaiC (Ser-431 and Thr-432) were replaced with Glu. However, the unstable rhythm observed in the mutant was not robust under different culture conditions and different ambient temperatures (6). In eukaryotic model organisms, the core process that generates and maintains self-sustaining circadian oscillations is reported to be driven by transcription/translation feedback loops (7). However, it was recently demonstrated in the pico-eukaryotic alga Ostreococcus tauri and human red blood cells that the oxidation of 2-Cys peroxiredoxin (PRX) proteins undergo ≈24-h modification cycles without transcription (8, 9). Therefore, we now require a more general understanding of the mechanisms by which posttranslational oscillators control overt physiological rhythms, such as transcription cycles.
Previously we performed DNA microarray experiments in Synechococcus under LL and DD conditions (2). In the WT strains, more than 30% of transcripts exhibited significant circadian rhythms under LL, peaking at subjective dawn and dusk. When the cells were transferred from the light to DD, the expression of not only the kai genes but also most other genes was rapidly suppressed, regardless of whether they were regulated continuously or in a circadian manner under LL conditions, and the total transcript levels were dramatically reduced in the dark, reaching ≈20% within 12 h. Because the ATP/(ADP+ATP) ratio falls dramatically under dark conditions (10), immediate genome-wide transcriptional suppression in the dark might function as an energy-saving process. In contrast, the Kai proteins are able to drive the KaiC phosphorylation cycle with only 15–25 ATP/molecule per day in vitro (11), consistent with the nutrition-compensated phosphorylation rhythms that occur under DD conditions (3). Nevertheless, a minor subset of genes was up-regulated after 4 h in the dark and did not show a clear circadian accumulation rhythm when the previously applied filtration method was used (2). On the basis of these observations, we proposed that the Synechococcus clock cannot drive the circadian transcriptional output in the dark (2, 3). However, it remains possible that dark-induced transcription could be modified in Synechococcus by the circadian clock, even in the absence of de novo kai gene expression.
Here we show that the dark-induced expression profiles of two representative genes were dramatically altered in the kaiABC-null Synechococcus strain: hspA, encoding a small heat-shock protein, and digA, encoding a hypothetical protein. Consistent with this, their dark-induced profiles were dependent on the circadian time (CT), indicating the circadian gating of the dark induction of these genes. For a more comprehensive insight, we performed a DNA microarray analysis, which demonstrated that 167 dark-induced transcripts, corresponding to 7% of the genes in the genome, are kai gene dependent, and these were classified into four differentially regulated groups. Further analysis revealed that all of the tested genes in each group exhibited time of day-dependent dark-induced profiles. Interestingly, kai-dependent transcriptional modification in the dark was exclusively observed when the cells were transferred to the dark at subjective dusk. Moreover, we found that the oscillation of digA expression was damped under DD conditions, which was temperature compensated and abolished in the kaiABC-disrupted strains. Interestingly, when a group 2 σ factor gene rpoD5 was genetically nullified, the amplitudes of the damped oscillations in digA and pilH/rre7 expression partly increased under DD and peaked at subjective dusk and dawn, respectively. These rhythms were affected in a kaiC short period mutant strain, further supporting the notion that the damped transcriptional rhythms are under the control of the Kai protein-based oscillator. Together, these observations indicate that the posttranslational KaiABC-based circadian oscillator regulates the transcriptional output, even in the dark, without the de novo transcription/translation of the kaiABC genes.
Results and Discussion
Differential Effects of the kai Genes on Dark-Expression Profiles Even Without de Novo kai Gene Transcription.
