Abstract
Members of the IclR family control bacterial genes involved in a number of physiological processes. The IclR-family member TtgV crystallizes as a tetramer, with each TtgV monomer consisting of two domains—a DNA binding domain and an effector recognition domain, which are interconnected by an extended α-helix. When bound to DNA, a kink is introduced so that the extended helix is split in two α-helices (helix-4 and -5). Differential scanning calorimetry studies revealed that TtgV unfolds in a single event, suggesting that the two domains unfold cooperatively. When mutations are introduced in helix-5 that disrupt interactions between Arg98 and Glu102, the thermal unfolding of the TtgV domains becomes uncoupled without compromising effector binding. Two of these mutants (TtgVE102R and TtgVE102A) showed impaired release from target DNA, suggesting that these mutations alter signal transmission. By combining various mutants, we found that the mutations in the connecting α-helix exhibited a dominant effect over mutations in DNA binding and effector binding domains. We propose a model in which the loss of cooperativity of unfolding of TtgV reflects perturbed interdomain communication, and that the transition from the continuous to discontinuous helix may mediate interdomain communication necessary for the proper functioning of TtgV.
Keywords: efflux pump, transcriptional repressor, two-domain proteins, gene regulation
Adaptive responses in bacteria are most often mediated by transcriptional regulators that trigger specific transcriptional processes in response to signals (1, 2). Families of transcription factors have been identified on the basis of their conserved motifs and their modes of DNA binding. A wide set of structural analyses have revealed that the helix-turn-helix (HTH) signature is the most recurrent motif used to bind DNA in prokaryotic transcription factors, and that this signature contains a sufficient level of discriminative sequences to permit the delineation of distinct families of regulators (3, 4).
We have concentrated our attention on the IclR family of regulators because members of this family are widely distributed among Gram-positive and Gram-negative bacteria and Archae. Regulators of this family can function as repressors or activators and are involved in processes including the control of carbon metabolism, regulation of multidrug efflux pumps, sporulation, and phytopathogenicity (5). Multisequence alignment of around 1,000 IclR family members showed conserved sequence identity along the entire length of these proteins (6) and a search for these domains revealed that members of this family have a wHTH DNA-binding domain at their N-terminal region and a highly conserved C-terminal region corresponding to the effector-binding domain (5–7). Because of the highly conserved domain organization of IclR family members, the 3D structure of an IclR-family member would be expected to provide insight into the general mode of action for members of this family. To achieve this, we chose to characterize the TtgV IclR family member, a Pseudomonas putida regulator that modulates the expression of the ttgDEF and ttgGHI operons, which encode multidrug efflux pumps with wide-substrate specificity. TtgV operates according to an effector-mediated derepression mechanism; namely, in the absence of effector the protein is bound to the promoter region, thereby repressing transcription (8). With effector bound TtgV is released from target sequences. Efficient TtgV effectors are 1-naphthol and 4-nitrotoluene (8, 9). Effector binding to the TtgV/DNA complex was proposed to produce an intramolecular stimulus that is transmitted to the DNA-binding domain, causing dissociation of TtgV from the operator.
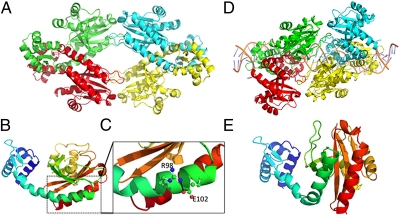
We have solved the 3D structure of apo–TtgV and TtgV complexed with its target operator at the ttgG promoter (10). The 3D structure of apo–TtgV revealed that the protein crystallized as a tetramer that adopts a diamond shape (Fig. 1A). The TtgV monomer is composed of an N-terminal DNA-binding domain, which is connected via a long α-helical linker to the effector binding domain (Fig. 1B) with little contact between them. The 3D structure of TtgV complexed with its operator (Fig. 1D), revealed that the regulator induces a 60° bend in the DNA. Upon binding to its operator, TtgV undergoes large conformational changes at the monomeric, dimeric, and tetrameric levels (10) such that the wHTH DNA of each monomer contacts the target DNA and produces a large footprint that extends over 42 bp (8, 11). In the TtgV/DNA complex structure, unlike to the apo–TtgV structure, the connecting linker is now discontinuous and exhibits a kink that generates two α-helices called α-helix4 and α-helix5, respectively (Fig. 1 B and E). This transition is key for TtgV's ability to stably bind to its target operator, and likewise, the transition from the discontinuous helix in the complex structure to the continuous helix in the apo structure is key for its release from the DNA operator.
