Abstract
The de novo and salvage dTTP pathways are essential for maintaining cellular dTTP pools to ensure the faithful replication of both mitochondrial and nuclear DNA. Disregulation of dTTP pools results in mitochondrial dysfunction and nuclear genome instability due to an increase in uracil misincorporation. In this study, we identified a de novo dTMP synthesis pathway in mammalian mitochondria. Mitochondria purified from wild-type Chinese hamster ovary (CHO) cells and HepG2 cells converted dUMP to dTMP in the presence of NADPH and serine, through the activities of mitochondrial serine hydroxymethyltransferase (SHMT2), thymidylate synthase (TYMS), and a novel human mitochondrial dihydrofolate reductase (DHFR) previously thought to be a pseudogene known as dihydrofolate reductase-like protein 1 (DHFRL1). Human DHFRL1, SHMT2, and TYMS were localized to mitochondrial matrix and inner membrane, confirming the presence of this pathway in mitochondria. Knockdown of DHFRL1 using siRNA eliminated DHFR activity in mitochondria. DHFRL1 expression in CHO glyC, a previously uncharacterized mutant glycine auxotrophic cell line, rescued the glycine auxotrophy. De novo thymidylate synthesis activity was diminished in mitochondria isolated from glyA CHO cells that lack SHMT2 activity, as well as mitochondria isolated from wild-type CHO cells treated with methotrexate, a DHFR inhibitor. De novo thymidylate synthesis in mitochondria prevents uracil accumulation in mitochondrial DNA (mtDNA), as uracil levels in mtDNA isolated from glyA CHO cells was 40% higher than observed in mtDNA isolated from wild-type CHO cells. These data indicate that unlike other nucleotides, de novo dTMP synthesis occurs within mitochondria and is essential for mtDNA integrity.
Keywords: folate, one-carbon metabolism, thymidine, deoxyribouridine
Regulation of cellular dTTP pools is essential for faithful replication of nuclear and mitochondrial DNA (mtDNA), with both depletion and expansion of the pool affecting DNA integrity and human health (1). Depletion of de novo dTMP synthesis results in deoxyuridine misincorporation into nuclear DNA leading to genome instability (2), whereas depletion of mitochondrial dTTP pools, due to mitochondrial deoxyribonucleoside kinases, deoxyguanosine kinase, or thymidine kinase 2 (TK2) mutations are associated with mtDNA-depletion syndromes and severe mitochondrial dysfunction in humans (3). Elevations in dTTP pools, as observed in mitochondrial neurogastrointestinal encephalomyopathy (4), an autosomal recessive disease resulting from decreased levels of cytoplasmic thymidine phosphorylase, results in mtDNA depletion (5), deletions (6), and site-specific point mutations (6).
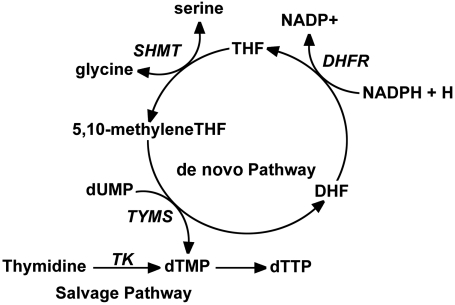
There are two distinct pathways for thymidine nucleotide synthesis (Fig. 1). The salvage pathway involves the conversion of thymidine to thymidylate, and occurs in the mitochondria and cytoplasm catalyzed by TK2 and thymidine kinase 1 (TK1), respectively (7). Salvage pathway synthesis of dTTP is not sufficient to sustain mtDNA replication (3) and therefore requires de novo dTMP synthesis, which involves the conversion of dUMP to dTMP, catalyzed by the tetrahydrofolate (THF)-dependent enzymes serine hydroxymethyltransferase (SHMT), thymidylate synthase (TYMS), and dihydrofolate reductase (DHFR) (8). In this pathway, 5,10-methyleneTHF, a one-carbon donor, is generated from serine by SHMT and used for the conversion of dUMP to dTMP in a reaction catalyzed by TYMS. The TYMS-catalyzed reaction generates dihydrofolate, which is converted to THF in an NADPH-dependent manner by DHFR. The regenerated THF can then be used for subsequent rounds of thymidylate biosynthesis.
Fig. 1.
De novo and salvage pathway synthesis of dTTP. There are two distinct pathways for dTTP synthesis. In the de novo pathway, SHMT catalyzes the conversion of serine and tetrahydrofolate (THF) to methyleneTHF and glycine. TYMS converts methyleneTHF and dUMP to dihydrofolate (DHF) and dTMP. DHF is converted to THF for subsequent rounds of dTMP synthesis in an NADPH-dependent reaction catalyzed by DHFR. In the salvage pathway, thymidine is phosphorylated by TK to form dTMP.
Both de novo and salvage dTMP synthesis have been assumed to occur in the cytoplasm to support both nuclear and mtDNA replication, but recent studies have demonstrated nuclear localization of TK1 (9) and the de novo dTMP biosynthesis pathway during S-phase and as a result of DNA damage (10, 11). The inability of the mitochondrial dTMP salvage pathway to support mtDNA synthesis, and recent studies indicating that de novo dTMP synthesis occurs in the nucleus, raised the possibility for a requirement for de novo thymidylate biosynthesis in mitochondria. De novo thymidylate synthesis has been shown to occur in plant mitochondria through the activities of an SHMT isozyme [mitochondrial serine hydroxymethyltransferase (SHMT2)] and a bifunctional enzyme containing both TYMS and DHFR (12). Interestingly, previous studies have localized TYMS to the mitochondria in mammalian cells (1, 13) and in neurospora crassa (14). In mammalian cells, mitochondria contain SHMT2, which is required for formate and glycine synthesis (15), but the third enzyme in the pathway, DHFR, does not localize to the mitochondria (16).