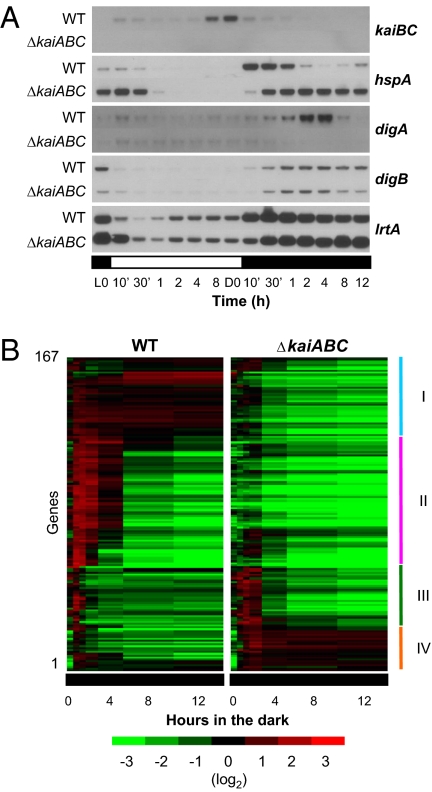
Initially, we selected four highly dark-inducible genes, Synpcc7942_2401/syc1704_d (hspA), Synpcc7942_1661/syc2427_c (hereafter designated digA, for dark-induced gene A), Synpcc7942_2540/syc1570_d (hereafter designated digB, for dark-induced gene B), and Synpcc7942_2352/syc1751_d (lrtA), on the basis of our previous microarray analysis (2). We then examined the effects of kaiABC nullification on the expression profiles of these genes in cells transferred to DD conditions after 12 h in the light. Our results demonstrate that the regulation of two transcripts, hspA and digA, was dependent on the kai genes, whereas the dark-induced profiles of the other genes were less affected by kaiABC nullification (Fig. 1A). The hspA gene encodes a small heat-shock protein that may be involved in the heat-stress response (12), and digA encodes a hypothetical protein of unknown function. In the WT strain, hspA expression was rapidly induced (within 30 min) in the dark and then down-regulated, reaching a nadir after 4–8 h, followed by a gradual increase. In contrast, the transcription of digA was induced more gradually, peaking after 4 h in the dark, followed by its down-regulation to the basal level after 12 h (Fig. 1A). Strikingly, the effects of kaiABC nullification also were opposite for the two genes. In the mutant strains, hspA was up-regulated rapidly in the dark, and its subsequent down-regulation was abolished. In contrast, the dark induction of the digA gene per se was abolished in the kaiABC-null strain. It is noteworthy that both hspA and digA are constitutively less expressed under LL, with no significant circadian accumulation rhythm (Fig. S1). Nevertheless, these results indicate that the Kai proteins are required for proper gene expression in the dark. Conversely, the dark-induced profiles of lrtA and digB expression were much less altered in the kaiABC-null strains (Fig. 1A). The lrtA genes in Synechocystis sp. PCC 6803 (13, 14) and Synechococcus sp. PCC 7002 (15) are known to be dark induced, although their functions remain unknown. The digB gene encodes a hypothetical protein that contains a potential copper-binding domain.
Fig. 1.
kaiABC nullification alters dark-induced gene expression. (A) Northern hybridization analysis of the temporal expression profiles of the kaiBC operon and four representative dark-induced genes under 12 h/12 h LD conditions. Wild-type and kaiABC-null (ΔkaiABC) mutant cells were collected at the indicated times. Note that hspA and digA are differentially regulated by the kai genes. (B) Of the genes in the genome, 167 (7%) were identified as kai-dependent dark-induced genes and were categorized into four groups, types I–IV. The colors for the microarray data represent the expression levels, in descending order from red to black to green.
For a more comprehensive analysis, we compared the whole-genome expression profiles of the WT and kaiABC-null Synechococcus strains transferred to the dark (for 30 min, 1 h, 2 h, 4 h, 8 h, and 12 h) after 12 h in the light (Fig. 1B and Fig. S2A), using previously designed DNA microarrays (2). According to two independent experiments, a tendency for global transcriptional repression in the dark was observed in both the kaiABC-null mutant and the WT strain (Fig. S2A). Under our experimental conditions, the reductions in the total amounts of mRNAs occurred slightly faster in the mutant than in the WT strain (Fig. S2B), which could be attributable to greater mRNA degradation and/or lower transcription rates. We then identified the dark-induced genes in both strains and confirmed that their expression profiles differed. Initially we identified 198 dark-induced genes from among the 2,515 genes examined. The dark-induced genes in this study were defined as those genes that are expressed more abundantly at at least one time point in the nocturnal period than at hour 0 in the dark (= after 12 h in the light; SI Materials and Methods). We next identified those genes among the 198 dark-induced genes that were regulated by the kai genes by comparing their expression profiles in the WT and kaiABC-null strains. To evaluate the kai dependence of these genes, we performed a correlation analysis to compare the waveforms of their expression profiles in the two strains and calculated the statistical differences between the temporal averages of the two expression profiles to examine the differences in the absolute expression levels of the genes (SI Materials and Methods). Fig. S2C shows the distributions of the two parameters for the 198 dark-induced genes tested. According to the statistical analysis, 167 genes, including hspA and digA, and 31 genes, including digB and lrtA, were categorized as kai-dependent and kai-independent dark-induced genes, respectively (Fig. S2C). The possible roles of the dark-induced genes involved many cellular functions (Fig. S2D). As expected, most (76 of 78) photosynthesis-specific genes, which do not directly share respiratory functions, were not induced by the dark, whereas more than half (four of seven) of the respiration-specific cytochrome c oxidase genes were induced by the dark. Notably, the ratio of the 113 dark-induced genes annotated as “unknown function” genes to 198 dark-induced genes (57%) is significantly higher than that of the total 1,094 unknown function genes to the total 2,515 tested genes (43%). This reflects the limited information available on the nocturnal life of Synechococcus, for which most physiological studies have been performed in the light.