Fig. 1.
The crystal structure of TtgV. (A) TtgV tetramer arrangement. Each monomer is colored differently. (B) Ribbon representation of TtgV monomer colored from N terminus (blue) to C terminus (red). (C) Proposed interaction between the lateral side chains of arginine 98 and aspartic acid 102 (Fig. S1 for further details). (D) TtgV tetramer bound to its target operator. (E) Detail of the kink in the connecting α-helix when TtgV is bound to its target operator. Other details can be found in Lu et al. (10).
We show here that although TtgV is composed of two different domains, the protein unfolds in a single event, indicative of cooperative unfolding and tight interdomain communication. Within the α-helical linker are residues E102 and R98, which interact to stabilize the regulator (Fig. S1). Replacement of glutamic acid 102 with arginine or alanine, and arginine 98 with glutamic acid or alanine abrogates cooperative unfolding. All of these mutants bind effectors with an affinity similar to that of the wild-type protein, and all but the TtgVR98E mutant are able to bind DNA. Release of TtgVE102R and TtgVE102A from its target DNA in response to effectors was partially impaired. These results provide insight into the importance of the linker domain within the context of interdomain crosstalk.
Results
Thermal Unfolding of TtgV Occurs in a Single Event.
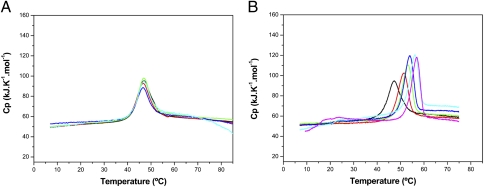
To explore the unfolding of the two domains of TtgV, we purified TtgV to homogeneity and submitted the protein to differential scanning calorimetry (DSC) assays, which revealed that TtgV unfolds in a single event at a melting temperature (Tm) 47 °C (Fig. 2A). Analysis of the normalized DSC profiles showed that Tm and ΔH did not change at different protein concentrations, suggesting that the TtgV oligomer is a stable tetramer. Rescans revealed that unfolding was an irreversible process, which does not depend on the protein concentration. Thermal unfolding of TtgV was also monitored by DSC in the presence of increasing concentrations of the effector molecules 1-naphthol (Fig. 2B) and 4-nitrotoluene (Fig. S2). As the effector concentration was increased the thermal stability of TtgV also increased, reaching a thermal stability plateau at saturating effector concentrations. In all cases, a single unfolding event was observed with a Tm shift of 4 °C for 4-nitrotoluene (Fig. S2), and 8 °C for 1-naphthol (Fig. 2B), with Tm of ∼51 °C and 55 °C, respectively. The increase in Tm was concomitant with increases in the unfolding enthalpic contribution of the system, as observed by the increase in the DSC peak area (Fig. 2). In control assays with TtgV and dimethyl sulfoxide (a compound used to prepare the effector stock solutions) no change in Tm was observed.
Fig. 2.
(A) Temperature dependency of the apparent heat capacity of TtgV. TtgV was prepared at different concentrations in 20 mM Pipes buffer: 0.17 mM (black trace), 0.1 mM (red trace), 0.06 mM (green trace), 0.03 mM (blue trace), and 0.017 mM (cyan trace). (B) Analysis of TtgV by DSC in the absence (black line) and in the presence of 1-naphthol. The protein was at a concentration of 30 μM, and ligand concentrations were 62.5 μM (red line), 125 μM (green line), 250 μM (blue line), 500 μM (cyan line), and 1,250 μM (magenta line).