In this study, a de novo dTMP biosynthesis pathway was identified in mammalian mitochondria that includes a unique mitochondrial isozyme of DHFR called DHFRL1, which was previously thought to be a pseudogene (17). De novo dTMP synthesis is shown to be necessary to prevent uracil accumulation in mtDNA.
Results
Mammalian Mitochondria Contain a De Novo dTMP Synthesis Pathway.
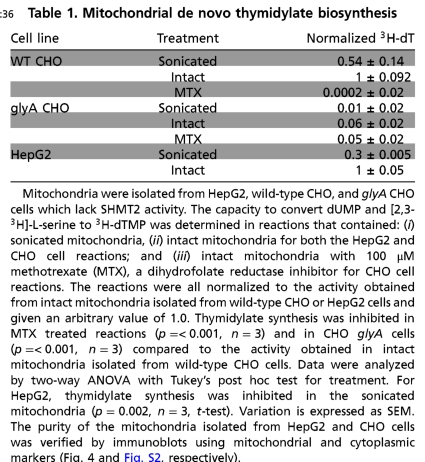
Purified intact mitochondria from Chinese hamster ovary (CHO) cells and HepG2 cells (Table 1 and Fig. S1) were capable of catalyzing the formation of 3H-dTMP from unlabeled dUMP, NADPH, and [2,3-3H]-L-serine in vitro. This activity is a unique demonstration of folate-dependent de novo dTMP biosynthesis in mammalian mitochondria. Methotrexate inhibited dTMP production in mitochondria isolated from wild-type CHO cells. Mitochondria isolated from glyA CHO cell mutants, which lack SHMT2, exhibited a 94% reduction in dTMP synthesis capacity, indicating the essentiality of SHMT2 within mitochondria for mitochondrial de novo dTMP synthesis. Disruption of mitochondria by sonication-reduced de novo dTMP synthesis activity, indicating that compartmentalization within mitochondria, or the formation of complexes within mitochondria, may be important for de novo dTMP synthesis. Alternatively, the reduced activity may reflect dilution or loss of the endogenous folate cofactor pool following sonication.
Table 1.
Mitochondrial de novo thymidylate biosynthesis
Mitochondria were isolated from HepG2, wild-type CHO, and glyA CHO cells which lack SHMT2 activity. The capacity to convert dUMP and [2,3-3H]-L-serine to 3H-dTMP was determined in reactions that contained: (i) sonicated mitochondria, (ii) intact mitochondria for both the HepG2 and CHO cell reactions; and (iii) intact mitochondria with 100 μM methotrexate (MTX), a dihydrofolate reductase inhibitor for CHO cell reactions. The reactions were all normalized to the activity obtained from intact mitochondria isolated from wild-type CHO or HepG2 cells and given an arbitrary value of 1.0. Thymidylate synthesis was inhibited in MTX treated reactions (p = < 0.001, n = 3) and in CHO glyA cells (p = < 0.001, n = 3) compared to the activity obtained in intact mitochondria isolated from wild-type CHO cells. Data were analyzed by two-way ANOVA with Tukey’s post hoc test for treatment. For HepG2, thymidylate synthesis was inhibited in the sonicated mitochondria (p = 0.002, n = 3, t-test). Variation is expressed as SEM. The purity of the mitochondria isolated from HepG2 and CHO cells was verified by immunoblots using mitochondrial and cytoplasmic markers (Fig. 4 and Fig. S2, respectively).
Identification of DHFRL1 as an Expressed Gene.
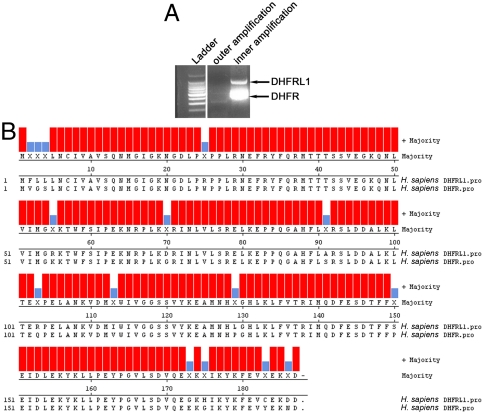
To determine if alternative human DHFR transcripts are expressed that encode a mitochondrial leader sequence, 5′ rapid amplification of cDNA ends (RACE) was conducted using total mRNA isolated from HepG2 cells. Two prominent bands were observed from the 5′ RACE experiment. Upon sequencing the two bands, both DHFR and DHFRL1 were identified (Fig. 2A). DHFRL1 is annotated as a mammalian pseudo gene in Entrez Gene, and expressed sequence tags are present (GenBank accession no. DN994912). The DHFR and DHFRL1 primary sequences are 92% identical (Fig. 2B). An energy minimized three-dimensional model of DHFRL1 was constructed using the crystal structure of human DHFR [Protein Data Bank (PDB) ID code 3GHW] as a template. The model of DHFRL1 structure and DHFR structure are highly similar with an rms value of 0.0034 (Fig. S2).