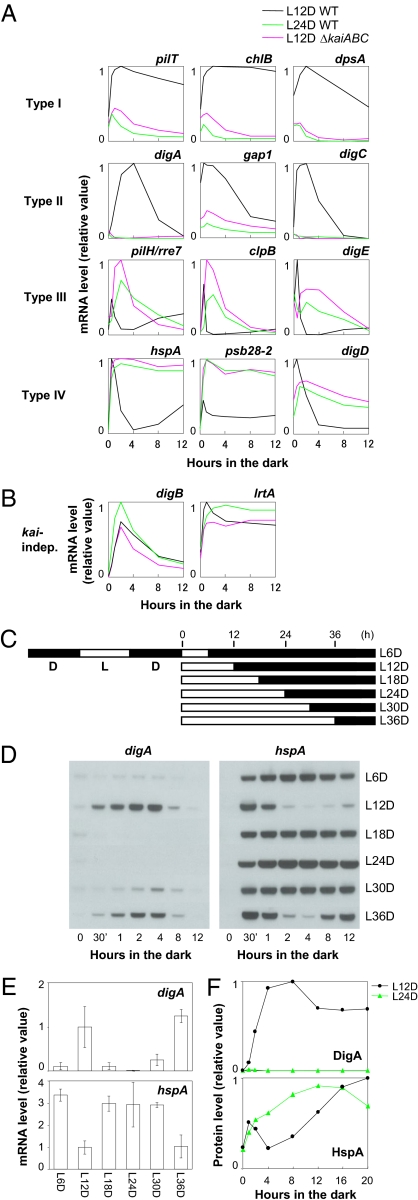
The 167 kai-dependent dark-induced genes were classified into four groups (Fig. 1B), which were validated by the Northern blotting analysis of three representative genes from each group. These exhibited similar patterns to those of the microarray profiles (Fig. S3A). As shown in Fig. 2A, type I genes, represented by pilT, chlB, and dpsA, were induced and accumulated constitutively after the cells were transferred to the dark after 12 h in the light in the WT strain (black lines), in contrast to their much lower expression in the kaiABC-null mutant (red lines). Type II genes, such as digA, gap1, and Synpcc7942_2590/syc1520_c (hereafter designated digC, for dark-induced gene C), exhibited transient dark induction in the WT, peaking after approximately 4 h in the dark but were not induced in the kaiABC-null mutant. Type III genes, represented by pilH/rre7, clpB, and Synpcc7942_1153/syc0397_d (hereafter designated digE, for dark-induced gene E), showed a transient dark-induction peak within 30 min, which was rapidly suppressed in the WT strain, whereas this suppression was delayed by kaiABC nullification, showing induction peaks after 2 h in the dark. Type IV genes, represented by hspA, psb28-2, and Synpcc7942_0316/syc1197_d (hereafter designated digD, for dark-induced gene D), were even more transiently and rapidly induced in the dark, peaking at approximately 30 min, whereas kaiABC nullification caused the constitutive accumulation of their transcripts. These results suggest that the kai genes are necessary for the production of the proper differential gene expression profiles in the dark.
Fig. 2.