Characterization of a Set of TtgV Mutants.
The 3D structure of TtgV revealed a long α-helix linking the effector binding domain with the DNA binding domain (Fig. 1C). When TtgV is bound to DNA, a kink at residue Q86, within the α-helix, is introduced, splitting the helix into two helices named α-helix4 and α-helix5 (Fig. 1D). Upon thorough inspection of the 3D structure, we identified interactions between the side chain of Glu102 and Arg98 in α-helix5 (Fig. S1); two amino acids that are relatively well conserved within IclR-family members (Fig. S3). Previous work has shown that Glu–Arg salt bridges often stabilize α-helixes (12, 13), leading us to believe that this specific salt bridge may act as a conformational switch important for signal transmission within TtgV. To explore the importance of these residues, we generated mutants in which Glu102 was replaced with Ala (E102A) or Arg (E102R), and Arg98 was replaced by Glu (R98E) or Ala (R98A). All mutant alleles were cloned into pET28b(+) (SI Experimental Procedures) and expressed as His-tagged proteins, which were purified to homogeneity. We determined the oligomerization state of the mutant proteins and the TtgV wild-type protein by dynamic light scattering and found that the hydrodynamic radius of all proteins was in the range of 6.2–6.6 nm (Fig. S4), which is consistent with a tetrameric quaternary structure in agreement with findings by Guazzaroni et al. (14).
We then tested the four TtgV mutants for effector and DNA binding. We found that all mutants were able to bind 1-naphthol with an affinity equivalent (15–20 μM) to that of the wild-type protein (Table 1). For DNA binding, we found that all mutants, except R98E were able to bind DNA. For the mutants able to bind DNA, we tested whether effector binding in the presence of target DNA occurred. With this aim we used a double-stranded 63-bp oligonucleotide spanning the TtgV target sequences in PttgD (SI Experimental Procedures). TtgV–DNA and mutant TtgV (E102A, E102R, and R98A)–DNA complexes were then titrated with effectors revealing that the affinity of E102A, E102R, and R98A mutants for 1-naphthol was in the range of 14–22 μM, which is similar to that of the wild-type protein (Table 1). Therefore, the affinity of both the uncomplexed and DNA-complexed TtgV mutants for effector molecules was unchanged.
Table 1.
Thermodynamic parameters for binding of 1-naphthol to TtgV free and complexed to its promoter ttgDEF
| KD | KA | ΔH | TΔS | ΔG | ||
| Protein-DNA | Effector | μM | M−1 | kcal/mol | kcal/mol | kcal/mol |
| WT | 1-naphthol | 18.7 ± 1.3 | (5.4 ± 0.38) × 104 | −40.4 ± 8.1 | −33.9 ± 8.1 | −6.5 ± 0.04 |
| WT-DEF | 1-naphthol | 16.8 ± 0.4 | (5.9 ± 0.15) × 104 | −23.9 ± 1.3 | −17.4 ± 1.3 | −6.5 ± 0.02 |
| Q51A | 1-naphthol | 17.5 ± 1.3 | (5.7 ± 0.43) × 104 | −41.0 ± 8.3 | −34.5 ± 8.3 | −6.5 ± 0.05 |
| Q51A-DEF | 1-naphthol | 16.5 ± 1.5 | (6.1 ± 0.56) × 104 | −4.9 ± 0.3 | 1.6 ± 0.3 | −6.5 ± 0.06 |
| E102A | 1-naphthol | 18.3 ± 1.1 | (5.5 ± 0.34) × 104 | −41.4 ± 6.5 | −35.0 ± 6.5 | −6.5 ± 0.04 |