Fig. 2.
Identification of DHFRL1 and alignment to DHFR. (A) 5′ RACE was performed to identify DHFR transcripts that encoded a mitochondrial DHFR isozyme. Using a primer complementary to DHFR sequence, two bands were obtained following electrophoresis. The bands were isolated, cloned, and sequenced revealing DHFRL1 as an expressed transcript. (B) The sequence of DHFR and DHFRL1 were aligned using ClustalW in Lasergene Megalign software. The sequences are 92% identical.
Localization of DHFRL1 and TYMS to Mitochondria.
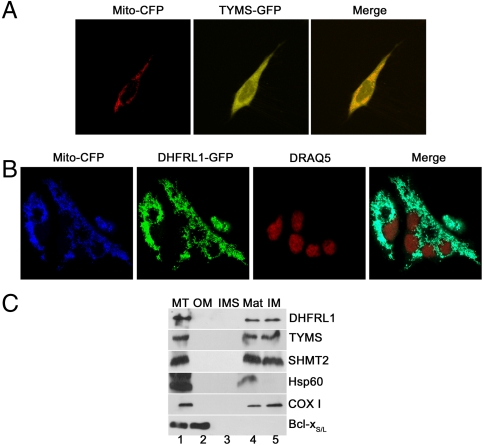
The subcellular localization of DHFRL1 and TYMS were determined by confocal microscopy following the transfection of pCMV6-AC-DHFRL1-GFP and pCMV6-AC-TYMS-GFP constructs into HeLa cells. The mitochondrial control vector pTagCFP-mito was also transfected to visualize mitochondria, and DRAQ5 nuclear stain was used to visualize nuclei. Colocalization of the mitochondrial control protein, and both TYMS-GFP and DHFRL1-GFP fusion proteins, was observed. TYMS-GFP localized to the cytoplasm, mitochondria, and nuclei (Fig. 3A), whereas DHFRL1-GFP was present exclusively within mitochondria (Fig. 3B). DHFRL1, SHMT2, and TYMS endogenous to HepG2 mitochondria were localized to the mitochondrial matrix and inner membrane in fractionated mitochondria (Fig. 3C), consistent with previous localization studies of SHMT2 (18, 19). Using MITOPRED (20, 21), a bioinformatics program for prediction of mitochondrial proteins, no canonical mitochondrial leader sequence was identified for DHFRL1. The primary sequence of DHFRL1 shown in Fig. 2C was sufficient for mitochondrial import. These data demonstrate that both TYMS and DHFRL1 are present in human mitochondria, and indicate that mitochondria contain a complete pathway to conduct de novo thymidylate biosynthesis.
Fig. 3.
TYMS and DHFRL1 localize to mitochondria. TYMS-GFP fusion proteins (A) and DHFRL1-GFP fusion proteins (B) were expressed in HeLa cells, as well as a mitochondrial marker mito-CFP. (A) TYMS-GFP localizes to the nucleus, cytoplasm, and colocalizes with the mito-CFP mitochondrial marker. (B) DHFRL1-GFP localizes exclusively to mitochondria. (C) TYMS, DHFRL1, and SHMT2 proteins localize to the matrix and inner membrane of fractionated HepG2 cells. Lane 1, mitochondrial extract; lane 2, outer membrane fraction; lane 3, inner membrane space; lane 4, matrix; lane 5, inner membrane. Hsp60 is the inner membrane/matrix marker, Cox I is an inner membrane marker, and Bcl-xS/L is an outer membrane marker.
DHFRL1 is a Functional Enzyme in Mitochondria.
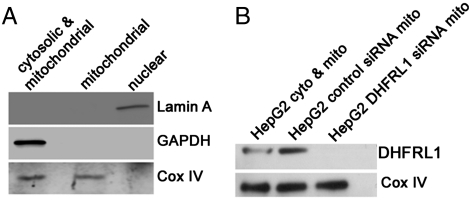
The presence of DHFR activity in mitochondria was investigated in human HepG2 cells treated with scrambled control siRNA, and siRNA specific to the DHFRL1 5′ untranslated region and not present in the DHFR transcript. The DHFR activity in extracts from mitochondria isolated from the control siRNA (32.4 ± 4.0 nmol/ min /mg protein) and DHFRL1-specific siRNA treatments (0.17 ± 0.008 nmol/ min /mg protein) were significantly different with a p value of 0.0002 (n = 3) where error is expressed as SEM. The mitochondria isolated for these assays exhibited no nuclear or cytosolic contamination as determined by immunoblotting against Lamin A and GAPDH, respectively (Fig. 4A). In the samples in which DHFRL1 was knocked down, no DHFRL1 protein was present in the mitochondrial extracts. COX-IV immunoblotting was used as a positive mitochondrial control. DHFR activity in the cytosolic extracts was unaffected by both scrambled and DHFRL1-specific siRNA treatment with specific activities of 4.5 + 2.6 and 4.5 ± 2.8 nmol/ min /mg protein, respectively, indicating that DHFRL1 knockdown did not affect DHFR expression. For comparison, DHFR activity in purified nuclei from HepG2 cells exhibited a specific activity of 14.3 ± 1.0 nmol/ min /mg protein.
Fig. 4.