Circadian gating of dark-induced gene expression. (A) Day-length dependence of dark-induced profiles. Northern blotting analysis of three representative genes from each group of kai-dependent dark-induced genes (Fig. 1B) under the dark. Type I: pilT, chlB, and dpsA; type II: digA, gap1, and digC; type III: pilH/rre7, clpB, and digE; and type IV: hspA, psb28-2, and digD. When the cells were transferred to the dark after 12 h in the light, the expression profile of each gene was quite different in the WT (black line) and kaiABC-null mutant (red line) strains. When the WT cells were transferred to the dark after 24 h in the light, the expression profiles (green line) changed, becoming essentially similar to those in the mutant strain. (B) Dark-induced profiles of the digB and lrtA genes, which are categorized as kai-independent genes (kai-indep). Data are shown as in panel A. (C) Schematic representation of the experimental schedule. After the cells were entrained in two 12 h/12 h LD cycles, they were placed in the light for the indicated times, ranging from 6 h to 36 h in increments of 6 h, and were then transferred to the dark conditions. (D) Northern blotting analysis of the digA and hspA genes in the dark when they had been transferred after 6 h (L6D) to 36 h (L36D) under LL. The cells were collected at hours 0, 0.5 (30 min), 1, 2, 4, 8, and 12 in the dark. (E) Time of day-dependent induction levels of digA and hspA mRNAs after 4 h in the dark. The mean values and SDs of four independent experiments are shown. These data were initially normalized to the level at hour 0 in the light; the average level after 4 h in the dark under L12D conditions was then deemed to be 1. (F) Densitometric data for the time of day-dependent DigA and HspA protein accumulation in the dark, when the cells had been transferred to the dark after 12 h (L12D) or 24 h (L24D) in the light, analyzed by Western blotting.
Subjective Night-Specific Gating of Dark-Induced Gene Expression by the Circadian Clock.
The kai-gene dependence of the dark-induced expression profiles does not always indicate that these profiles are under the control of the circadian clock, because the kai genes may have functions other than those in the circadian timing mechanism. Therefore, we examined whether the kai-dependent dark-induced profiles of the digA (type II) and hspA (type IV) genes were dependent on CT when the cells were transferred to the dark from LL after two 12 h/12 h light/dark (LD) cycles, on the experimental schedule shown in Fig. 2C. The dark induction of digA and the dark suppression of hspA, respectively, were only observed when the cells were transferred to the dark from subjective dusk (Fig. 2 D and E). Therefore, the Kai-based clock seems to regulate the timing of digA induction and hspA attenuation that occur after 2–8 h in the dark, when the cells are transferred to the dark from the light at subjective dusk. The translation rate of S. elongatus, as well as its transcriptional activity, is also largely reduced by dark acclimation (16). Therefore, transcriptional activation in the dark may not always be accompanied by the subsequent translational induction of the encoded proteins. We analyzed the strains in which each DigA or HspA protein was expressed by immunoblotting of the FLAG epitope (SI Materials and Methods). Consistent with the mRNA profiles, the induction of DigA protein and the transient repression of HspA expression in the dark were observed when the cells were transferred from subjective dusk (after 12 h in the light) to the dark and were abolished in cells transferred from subjective dawn (after 24 h in the light) to the dark (Fig. 2F). Note that the expression of FLAG-tagged DigA or HspA did not affect kaiBC expression rhythms under LL conditions (Fig. S4). These results indicate that the KaiABC-based clock regulates not only the transcription but also the translation of the two genes in the dark without the de novo expression of the clock genes.
For a more comprehensive analysis, we examined the dawn/dusk dependence of the dark induction of type I–IV kaiABC-dependent genes. Interestingly, regardless of the gene type, the dark-accumulation profile of each of the genes in the WT cells, which were transferred to the dark after 24 h in the light, was essentially similar to that in the kaiABC-null mutant cells, which were incubated in the dark after 12 h in the light (Fig. 2A). Conversely, negligible time of day-dependent regulation was observed for the digB and lrtA genes, whose dark-induction profiles were not dependent on the kai genes (Fig. 2B). We also confirmed that the time of day-dependent dark-induction of digA and hspA was abolished when the kaiABC-null mutant cells were transferred from the light to the dark, regardless of whether it was at subjective dusk or subjective dawn (Fig. S3C). These results further demonstrate that the Kai-based clock system gates the timing of most dark-induced gene expression and that this kai-dependent gating is triggered when the cells are transferred from the light to the dark at subjective dusk. It seems possible that as-yet-unknown transcription factors mediating this circadian gating are induced exclusively during subjective dusk in the light and are activated after dark acclimation. Alternatively, the transcription regulators could be constitutively expressed or induced in the dark, independent of the kai genes, and then activated by unknown subjective dusk-specific, clock-controlled posttranslational signals.