| E102A-DEF | 1-naphthol | 15.2 ± 0.7 | (6.6 ± 0.33) × 104 | −26.4 ± 2.8 | −19.8 ± 2.8 | −6.6 ± 0.03 |
| E102R | 1-naphthol | 20.3 ± 0.7 | (4.9 ± 0.17) × 104 | −50.6 ± 7.7 | −44.2 ± 7.6 | −6.4 ± 0.02 |
| E102R-DEF | 1-naphthol | 12.4 ± 0.9 | (8.1 ± 0.64) × 104 | −36.6 ± 3.8 | −29.8 ± 3.8 | −6.7 ± 0.05 |
| V223A | 1-naphthol | 5.4 ± 0.7 | (18.4 ± 2.30) × 104 | −14.8 ± 1.7 | −7.6 ± 1.7 | −7.2 ± 0.07 |
| V223A-DEF | 1-naphthol | 11.3 ± 0.3 | (8.8 ± 0.26) × 104 | −20.4 ± 0.9 | −13.7 ± 0.9 | −6.7 ± 0.02 |
| R98E | 1-naphthol | 10.3 ± 1.0 | (9.7 ± 0.96) × 104 | −41.4 ± 8.3 | −34.6 ± 8.3 | −6.8 ± 0.06 |
| R98A | 1-naphthol | 19.9 ± 1.3 | (5.0 ± 0.32) × 104 | −20.5 ± 0.1 | −14.1 ± 1.2 | −6.4 ± 0.04 |
| R9A-DEF | 1-naphthol | 13.1 ± 1.3 | (7.7 ± 0.68) × 104 | −33.0 ± 1.0 | −26.3 ± 1.0 | −6.7 ± 0.05 |
ITC assays are described in SI Experimental Procedures. Titration curves were fitted by a nonlinear least squares method to a function for the binding of a ligand to a macromolecule. From the curve fitted, the parameters ΔH and KA were determined. The change in free energy (ΔG) and in entropy (ΔS) were calculated using the equation: ΔG = −RT LnKA = ΔH − TΔS, where R is the universal molar gas constant and T is the absolute temperature.
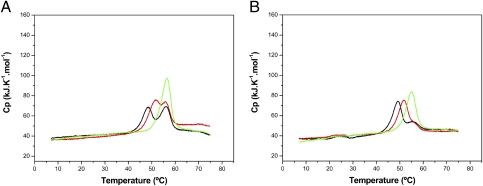
We then studied the unfolding of the mutant proteins by DSC. We found that, whereas the R98A TtgV unfolded in a single event similar to wild-type TtgV, the mutants E102R, E102A, and R98E exhibited an altered denaturation pattern, with unfolding occurring in two events (Fig. 3 and Fig. S5). Therefore, mutations at both residues 98 and 102 appear to be able to uncouple the cooperative unfolding of both domains observed in the wild-type protein.
Fig. 3.
Effect of 1-naphthol on the thermal unfolding of TtgVE102R (A) and TtgVE102A (B). For both panels, the DSC experiments of the TtgV mutants were performed in the absence of effector (black line) and in the presence of 33 μM (red line) and 250 μM (green line) 1-naphthol.
To test whether the two unfolding events in the three mutants represented the consecutive unfolding of the two domains, we tested whether the presence of effectors influenced the thermal stability of any of the two events seen with E102R, E102A, and R98E. In the first series of assays we used 33 μM 1-naphthol and found that first unfolding event was stabilized with an increased denaturation temperature, whereas the second unfolding event showed an unaltered Tm value (Fig. 3 A and B). This indicates that the first event represents the unfolding of the effector binding domain, whereas the second event corresponds to the unfolding of the DNA binding domain. It should be noted that at the highest effector concentration a single peak is seen; this is not the result of a single unfolding event but rather the area of this single peak is almost equal to the area of the two peaks and hence it seems to represent maximal stabilization of the protein after effector binding.