Knockdown of DHFRL1 inhibits DHFR activity in mitochondrial extracts. Scrambled siRNA and siRNA against DHFRL1 were transfected into HepG2 cells and DHFRL1 protein and activity was measured. (A) Mitochondria from HepG2 cells that were purified for de novo dTMP synthesis assays (Table 1), and DHFRL1 activity experiments exhibited no nuclear or cytosolic contamination using immunoblots against GAPDH and Lamin A. (B) No DHFRL1 was present in mitochondria of HepG2 cells transfected with siRNA against DHFRL1.
De Novo Thymidylate Biosynthesis is Required to Prevent Uracil Accumulation in mtDNA.
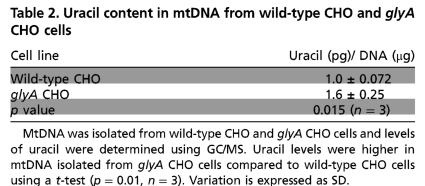
mtDNA was isolated from both wild-type and glyA CHO cells and uracil content in mtDNA was quantified. GlyA CHO cells lack SHMT2 activity and are auxotrophic for glycine (22, 23). mtDNA isolated from wild-type cells contained 40% less uracil than mtDNA from glyA CHO cells (Table 2). These data confirm the importance of SHMT2 activity within mitochondria for the production of dTMP, and demonstrate the importance of mitochondrial dTMP de novo biosynthesis in protecting mtDNA from uracil misincorporation.
Table 2.
Uracil content in mtDNA from wild-type CHO and glyA CHO cells
MtDNA was isolated from wild-type CHO and glyA CHO cells and levels of uracil were determined using GC/MS. Uracil levels were higher in mtDNA isolated from glyA CHO cells compared to wild-type CHO cells using a t-test (p = 0.01, n = 3). Variation is expressed as SD.
DHFRL1 Expression Rescues the Glycine Auxotrophy in glyC CHO Cells.
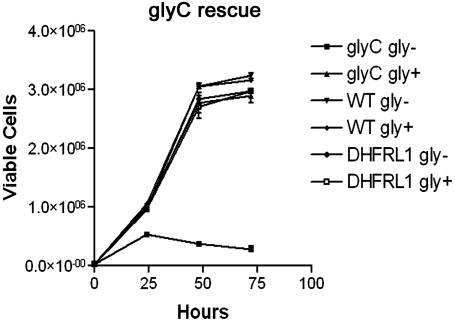
To verify the function of DHFRL1 in mitochondrial folate metabolism, the ability of DHFRL1 to rescue the glycine auxotrophy caused by disrupted mitochondrial folate-metabolism in a previously uncharacterized CHO cell line, glyC, was determined (Fig. 5). Four stable transfectants were selected for G418 resistance from glyC cells electroporated with either an empty pCMV6-AC-GFP plasmid, or a plasmid containing the DHFRL1 cDNA with a 5′ CMV promoter. Stable transfectants containing the pCMV6-AC-GFP empty vector did not rescue the glycine auxotrophy. However, the glycine auxotrophy was rescued in all selected cell lines transfected with the vector that contained the DHFRL1 cDNA. These results confirm that DHFRL1 localizes to mitochondria and can rescue mitochondrial one-carbon metabolism.
Fig. 5.
DHFRL1 cDNA rescues glycine auxotrophy in glyC CHO cells. CHO glyC cells were transfected with cDNAs encoding human DHFRL1-GFP, or a GFP-empty vector control. Stable cells lines were selected for G418 resistance in the presence of 200 μM glycine and 10 mg/L thymidine. For growth assays, cells were cultured with and without glycine and trypan blue exclusion was used to quantify viable cells. Four independent lines were assayed per transfection and experiments were done in triplicate. There was no significant difference in growth among the cells transfected with DHFRL1-GFP with or without glycine. CHO glyC cells transfected with GFP-empty vector and cultured without glycine failed to proliferate over the 72 h time period.
Discussion
This study demonstrates that human mitochondria contain an active de novo thymidylate biosynthesis pathway, composed of SHMT2, TYMS and DHFRL1. This study supports two previous studies that have observed TYMS protein localized within mammalian mitochondria, although the functional significance of these observations was never established (1, 13). Folate-activated one-carbon units for mammalian mitochondrial thymidylate biosynthesis are derived from serine through the catalytic activity of SHMT2; mitochondria lacking functional SHMT2, isolated from glyA CHO cells, exhibited impaired de novo thymidylate synthesis. The discovery of DHFRL1 as the functional mitochondrial enzyme completes the de novo thymidylate synthesis cycle. Inhibition of DHFRL1 expression by siRNA significantly reduced DHFR activity in mitochondria isolated from HepG2 cells. DHFRL1 was previously thought to be a pseudogene and was mapped to chromosome 3q11.2 (17). The mechanism whereby TYMS and DHFRL1 are localized to mitochondria remains to be established as neither protein contains a canonical mitochondrial leader sequence at its amino terminus.
Disruption of mitochondrial folate metabolism results in a glycine auxotrophy due to loss of SHMT2 function (22). In this study, we show that DHFRL1 complements the previously uncharacterized CHO glycine auxotrophic mutant, glyC. Previous reports have observed that glyC mutants have normal SHMT levels in both the cytosol and mitochondria (15, 24). Because the mitochondrial and cytoplasmic folate pools are not in equilibrium, loss of DHFR activity in mitochondria is expected to result in an accumulation of mitochondrial folate as dihydrofolate, thereby depleting THF pools for the SHMT2-catalyzed conversion of serine to glycine.