Kai-Based Posttranslational Oscillator Drives Damped Oscillation of digA and pilH/rre7 Transcription in the Dark Without de Novo kai Gene Expression.
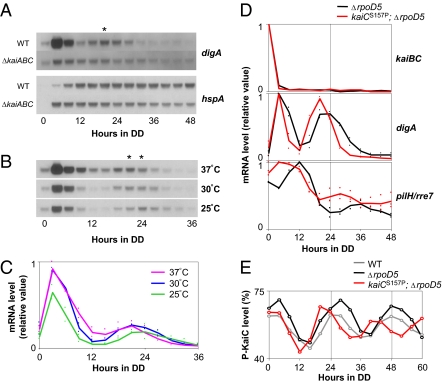
We also found that under extended DD conditions, digA mRNA showed an additional transient peak after approximately 20–24 h under DD after its initial expression peak after 4–8 h in the dark, when the cells had been transferred to the dark after 12 h in the light (Fig. 3A). The second peak was usually smaller than the first peak, forming a damped oscillation pattern. Notably, no third peak was observed when the DD conditions were further extended. In the kaiABC-null strains, a faint rapid induction in the dark was followed by a gradual reduction in digA transcript levels, and no second peak was observed. Therefore, this damped oscillation in the dark seemed to be under the control of the Kai-based clock. The second peak always appeared after approximately 20–24 h in the dark under different continuous-temperature conditions (25 °C, 30 °C, or 37 °C), further supporting the idea that the temperature-compensated clock system regulates the cyclic induction of digA in the dark, even with no de novo expression of the clock genes (Fig. 3 B and C). Our previous analysis, in which we identified clock-controlled transcripts, was based on cosine curve fitting, with a period within the circadian range (2). This neglected the damped oscillation of digA, which had a shorter peak-to-peak interval and a significantly lower induction level at the second peak. In contrast to digA, under extended DD conditions, hspA showed a constitutive accumulation profile after 12 h in the dark, and no damped oscillation was observed (Fig. 3A).
Fig. 3.
Temperature-compensated, kaiC-dependent damped oscillations in digA and pilH/rre7 expression under DD. (A) Temporal expression profiles of the digA and hspA genes in the WT and kaiABC-null (ΔkaiABC) strains under DD after the cells were transferred from 12 h in the light (L12D). Asterisk shows the second peak of the damped digA mRNA cycle observed in the WT strain. (B) Temporal digA expression profile under DD at different temperatures (37 °C, 30 °C, and 25 °C). Note that the standard temperature in all other experiments described in the text was 30 °C. (C) Densitometric analysis of the temporal digA expression profile under DD at different temperatures in two independent experiments. (D and E) The kaiBC, digA, and pilH/rre7 mRNA expression profiles (D) and the KaiC phosphorylation cycles (E) in the rpoD5-null (ΔrpoD5) mutant strain and the kaiCS157P;ΔrpoD5 double mutant strain. For the experiments shown in A and B, the short-period mutant strain was transferred to the dark at hour 10.5 (estimated subjective dusk = CT 12, considering its period length of 21 h) from the light, after two 12 h/12 h LD cycles, whereas the WT strain was transferred to the dark at hour 12 (CT 12).