From a mechanistic point of view, TtgV achieves up-regulation of gene expression via effector-mediated dissociation from its operator (11, 14, 15). We used EMSA to study the ability of TtgV and its mutants to be released from the ttgD operator in response to effector. In the first series of assays, we used 50 nM of a 295-bp fragment bearing the ttgD promoter, and 50 nM of wild-type or mutant TtgV (E102R, E102A, and R98A). The assays were done in the absence or presence of 1 mM 1-naphtol—a concentration that is 50 times the KD. The densitometric analysis of the gels revealed that, as expected, wild type and the three TtgV mutants are able to bind to the ttgD promoter in the absence of effector, with the amount of DNA retarded being around 80% in all cases (Fig. S6). In the presence of effector, both wild type and TtgVR98A were released (such that only 30% of the DNA was retarded), whereas the E102A and E102R mutants were compromised in their ability to be released (Fig. S6). Because these two mutants are able to recognize and bind effectors, but are less efficiently released from the operator, we hypothesized that defective effector-mediated protein release in these mutants may arise from failed intramolecular crosstalk between the effector recognition domain and the DNA-binding domain. To test this hypothesis, we titrated the release of wild-type and mutant TtgV proteins in response to a range of 1-naphthol concentrations. We found that higher concentrations of 1-naphthol were needed to achieve 50% release of E102A and E102R mutants versus wild type (Fig. 4). This suggests that interdomain signal transmission may be altered in the mutant variants. In this regard, we have previously shown that TtgV mutants Q51A (11), in the DNA binding domain, and V223A (15), in the effector binding domain, were able to bind target DNA; however, in contrast with the wild type, their liberation from the target promoters in response to effectors was also partially impaired. Guazzaroni et al. (14) suggested that these two residues could be part of the interdomain communication system, but they did not test whether it was due to defects in effector binding. We have done isothermal titration calorimetry (ITC) assays with mutants Q51A and V223A and the results (Table 1) show that these two mutants recognized effectors with a similar affinity to wild type whether in solution or complexed with DNA (Table 1). Subsequently, the unfolding of these mutant proteins was assessed using DSC assays. Mutants Q51A and V223A unfolded in a single event with thermal parameters similar to those of wild-type TtgV, both in the absence and in the presence of effectors (Fig. S7 and Table 1). Because the liberation of mutants Q51A and V223A from target DNA is partially impaired, we suggest that it may be related with a deficient initiation of communication between the domains and that this deficiency does not alter the of linker structure (and subsequently protein unfolding), but influences the linker functioning. This hypothesis needs to be corroborated by resolving the 3D structure of the mutants.
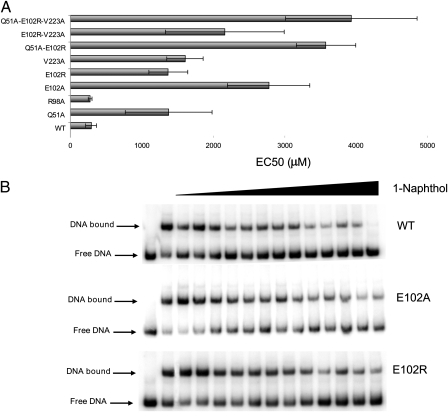
Fig. 4.
Release of TtgV mutant variants bound to the ttgD operator in presence of increasing concentrations of 1-naphthol. EMSAs were carried out and densitometric analyses were performed (using Quantity One software) to determine the amount of shifted DNA released in presence of increasing concentration of 1-naphthol with respect to the total shifted DNA in absence of ligand. (A) The EC50 of each protein (corresponding to the ligand concentration for which 50% of the shifted DNA is released) was determined by fitting each titration curve with ORIGIN software (MicroCal). (B) EMSAs were carried out with 1 nM of the indicated ttgDEF operator (295-bp fragment) and 50 nM wild-type or mutant TtgV and incubated with increasing concentration of 1-naphthol (25, 50, 100, 200, 300, 400, 800, 1,200, 2,000, 2,500, 3,000, 4,000, and 4,500 μM).