De novo thymidylate synthesis appears to be required to maintain mtDNA integrity, as glyA cells were observed to have elevated levels of uracil in DNA. During nuclear DNA replication, dTTP is the only deoxyribonucleotide that is not essential for DNA synthesis as dUTP can be incorporated into DNA when dTTP levels fall (2). Results from these studies indicate that impaired de novo thymidylate synthesis in mitochondria also results in deoxyuridylate incorporation in mtDNA. A recent report observed increases in mtDNA deletions in mice lacking the Ung gene undergoing folate deprivation compared to wild-type controls (25). Other studies support a role for de novo thymidylate biosynthesis in maintaining DNA integrity. Patients with the mitochondrial disorder, Friedrich’s ataxia, which leads to large increases in mtDNA damage, exhibited elevated DHFRL1 mRNA (26). Elevated DHFRL1 expression may help limit some deleterious changes in mtDNA. Other studies have demonstrated that TYMS inhibition leads to caspase-dependent apoptosis and changes in mitochondrial membrane potential (27, 28). mtDNA damage has been shown to initiate apoptosis rapidly, independent of deleterious mutations (29). Mitochondrial DNA damage due to inhibition of mitochondrial dTMP synthesis could explain the increases in apoptosis observed (27). Further investigation should be undertaken to determine if TYMS inhibition-dependent apoptosis is due to uracil misincorporation within mtDNA and/or reduced mitochondrial thymidylate biosynthesis.
Unlike the synthesis of purine and cytosine deoxyribonucleotides, dTMP synthesis is compartmentalized to the nucleus and mitochondria, both sites of DNA replication. Elevated uracil content in DNA results from both nutritional deficiency of folate (2), and from impaired expression of the thymidylate biosynthesis pathway genes (30); genetic disruption of SHMT1 in mice increases uracil levels in nuclear DNA (31). To our knowledge, impairment of de novo thymidylate biosynthesis has not been shown previously to elevate uracil in mtDNA. These data also indicate that a high level of organization within the mitochondria may be required for efficient dTMP biosynthesis, such as the proposed complex in nuclei for dTMP biosynthesis (11, 32, 33). As observed previously in nuclei, disruption of mitochondria by sonication inhibited thymidylate biosynthesis (11). Others have observed that SHMT2 cross-links to mtDNA in HeLa cells upon formaldehyde treatment (34, 35). Collectively, these data suggest that a multienzyme complex may exist within mitochondria at mtDNA replication forks. Ongoing studies will address the biological necessity for thymidylate biosynthesis within both the nuclei and mitochondria, whereas purine biosynthesis occurs in the cytosol (11, 36).
Methods
Cell Lines and Culture.
HeLa and HepG2 cells were acquired from American Type Culture Collection (ATCC). GlyA, a CHO cell mutant lacking SHMT2 activity, was obtained from Larry Thompson, Lawrence Livermore Labs, Livermore, CA. GlyC, an uncharacterized CHO cell mutant which is auxotrophic for glycine was obtained from Barry Shane, University of California, Berkeley, CA. All cells were cultured at 37 °C in a 5% CO2 atmosphere. HeLa and HepG2 cells were maintained in minimal essential medium (α-MEM) (Hyclone) with 10% fetal bovine serum (Hyclone) and penicillin/streptomycin (Gibco). GlyA and GlyC cells were cultured in defined minimal essential medium which lacks glycine and supplemented with 10% dialyzed and charcoal treated fetal bovine serum (dDMEM), pencillin/streptomycin, and 20 nM leucovorin (Sigma). For the complementation assays, positive growth controls included 200 μM glycine.
5′ Rapid Amplification of cDNA Ends (5′ RACE) Identifies DHFRL1 Transcript.
Polyadenylated mRNA was isolated from HepG2 cells using the Dynabeads mRNA purification kit (Invitrogen) as per manufacturer’s instructions. The isolated mRNA was then used for 5′ RACE using the FirstChoice RNA Ligase Mediated-RACE kit (Ambion, Inc.) as per manufacturer’s instructions. For the outer and inner amplifications the primers used were 5′-TGG AGG TTC CTT GAG TTC TCT GCT-3′ and 5′- AGG TCG ATT CTT CTC AGG AAT GGA GA-3′, respectively. The primers were made to detect DHFR but also identified DHFRL1. The outer and inner primers are made to nucleotides 229–252 and 176–201 of both DHFR and DHFRL1 coding sequence. The outer primer has a sequence identity of 96% to DHFRL1. The inner primer has 100% sequence identity to DHFRL1. The nested PCR products were run on a 2% agarose gel. Bands were cut from the gel and purified using the QIAquick gel extraction kit (Qiagen). The isolated cDNA was then Topo cloned using the pCR-Blunt II-Topo vector (Invitrogen) as per manufacturer’s instructions. Topo clones were picked and grown overnight at 37 °C for DNA isolation. Plasmid DNA was isolated using a Miniprep kit from Qiagen as per manufacturer’s instructions. The isolated plasmid DNA was sent to sequencing at the Biotech Resource Center at Cornell University. Primers against the T7 promoter priming site present in the pCR-Blunt II-Topo vector was used for sequencing.
Homology Modeling and Alignment of DHFRL1.