If the damped oscillation in digA expression under DD is controlled by the Kai-based oscillator, it should be affected by period mutations mapped to the kai genes. Because of the rapid damping property, it was difficult to estimate its convincing period length. We found that the magnitude of the second peak of the digA transcripts was much higher than the first peak, in Synechococcus strains in which the rpoD5 gene was genetically nullified, although no third peak was observed (Fig. 3D, black lines, and Fig. S5A). The rpoD5 gene (also known as sigC) encodes a group 2 σ factor of RNA polymerase and is reported to modulate the circadian transcriptional outputs in Synechococcus under LL, although the detailed underlying mechanism remains unknown (17). It is noteworthy that rpoD5 nullification did not affect the immediate suppression of the kaiBC transcripts in the dark (Fig. 3D and Fig. S5A), the self-sustaining KaiC phosphorylation cycle under DD for 60 h (Fig. 3E), or the kaiBC expression rhythm under LL, monitored with a bioluminescence reporter (Fig. S5B) (17). Therefore, the core oscillatory mechanism remained intact after the removal of the rpoD5 gene. We found that the transcript of pilH/rre7 also showed a damped oscillation in the rpoD5-null mutant strains, whereas this was not obvious in the WT strains (Fig. 3D and Fig. S5A). pilH/rre7 encodes a CheY-type response regulator of unknown function: we have previously shown that its genetic nullification does not affect circadian kaiBC gene expression profile in LL (18). Because its expression peak at hour 12 is kai dependent (Fig. 2A), pilH/rre7 provides another example of the genes periodically controlled under DD, like digA, although digA and pilH/rre7 peaked differentially at subjective dusk and dawn, respectively. We then examined whether the expression profiles of digA and pilH/rre7 in the rpoD5-null background were affected by the kaiCS157P mutation in the kaiC gene. The kaiCS157P mutation shortens the free-running period by ≈3 h under LL conditions (Fig. S5B) (4) and in vitro (4, 11). We confirmed that the short-period phenotype in the kaiBC expression rhythm under LL was not affected by rpoD5-nullification (Fig. S5B), and the kaiCS157P;rpoD5-null double mutant strain showed a shorter period oscillation in KaiC phosphorylation under DD, as expected (Fig. 3E). Consistent with this, the peaks of the digA and pilH/rre7 expression rhythms were advanced by the short-period mutation under DD compared with those in the intact kaiC background (Fig. 3D). On the basis of these observations, we conclude that the damped oscillations of digA and pilH/rre7 transcripts under DD were driven by the Kai-based posttranslational oscillator without any de novo expression of the kai genes. The mechanism by which rpoD5 nullification enhances the expression of these genes remains to be resolved. Importantly, a tendency to accumulate more transcripts under DD from hours 12 to 36 was observed not only for the digA (type II) and pilH/rre7 (type III) genes but also for the pilT (type I) and hspA (type IV) genes, and even the kai-independent digB and lrtA genes (Fig. S5A). Therefore, RpoD5 may function directly or indirectly for the transcriptional repression or destabilization of the dark-induced transcripts in the dark in a gene-nonspecific manner, and its genetic nullification would restore the higher-amplitude digA and pilH/rre7 circadian transcription cycles.
Under LL conditions, a two-component regulatory system composed of a KaiC-interacting sensory histidine kinase, SasA, and its cognate response regulator, RpaA, is the main output component, mediating time signals from the Kai-based oscillator to genome-wide circadian gene expression via phosphotransfer activities in Synechococcus (18, 19). The phosphoryl transfer activity rhythm has been observed from SasA to RpaA in the in vitro reconstituted KaiC phosphorylation cycle system (18). In both the sasA- and rpaA-null mutant strains, the kaiA and kaiBC genes are also dramatically down-regulated and conditionally arrhythmic under LL, indicating a secondary feedback to the core timing mechanism (18, 19). It is of interest to ask whether the SasA-RpaA system is also involved in circadian output in the dark. If any change in the dark-induction profiles is observed in these mutants, however, it would be difficult to interpret whether it is due to defects in the output pathways or that in the oscillator per se. Although the SasA-RpaA system may be involved at least indirectly in the damped transcriptional oscillation in the dark, it seems insufficient to explain the mechanism by which digA, a “low-amplitude” or “arrhythmic” gene (defined from their expression profiles in LL; Fig. S1) specifically turned to be one of exceptionally rhythmic genes in the dark, where most of “high-amplitude” genes defined in LL become arrhythmia.