Because mutations at different locations yielded TtgV proteins with defects in release from its target promoter, and because these mutants behave differentially with respect to thermal stability, we decided to combine mutations in each of the domains and one mutation in the connecting helix. We constructed double TtgV mutants Q51A/E102R, and E102R/V223A, and a triple mutant Q51A/E102R/V223A and submitted the mutant proteins to several assays. For thermal unfolding assays we used 33 μM of the double and triple mutants. We found that the double mutants and the triple mutant showed two thermal unfolding events as seen with the single E102R mutant. Furthermore, as expected, the unfolding event corresponding to the effector binding domain was stabilized in the presence of effector molecules in the double and triple mutant. We also tested DNA binding of the double mutants (Q51A/E102R and E102R/V223A) and the triple mutant (Q51A/E102R/V223A) to the target PttgD operator and its subsequent release from DNA in response to effectors. We found that the liberation of the double mutants and the triple mutant was partially impaired and that 10- to 15-fold higher 1-naphthol concentrations were needed to achieve 50% release (Fig. 4).
Discussion
The published DSC data available for multidomain proteins show that often the individual domains of these proteins unfold independently in a consecutive manner (16, 17). However, transcriptional regulators, which are typically composed of an effector binding domain and a DNA binding domain, do not appear to follow this trend. Examples include the two-domain–containing regulators NmrA (18), TetR (19), and Crp (20), which belong to three different families of regulators—all of which unfold in a single event. Similarly to these regulators, the thermal unfolding of TtgV, a member of the IclR family, is also characterized by a single unfolding event (Fig. 2). Therefore, it follows that the tendency these two-domain transcriptional regulators to have a single unfolding event can be considered a result of the tight functional communication between domains and that intercommunication of domains is relevant for protein function. For TtgV of P. putida our DSC data also showed that the binding of the effector 1-naphthol stabilized TtgV structure by 8 °C, although denaturation also occurred as a single event (Fig. 2), confirming the cooperativity that exists between the domains of TtgV.
Protein unfolding and interdomain communication are complex processes that are still poorly understood. The potential existence of intramolecular domain communication in TtgV was first revealed through analysis of point mutants in valine 223, located in the effector binding pocket and adjacent to the β-strand at the transdimer interface (10), which showed compromised ability to dissociate from DNA in the presence of effectors (14, 15), mimicking the defects observed in the mutants at glutamine 51, located in the DNA-binding domain (11). The 3D structure of TtgV showed little physical interaction between the effector binding domain and the DNA binding domain, and it was hypothesized that the linker that bridges them could be part of the intramolecular signaling system (10). Here, we demonstrate that in TtgV the function of the linker is not simply to physically bridge the two protein domains, but to mediate domain communication because mutations in the linker led to defects in liberation from its target promoter (Fig. 4). Deficiencies in interdomain communication that result in defects in the release of a repressor from its target promoter have been reported for other regulators (e.g., FapR). Schujman et al. (21) proposed that binding of the malonyl-CoA effector to the effector binding domain of FapR provoked a disorder-to-order transition of a loop that modified the orientation of the α-helix linker that bridges this domain and the DNA binding domain, such that ligand binding induced modifications that impaired binding of the HTH to the operator region. It is worth mentioning that mutations in the connecting α-helix that provoked loss of protein flexibility resulted in FapR mutants being deficient in DNA binding (21). We have found that the sequence of the interconnecting linker of a number of well-studied members of the IclR family is relatively well conserved when sequences are multialigned, and that the prediction of their secondary structure revealed this region is likely organized as α-helices (Fig. S3). Regarding the interconnecting α-helix in TtgV, it is worth noting that we have identified two charged residues, R98 and E102 (Fig. S1), which interact through their side chains; these residues are relatively well conserved in the aligned sequences and our findings could be relevant to the understanding of interdomain signal transmission in the IclR family. The importance of these two residues in TtgV is apparent from the fact that replacement of Glu102 by Arg or Ala or Arg98 by Glu caused a significant uncoupling of the thermal unfolding of the two domains of TtgV leading to a two-stage unfolding event. However, ITC experiments demonstrated that all mutant proteins recognized effectors with binding affinities similar to the wild-type protein (Table 1). In terms of protein evolution, this observation is consistent with mutations in the linker having primarily an effect on the mechanism of interdomain communication rather than on the recognition of effectors. Still it should be noted that these mutations alter the interaction pattern with the ttgD promoter and, whereas the R98E TtgV mutant did not bind to DNA, the E102R, E102A, and R98A mutants bound to target operators, although the E102A and E102R mutants did not liberate from DNA as efficiently as the wild-type protein. We propose that this could be due to defects in signal transmission (Fig. 4). Using double mutants that combine a mutation in the linker α-helix with mutations in the effector binding pocket (i.e., E102R/V223A) and the DNA binding domain (i.e., Q51A/E102R), we found that the mutant proteins unfolded in two steps, substantiating the independent nature of the unfolding of the two domains and the essential role of the interconnecting α-helix in the response to effectors. Double mutants either conserved or exhibited an exacerbated defect in DNA liberation, indicating possible additive effects due to accumulation of deficiencies in the chain of signal transmission.