The primary protein sequence of DHFRL1 (GenBank accession no. AAH63379.1) was modeled against the structure of human DHFR (PDB ID code 3GHW) using DeepView/Swiss-PDB viewer 3.7. The Iterative Magic Fit of all atoms tool was used to generate a structural alignment. The structure model was then energy minimized using GROMOS96 energy minimization tool. PyMOL was used to generate three-dimensional structures and the RMS was determined using the align action within PyMOL. Primary protein sequences alignments between DHFR (GenBank accession no. AAH71996.1) and DHFRL1 were generated using the ClustalW tool in Lasergene Megalign software.
Localization of TYMS and DHFRL1 by Confocal Microscopy in HeLa Cells.
TYMS and DHFRL1 cDNAs were fused to the N terminus of GFP within the pCMV6-AC-GFP vector (Origene). A vector expressing a mitochondrial marker, pTagCFP-mito (Evrogen) was used for control transfections to identify mitochondria. Plasmids were transfected into HeLa cells using the Nucleofector II and the nucleofection kit R (Lonza) as per manufacturer’s protocol. The DNA binding dye, Draq5 (Biostatus Limited) was used to visualize nuclei. Confocal fluorescence microscopy (Zeiss 710 Confocal system) was used to image all cells at the Cornell Microscope and Imaging Facility.
Rescue of the Glycine Auxotrophy in CHO GlyC Cells.
GlyC CHO cells were cultured in dDMEM supplemented with 200 μM glycine and 20 nM leucovorin. pCMV6-DHFRL1-GFP and pCMV6-GFP vectors were linearized using ScaI (New England Biolabs), electroporated into CHO GlyC cells, and selected as previously described (11). Trypan blue exclusion assays were completed in media with and without glycine as previously reported (37).
Knockdown of DHFRL1 and Dihydrofolate Reductase Activity in Mitochondria.
DHFRL1-specific siRNA and control scrambled siRNA (Qiagen) were transfected into HepG2 cells using Nucleofection (Lonza) as per manufacturer’s instructions. After a 24 h incubation, mitochondria were isolated using the same protocol described below for the mitochondrial dTMP biosynthesis assays. Mitochondria were lysed using mammalian protein extraction reagent (Pierce) supplemented with 1 mM EDTA, 10 mM PMSF (Alexis Biochemicals), and 1∶1,000 dilution of protease inhibitor (Sigma). DHFR enzyme activity was determined in isolated mitochondria from both control siRNA-treated and DHFRL1 siRNA-treated HepG2 cells using the DHFR activity kit (Sigma) as per manufacturer’s instructions. To control for cytoplasmic and nuclear contamination, immunoblotting was performed as previously reported (11). To determine if DHFRL1 was present in mitochondria and knocked down in the siRNA-treated samples, immunoblots were performed using a mouse polyclonal antibody toward DHFRL1 (Abnova) at a concentration of 1∶5,000. Goat anti-mouse HRP (Pierce) conjugated secondary was used at a concentration of 1∶10,000. Cox IV antibody was used as a mitochondrial control as previously described (11).
Mitochondrial Thymidylate Biosynthesis Assay.
Mitochondria were isolated from 109 cells of each HepG2, wild-type CHO and glyA CHO cells. Cells were grown in Hyperflasks (Corning) in DMEM (Gibco) supplemented with 10% fetal bovine serum (Hyclone) and penicillin/streptomycin (Gibco). Cells were trypsinized and collected in 50 mL conical tubes and centrifuged at 2,000 rpm for 5 min. The cell pellets were washed twice with phosphate buffered saline with centrifugation at 2,000 rpm between washes. Isolation of mitochondrial fractions were completed as previously reported (38). The mitochondrial pellets were resuspended in buffer containing 0.3 M sucrose, 1.0 mM EGTA, 5.0 mM MOPS, 5.0 mM K2PO4, and 0.1% BSA, and adjusted to pH 7.4 with KOH. The mitochondria were then distributed equally into 1.5 mL tubes, nine fractions for each of wild-type CHO cells and glyA CHO cells and six fractions for HepG2 cells. These tubes were centrifuged at 5,000 × g for 10 min at 4 °C. The pelleted mitochondria were then resuspended in 600 μL of thymidylate reaction buffer containing 5 mM NADPH (Sigma), 100 mM β-mercaptoethanol (Sigma), 50 mM Hepes, pH 7.4, 2 mM MgCl2, 50 mM KCl, 25 mM K2HPO4, 1 mM dUMP, and 0.3 M sucrose. The mitochondria used for the sonicated reactions were then subjected to five 10 s pulses with 10 s rests between pulses on ice. The small probe on a Branson sonifier was used at 20% power. O2 gas was bubbled into each reaction vessel for 3 min. prior to addition of 8 µL [2,3-3H]-L-serine (Moravek). The reactions were put in a shaking incubator at 300 rpm and 37 °C for 12 h. The samples were then centrifuged at 5,000 × g for 10 min to pellet mitochondria and the supernatant was collected. The mitochondria were then lysed using 500 µL 1% SDS and sonication as previously stated. The lysate was then analyzed using HPLC as previously reported (11).
mtDNA Purification and Determination of Uracil Misincorporation in glyA and Wild-Type CHO Cells.