Conclusion
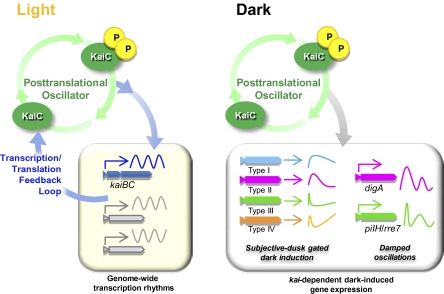
Our study demonstrates that the posttranslational Kai-based circadian oscillator regulates the transcriptional output in the dark, even when de novo kai gene expression is inhibited. Before our study, several lines of previous studies have suggested in Synechococcus that the circadian kaiBC promoter activity is not always an essential requirement for driving its own transcription cycles. The rhythm of the kaiBC promoter activity has been observed when a moderate level of the KaiC protein was constitutively induced under the control of the inducible trc promoter under LL (20–22). Moreover, when the kaiBC operon is under the control of the purF promoter, peaking at subjective dawn, the subjective dusk peaking kaiBC promoter activity, monitored with a bioluminescence reporter, is restored, suggesting that the phase of clock gene expression does not dictate the phase of the kaiBC promoter activity rhythm (23). However, these studies did not report rhythmic transcription in the dark or in the absence of de novo clock gene expression in Synechococcus. Our results further established that the kaiBC promoter is not the only primary transcriptional target of the clock, although most previous transcriptional studies of the Synechococcus clock system have involved the kaiBC promoter/mRNA rhythms. Because most physiological studies of Synechococcus have been performed exclusively under LL, the functions of most dark-induced genes are largely unknown. This study should be the basis for further analysis of the unknown nocturnal life of this representative obligately photoautotrophic model strain. More generally, the results shown here provide insight into how the core posttranslational oscillator is coordinated with transcription/translation feedback loops for clock gene expression. In Synechococcus under LL conditions, the core phosphorylation/ATPase oscillator is coupled to the transcription/translation feedback loop of kai gene expression, in which the KaiC protein negates its own transcription and drives genome-wide expression rhythms. Under the DD condition, the expression of most genes, including kaiBC transcription/translation, is strongly suppressed, whereas the Kai-based oscillator keeps time, gates the expression of the dark-induced genes, and drives damped oscillations in a subset of genes without the de novo expression of the clock genes (Fig. 4). The coordination of the posttranslational oscillator and the transcription/translation feedback loops has recently also been suggested to occur in the mammalian (8) and Ostreococcus circadian systems (9). In both cases, transcription/translation feedback loops regulated by transcription-related proteins (such as the CLOCK and BMAL proteins in mammals and the CCA1 and TOC1 proteins in Ostreococcus) are important in driving the transcriptional rhythms. They are probably coupled to posttranslational oscillators driving the rhythmic oxidation/oligomerization of PRX, which is sustained without de novo transcription/translation (8, 9). It is noteworthy that the Ostreococcus circadian system, in particular, shares some important properties with that of Synechococcus: (i) high-amplitude transcription cycles are observed under LL; (ii) general transcription activity, including the de novo expression of known clock genes, is strongly inhibited in the dark; and (iii) some posttranslational oscillations are sustained to keep time under DD, so that the phase of the transcriptional rhythms can be adjusted after light returns (3, 8). Remaining questions of interest are how the Kai-based posttranslational oscillator regulates transcription under DD and whether the posttranslational oscillator regulates transcription cycles independent of the classical transcription/translation feedback loops in eukaryotic systems.
Fig. 4.
Schematic representation of the differential coordination of the Kai protein-based posttranslational oscillator and the transcription/translation feedback process under light and dark conditions.
Materials and Methods
Bacterial Strains.
Synechococcus elongatus PCC 7942 and its derivatives used in this study are listed in Table S1. Detailed information on the strains is given in SI Materials and Methods.
Culture Conditions.
For the microarray, Northern, and Western blotting analyses, Synechococcus cells were grown in BG-11 medium in a continuous culture system in a 1.5-L flat glass bottle for algal culture (Fujimoto Rika; W 160 mm, D 40 mm, H 500 mm), at an optical density at 730 nm (OD730) of ≈0.3 at 30 °C under white fluorescent light at 40 μmol m−2 s−1, illuminating the lateral flat surface of the bottle. To synchronize the circadian clock, the culture was acclimated to two LD cycles (12 h/12 h), transferred to the light for the indicated periods, and then removed to DD conditions.
Northern Blotting Analysis.
Northern blotting analysis was performed by loading 2 μg of total RNA into each lane, as described previously (3).
DNA Microarray Analysis.