In summary, we present relevant results for the IclR family of regulators, showing that the two domains of TtgV are tightly associated but can be uncoupled via mutations in the connecting linker. Furthermore, this uncoupling does not necessarily result in loss of function but leads to differential responses to ligands, illustrating the importance of intramolecular domain crosstalk to TtgV function.
Experimental Procedures
Bacterial Strains, Plasmids, Culture Medium, and Protein Purification.
The bacterial strains and plasmids used in this study are listed in Table S1. Escherichia coli BL21(DE3) bearing appropriate plasmids was used for expression and purification of TtgV proteins (14).
Site-Directed Mutagenesis, Electrophoresis Mobility Shift Assay, and Isothermal Titration Calorimetry.
Assays were carried out as described in SI Experimental Procedures (14, 15, 22).
Differential Scanning Calorimetry.
Differential scanning calorimetry experiments were carried out with a VP-DSC (Valerian-Plotnikov differential scanning calorimeter) capillary-cell microcalorimeter from MicroCal at a scan rate of 60 °C h−1. Protein solutions for the calorimetric experiments were prepared by exhaustive dialysis against a buffer with 20 mM Pipes, pH 7.2, 8 mM magnesium acetate, 150 mM KCl, and 1 mM TCEP. The buffer from the last dialysis step was used in the reference cell of the calorimeter. Calorimetric cells (operating volume 0.137 mL) were kept under an excess pressure of 60 psi bar to prevent degassing during the scan and also to permit the scans to be performed at a temperature up to 80 °C. Several buffer–buffer baselines were obtained before each run with protein solution to ascertain proper equilibration of the instrument. Reheating runs were carried out to determine the calorimetric reversibility of the denaturation process.
Supplementary Material
Acknowledgments
We thank Benjamin J. Pakuts for critical reading of the manuscript and M. Mar Fandila for secretarial assistance. This study was supported by Fondos Europeos del Desarollo (FEDER) grants from the Junta de Andalucia (Proyecto de Excelencia, CVI191), ADHERS (BIO2008-04419-E/) from the Pathogenomic European Research Area Network (ERANET) program, and BIO2006-05668BIO2010-17227 from the Spanish Ministry of Science and Innovation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018894108/-/DCSupplemental.