Mitochondria were isolated from both CHO wild-type and CHO glyA cells. The mitochondrial pellet was resuspended in 50 mM glucose (Sigma), 20 mM Tris·HCl pH 8.0 (Fisher), and 10 mM EDTA (Fisher) at 1 mL buffer per 500 μg mitochondria. Lysozyme (Sigma) was added at a concentration of 2 mg/mL and incubated on ice for 20 min. Two milliliters of 0.2 M NaOH (Fisher) containing 1% SDS (Fisher) were added and incubated on ice for 5 min. One and a half milliliters of 3 M potassium acetate dissolved in 2 M acetic acid was used to neutralize the sample to pH 7.0. The samples were incubated on ice for 1 h. The samples were then centrifuged at 11,000 × g for 30 min. The supernatant was decanted and then treated with 1/10 the volume of 3 M sodium acetate and two volumes of ethanol. The mtDNA was precipitated for 2 h at -20 °C. The samples were centrifuged at 10,000 × g for 20 min at 4 °C. The pellet was resuspended in 0.6 mL of 10 mM Tris/1 mM EDTA, pH 8.0 containing 0.1 mg/mL RNaseA. The mtDNA was purified using 1 mL of the Wizard Plus Resin from the Wizard Plus Miniprep kit (Promega) as per manufacturer’s instructions. Analysis of uracil levels was performed as previously reported with modifications; a Shimadzu QP2010 Plus GC/MS was used for analysis (31).
Mitochondrial Subfractionation.
Purified mitochondria from HepG2 cells were subfractionated into matrix, outer membrane, and inner membrane fractions as previously reported (39) and immunoblots were performed on each fraction to determine submitochondrial localization of DHFRL1, TYMS, and SHMT2. DHFRL1 immunoblots were performed as mentioned above. TYMS and SHMT2 immunoblots were performed as previously reported (11). Mouse anti-hsp60 (inner membrane/matrix marker), mouse anti-Cox I (inner membrane marker), and mouse anti-Bcl-xS/L (outer membrane marker) were purchased from Santa Cruz Biotechnology and were used at a dilution of 1∶200. HRP conjugated goat anti-mouse (Pierce) secondary antibodies were used at a dilution of 1∶10,000.
Supplementary Material
Acknowledgments.
This work was supported by Public Health Service Grant DK58144 (to P.J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103623108/-/DCSupplemental.
References
- 1.Samsonoff WA, et al. Intracellular location of thymidylate synthase and its state of phosphorylation. J Biol Chem. 1997;272:13281–13285. doi: 10.1074/jbc.272.20.13281. [DOI] [PubMed] [Google Scholar]
- 2.Blount BC, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: Implications for cancer and neuronal damage. Proc Natl Acad Sci USA. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou X, et al. Progressive loss of mitochondrial DNA in thymidine kinase 2-deficient mice. Hum Mol Genet. 2008;17:2329–2335. doi: 10.1093/hmg/ddn133. [DOI] [PubMed] [Google Scholar]
- 4.Nishino I, Spinazzola A, Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283:689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- 5.Pontarin G, et al. Mitochondrial DNA depletion and thymidine phosphate pool dynamics in a cellular model of mitochondrial neurogastrointestinal encephalomyopathy. J Biol Chem. 2006;281:22720–22728. doi: 10.1074/jbc.M604498200. [DOI] [PubMed] [Google Scholar]
- 6.Nishigaki Y, Marti R, Hirano M. ND5 is a hot-spot for multiple atypical mitochondrial DNA deletions in mitochondrial neurogastrointestinal encephalomyopathy. Hum Mol Genet. 2004;13:91–101. doi: 10.1093/hmg/ddh010. [DOI] [PubMed] [Google Scholar]
- 7.Pontarin G, Gallinaro L, Ferraro P, Reichard P, Bianchi V. Origins of mitochondrial thymidine triphosphate: Dynamic relations to cytosolic pools. Proc Natl Acad Sci USA. 2003;100:12159–12164. doi: 10.1073/pnas.1635259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitam Horm. 2008;79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- 9.Chen YL, Eriksson S, Chang ZF. Regulation and functional contribution of thymidine kinase 1 in repair of DNA damage. J Biol Chem. 285:27327–27335. doi: 10.1074/jbc.M110.137042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox JT, Shin WK, Caudill MA, Stover PJ. A UV-responsive internal ribosome entry site enhances serine hydroxymethyltransferase 1 expression for DNA damage repair. J Biol Chem. 2009;284:31097–31108. doi: 10.1074/jbc.M109.015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson DD, Stover PJ. SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS One. 2009;4:e5839. doi: 10.1371/journal.pone.0005839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neuburger M, Rebeille F, Jourdain A, Nakamura S, Douce R. Mitochondria are a major site for folate and thymidylate synthesis in plants. J Biol Chem. 1996;271:9466–9472. doi: 10.1074/jbc.271.16.9466. [DOI] [PubMed] [Google Scholar]
- 13.Brown SS, Neal GE, Williams DC. Subcellular distribution of some folic acid-linked enzymes in rat liver. Biochem J. 1965;97:34C–36C. doi: 10.1042/bj0970034c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossi M, Woodward DO. Enzymes of deoxythymidine triphosphate biosynthesis in Neurospora crassa mitochondria. J Bacteriol. 1975;121:640–647. doi: 10.1128/jb.121.2.640-647.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Appling DR. Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. FASEB J. 1991;5:2645–2651. doi: 10.1096/fasebj.5.12.1916088. [DOI] [PubMed] [Google Scholar]