DNA microarray analysis was performed using an Affymetrix GeneChip designed based on the Synechococcus genome (2), and the data deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE22468) were processed with statistical analysis to identify dark-induced genes and kai-dependent genes, as described in the SI Materials and Methods. The statistical data are provided in Fig. S2C and Dataset S1. The procedure for the normalization of the time course data is shown schematically in Fig. S6.
Western Blotting Analysis.
Proteins were extracted from each sample as described previously (3). Immunoblots were incubated in the presence of anti-FLAG antibody (Sigma) diluted 1:1,000 or 3:1,000, and the FLAG-tagged proteins were visualized with chemiluminescence reagents (GE Healthcare).
Bioluminescence Assays.
The bioluminescence profiles were measured with a photomultiplier tube on agar plates under LL after two cycles of 12 h/12 h LD alternations, as described previously (1).
Supplementary Material
Acknowledgments
We thank Ms. Hiroko Kushige (Waseda University) and Dr. Yoriko Murayama (Waseda University) for their helpful suggestions, and members of the Iwasaki Laboratory (Waseda University) for their valuable comments and advice. This study was supported in part by Grants-in-Aid 20370072, 22657043, 21010517, and 23687002 from the Japanese Society for the Promotion of Science, Waseda University Grant for Special Research Projects 2010A-503, the Asahi Glass Foundation, and the Mitsubishi Foundation (to H. Iwasaki); and the Waseda University Project and Program-Based Learning Graduate Student Program (N.H. and T.S.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) (accession no. GSE22468).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019612108/-/DCSupplemental.
References
- 1.Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 2.Ito H, et al. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci USA. 2009;106:14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 5.Kitayama Y, Nishiwaki T, Terauchi K, Kondo T. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 2008;22:1513–1521. doi: 10.1101/gad.1661808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin X, Byrne M, Xu Y, Mori T, Johnson CH. Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol. 2010;8:e1000394. doi: 10.1371/journal.pbio.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang EE, Kay SA. Clocks not winding down: Unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- 8.O'Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rust MJ, Golden SS, O'Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331:220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terauchi K, et al. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104:16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakthivel K, Watanabe T, Nakamoto H. A small heat-shock protein confers stress tolerance and stabilizes thylakoid membrane proteins in cyanobacteria under oxidative stress. Arch Microbiol. 2009;191:319–328. doi: 10.1007/s00203-009-0457-z. [DOI] [PubMed] [Google Scholar]
- 13.Imamura S, et al. Antagonistic dark/light-induced SigB/SigD, group 2 sigma factors, expression through redox potential and their roles in cyanobacteria. FEBS Lett. 2003;554:357–362. doi: 10.1016/s0014-5793(03)01188-8. [DOI] [PubMed] [Google Scholar]
- 14.Summerfield TC, Sherman LA. Role of sigma factors in controlling global gene expression in light/dark transitions in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2007;189:7829–7840. doi: 10.1128/JB.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samartzidou H, Widger WR. Transcriptional and posttranscriptional control of mRNA from lrtA, a light-repressed transcript in Synechococcus sp. PCC 7002. Plant Physiol. 1998;117:225–234. doi: 10.1104/pp.117.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doolittle WF. The cyanobacterial genome, its expression, and the control of that expression. Adv Microb Physiol. 1979;20:1–102. doi: 10.1016/s0065-2911(08)60206-4. [DOI] [PubMed] [Google Scholar]
- 17.Nair U, Ditty JL, Min H, Golden SS. Roles for sigma factors in global circadian regulation of the cyanobacterial genome. J Bacteriol. 2002;184:3530–3538. doi: 10.1128/JB.184.13.3530-3538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takai N, et al. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki H, et al. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 20.Murayama Y, Oyama T, Kondo T. Regulation of circadian clock gene expression by phosphorylation states of KaiC in cyanobacteria. J Bacteriol. 2008;190:1691–1698. doi: 10.1128/JB.01693-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: Roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 2003;22:2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakahira Y, et al. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proc Natl Acad Sci USA. 2004;101:881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ditty JL, Canales SR, Anderson BE, Williams SB, Golden SS. Stability of the Synechococcus elongatus PCC 7942 circadian clock under directed anti-phase expression of the kai genes. Microbiology. 2005;151:2605–2613. doi: 10.1099/mic.0.28030-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.