References
- 1.Ghosh T, Bose D, Zhang X. Mechanisms for activating bacterial RNA polymerase. FEMS Microbiol Rev. 2010;34:611–627. doi: 10.1111/j.1574-6976.2010.00239.x. [DOI] [PubMed] [Google Scholar]
- 2.Ishihama A. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol Rev. 2010;34:628–645. doi: 10.1111/j.1574-6976.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- 3.Pérez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramos JL, et al. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molina-Henares AJ, Krell T, Guazzaroni ME, Segura A, Ramos JL. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol Rev. 2006;30:157–186. doi: 10.1111/j.1574-6976.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- 6.Krell T, Molina-Henares AJ, Ramos JL. The IclR family of transcriptional activators and repressors can be defined by a single profile. Protein Sci. 2006;15:1207–1213. doi: 10.1110/ps.051857206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang HB, Wang LH, Zhang LH. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2002;99:4638–4643. doi: 10.1073/pnas.022056699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guazzaroni ME, Terán W, Zhang X, Gallegos MT, Ramos JL. TtgV bound to a complex operator site represses transcription of the promoter for the multidrug and solvent extrusion TtgGHI pump. J Bacteriol. 2004;186:2921–2927. doi: 10.1128/JB.186.10.2921-2927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guazzaroni ME, et al. The multidrug efflux regulator TtgV recognizes a wide range of structurally different effectors in solution and complexed with target DNA: Evidence from isothermal titration calorimetry. J Biol Chem. 2005;280:20887–20893. doi: 10.1074/jbc.M500783200. [DOI] [PubMed] [Google Scholar]
- 10.Lu D, et al. Crystal structure of TtgV in complex with its DNA operator reveals a general model for cooperative DNA binding of tetrameric gene regulators. Genes Dev. 2010;24:2556–2565. doi: 10.1101/gad.603510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fillet S, et al. TtgV represses two different promoters by recognizing different sequences. J Bacteriol. 2009;191:1901–1909. doi: 10.1128/JB.01504-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marqusee S, Baldwin RL. Helix stabilization by Glu-…Lys+ salt bridges in short peptides of de novo design. Proc Natl Acad Sci USA. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyu PC, Gans PJ, Kallenbach NR. Energetic contribution of solvent-exposed ion pairs to alpha-helix structure. J Mol Biol. 1992;223:343–350. doi: 10.1016/0022-2836(92)90735-3. [DOI] [PubMed] [Google Scholar]
- 14.Guazzaroni ME, et al. The transcriptional repressor TtgV recognizes a complex operator as a tetramer and induces convex DNA bending. J Mol Biol. 2007;369:927–939. doi: 10.1016/j.jmb.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 15.Guazzaroni ME, Gallegos MT, Ramos JL, Krell T. Different modes of binding of mono- and biaromatic effectors to the transcriptional regulator TTGV: Role in differential derepression from its cognate operator. J Biol Chem. 2007;282:16308–16316. doi: 10.1074/jbc.M610032200. [DOI] [PubMed] [Google Scholar]
- 16.Ruíz-Arribas A, Santamaría RI, Zhadan GG, Villar E, Shnyrov VL. Differential scanning calorimetric study of the thermal stability of xylanase from Streptomyces halstedii JM8. Biochemistry. 1994;33:13787–13791. doi: 10.1021/bi00250a032. [DOI] [PubMed] [Google Scholar]
- 17.Krell T, et al. Insight into the structure and function of the transferrin receptor from Neisseria meningitidis using microcalorimetric techniques. J Biol Chem. 2003;278:14712–14722. doi: 10.1074/jbc.M204461200. [DOI] [PubMed] [Google Scholar]
- 18.Lamb HK, et al. The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides. J Biol Chem. 2003;278:32107–32114. doi: 10.1074/jbc.M304104200. [DOI] [PubMed] [Google Scholar]
- 19.Kedracka-Krok S, Wasylewski Z. A differential scanning calorimetry study of tetracycline repressor. Eur J Biochem. 2003;270:4564–4573. doi: 10.1046/j.1432-1033.2003.03856.x. [DOI] [PubMed] [Google Scholar]
- 20.Blaszczyk U, Wasylewski Z. Interaction of cAMP receptor protein from Escherichia coli with cAMP and DNA studied by differential scanning calorimetry. J Protein Chem. 2003;22:285–293. doi: 10.1023/a:1025024604677. [DOI] [PubMed] [Google Scholar]
- 21.Schujman GE, et al. Structural basis of lipid biosynthesis regulation in Gram-positive bacteria. EMBO J. 2006;25:4074–4083. doi: 10.1038/sj.emboj.7601284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniels C, Daddaoua A, Lu D, Zhang X, Ramos JL. Domain cross-talk during effector binding to the multidrug binding TTGR regulator. J Biol Chem. 2010;285:21372–21381. doi: 10.1074/jbc.M110.113282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.