- 16.Wang FK, Koch J, Stokstad EL. Folate coenzyme pattern, folate linked enzymes and methionine biosynthesis in rat liver mitochondria. Biochem Z. 1967;346:458–466. [PubMed] [Google Scholar]
- 17.Anagnou NP, Antonarakis SE, O’Brien SJ, Modi WS, Nienhuis AW. Chromosomal localization and racial distribution of the polymorphic human dihydrofolate reductase pseudogene (DHFRP1) Am J Hum Genet. 1988;42:345–352. [PMC free article] [PubMed] [Google Scholar]
- 18.Da Cruz S, et al. Proteomic analysis of the mouse liver mitochondrial inner membrane. J Biol Chem. 2003;278:41566–41571. doi: 10.1074/jbc.M304940200. [DOI] [PubMed] [Google Scholar]
- 19.Cybulski RL, Fisher RR. Intramitochondrial localization and proposed metabolic significance of serine transhydroxymethylase. Biochemistry. 1976;15:3183–3187. doi: 10.1021/bi00660a004. [DOI] [PubMed] [Google Scholar]
- 20.Guda C, Fahy E, Subramaniam S. MITOPRED: A genome-scale method for prediction of nucleus-encoded mitochondrial proteins. Bioinformatics. 2004;20:1785–1794. doi: 10.1093/bioinformatics/bth171. [DOI] [PubMed] [Google Scholar]
- 21.Guda C, Guda P, Fahy E, Subramaniam S. MITOPRED: A web server for the prediction of mitochondrial proteins. Nucleic Acids Res. 2004;32:W372–374. doi: 10.1093/nar/gkh374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfendner W, Pizer LI. The metabolism of serine and glycine in mutant lines of Chinese hamster ovary cells. Arch Biochem Biophys. 1980;200:503–512. doi: 10.1016/0003-9861(80)90382-3. [DOI] [PubMed] [Google Scholar]
- 23.Stover PJ, et al. Molecular cloning, characterization, and regulation of the human mitochondrial serine hydroxymethyltransferase gene. J Biol Chem. 1997;272:1842–1848. doi: 10.1074/jbc.272.3.1842. [DOI] [PubMed] [Google Scholar]
- 24.Taylor RT, Hanna ML. Folate-dependent enzymes in cultured Chinese hamster ovary cells: Impaired mitochondrial serine hydroxymethyltransferase activity in two additional glycine-auxotroph complementation classes. Arch Biochem Biophys. 1982;217:609–623. doi: 10.1016/0003-9861(82)90543-4. [DOI] [PubMed] [Google Scholar]
- 25.Kronenberg G, et al. Folate deficiency increases mtDNA and D-1 mtDNA deletion in aged brain of mice lacking uracil-DNA glycosylase. Exp Neurol. 2011;228(2):253–258. doi: 10.1016/j.expneurol.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Haugen AC, et al. Altered gene expression and DNA damage in peripheral blood cells from Friedreich’s ataxia patients: Cellular model of pathology. PLoS Genet. 6:e1000812. doi: 10.1371/journal.pgen.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakoff JA, Ackland SP. Thymidylate synthase inhibition induces S-phase arrest, biphasic mitochondrial alterations and caspase-dependent apoptosis in leukaemia cells. Cancer Chemother Pharmacol. 2000;46:477–487. doi: 10.1007/s002800000164. [DOI] [PubMed] [Google Scholar]
- 28.Pritchard DM, Hickman JA. Genetic determinants of cell death and toxicity. In: Jackman AL, editor. Antifolate Drugs in Cancer Therapy. Totowa, NJ: Humana Press; 1999. pp. 437–451. [Google Scholar]
- 29.Ricci C, et al. Mitochondrial DNA damage triggers mitochondrial-superoxide generation and apoptosis. Am J Physiol Cell Ph. 2008;294:C413–422. doi: 10.1152/ajpcell.00362.2007. [DOI] [PubMed] [Google Scholar]
- 30.Goulian M, Bleile B, Tseng BY. Methotrexate-induced misincorporation of uracil into DNA. Proc Natl Acad Sci USA. 1980;77:1956–1960. doi: 10.1073/pnas.77.4.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacFarlane AJ, et al. Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J Biol Chem. 2008;283:25846–25853. doi: 10.1074/jbc.M802671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson DD, Woeller CF, Stover PJ. Small ubiquitin-like modifier-1 (SUMO-1) modification of thymidylate synthase and dihydrofolate reductase. Clin Chem Lab Med. 2007;45:1760–1763. doi: 10.1515/CCLM.2007.355. [DOI] [PubMed] [Google Scholar]
- 33.Woeller CF, Anderson DD, Szebenyi DM, Stover PJ. Evidence for small ubiquitin-like modifier-dependent nuclear import of the thymidylate biosynthesis pathway. J Biol Chem. 2007;282:17623–17631. doi: 10.1074/jbc.M702526200. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J Biol Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 35.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 36.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 37.Field MS, Anguera MC, Page R, Stover PJ. 5,10-Methenyltetrahydrofolate synthetase activity is increased in tumors and modifies the efficacy of antipurine LY309887. Arch Biochem Biophys. 2009;481:145–150. doi: 10.1016/j.abb.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham JM. Isolation of mitochondria from tissues and cells by differential centrifugation. Curr Protoc Cell Biol. 2001:3.3.1–3.3.15. doi: 10.1002/0471143030.cb0303s04. [DOI] [PubMed] [Google Scholar]
- 39.Bolusani S, et al. Mammalian MTHFD2L encodes a mitochondrial methylenetetrahydrofolate dehydrogenase isozyme expressed in adult tissues. J Biol Chem. 286:5166–5174. doi: 10.1074/jbc.M110.196840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.