Abstract
The respirasome is a multisubunit supercomplex of the respiratory chain in mitochondria. Here we report the 3D reconstruction of the bovine heart respirasome, composed of dimeric complex III and single copies of complex I and IV, at about 2.2-nm resolution, determined by cryoelectron tomography and subvolume averaging. Fitting of X-ray structures of single complexes I, III2, and IV with high fidelity allows interpretation of the model at the level of secondary structures and shows how the individual complexes interact within the respirasome. Surprisingly, the distance between cytochrome c binding sites of complexes III2 and IV is about 10 nm. Modeling indicates a loose interaction between the three complexes and provides evidence that lipids are gluing them at the interfaces.
Keywords: electron microscopy, oxidative phosphorylation
Oxidative phosphorylation (OXPHOS) in mitochondria is carried out by five multisubunit complexes (complexes I–V). They work in concert within the respiratory chain and catalyze the transfer of electrons from NADH to molecular oxygen. This electron flow is coupled to proton pumping over the inner—or crista—membrane, which generates a proton gradient utilized for ATP production by the ATP synthase (complex V). The five OXPHOS complexes are all catalytically active as single units, but ideas about their overall function changed after it was shown by blue-native gel electrophoresis (BN-PAGE) that they further associate into supercomplexes (1). The idea of their free occurrence within the crista—or inner—membrane became progressively replaced by the concept of dedicated supercomplexes, which were proposed to be stable interactions of single complexes. The I + III2 and the V2 supercomplexes were the first ones to be structurally characterized (2–4), providing additional proof of their existence. The I + III2 + IV1–4 supercomplex or the respirasome is one of the most intriguing supercomplexes, because it autonomously carries out the respiration steps from electron transfer from NADH to molecular oxygen. The exact reason for its presence, however, remains elusive. It was proposed that it could stabilize the single complexes, enhance the electron flow between these complexes, and prevent excess formation of oxygen radicals (5).
Single molecule averaging of EM projections of ice-embedded macromolecules can provide atomic resolution for large, water-soluble complexes (6). A common way to perform a 3D reconstruction of hydrophilic proteins is to use projections from randomly oriented particles. In amorphous ice layers, prepared on holey carbon grids, the distribution of particle orientations is usually close to fully random and projections can be averaged by the angular reconstitution technique after determination of the relative angular orientations of the projections (7). Cryoelectron EM (cryo-EM) studies of intrinsic membrane proteins have been hampered by the necessary presence of detergent to keep particles in a monodisperse state after purification. Because of the reduced surface tension, membrane proteins are difficult to prepare in an unsupported vitrified water layer on holey carbon grids. An additional thin carbon film is a suitable alternative, although this leads to preferential orientation of the complexes on the support structure, resulting in a limited range of covered orientation angles of the molecules. The random conical tilt data collection method could be used to compensate for the lack of randomness in orientations, by tilting the grids in the microscope to one specific angle (8). This method, however, requires the particles to be attached in one or several specific orientations to the support film. A powerful alternative data collection scheme that compensates for the lack of sampling over the full 3D space is electron tomography, in which the missing structural information is obtained from tilt series (9). Selected subvolumes of tomographic reconstructions (tomograms), containing specific protein complexes, can then be further averaged to enhance the signal and resolution (10, 11). In this study, tomographic subvolume averaging was applied on the I + III2 + IV supercomplex, further referred to as the respirasome, which is a highly abundant supercomplex in bovine heart mitochondria. Fitting high-resolution structures of its three components—dimeric complex III (12), monomeric complex IV (13), and monomeric complex I (14)—in a cryo-EM reconstruction at 2.2-nm resolution provides first insight into the unique interaction between these complexes within the respirasome.
Results
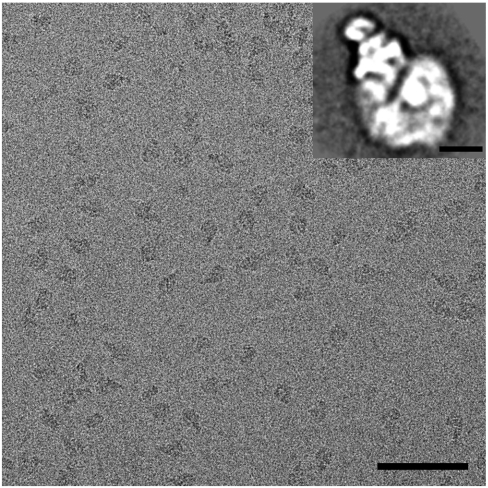
The bovine heart I + III2 + IV supercomplex was purified to homogeneity by BN-PAGE, followed by electroelution, a prerequisite to perform a cryoelectron microscopy study. Preparation of frozen hydrated cryo-EM grids by adsorption of the purified sample onto holey carbon film grids and plunge-freezing, however, resulted in inhomogeneously distributed particles in the grid holes. We therefore used holey carbon film grids that were covered with a thinner second carbon film, to which the particles could adsorb. This approach resulted in evenly distributed molecules (Fig. 1). Single particle analysis, including classification and averaging of two-dimensional projections, showed that the respirasome molecules were preferentially oriented in a few positions on carbon support film. The most abundant projection type has a triangular shape (Fig. 1, Inset), as also previously noticed in negatively stained samples (5). We performed electron tomography on these cryo-EM specimens and from 21 tilt series the 3D volumes of the samples were reconstructed. From these tomograms, 2,466 subtomograms containing single respirasomes were extracted for 3D averaging. Combination of XMIPP (X-Window-based Microscopy Image Processing Package) maximum likelihood (ML) global alignment (11) and local refinement by cross-correlation with AV3 toolbox (15) was used to create an average structure. The resolution of the respirasome is anisotropic (Fig. S1) and correlates to the angular sampling (Fig. S2). The highest achievable resolution of our average structure in the direction perpendicular to the incident beam is 1.7 nm and the lowest one is 2.55 nm parallel to the beam. We have filtered the entire reconstruction to 2.2 nm, which is at the first zero of the contrast transfer function.
Fig. 1.
Cryoelectron micrograph of isolated bovine respirasomes. (Scale bar: 100 nm.) (Inset) Two-dimensional projection map of the respirasome in a top view position from single particle image analysis. (Scale bar: 10 nm.)
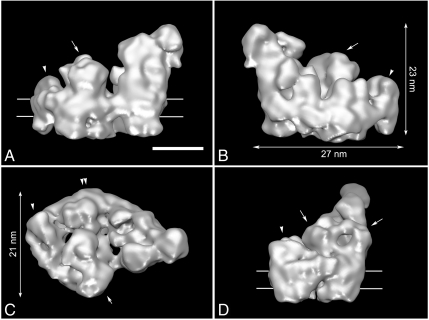
The reconstruction shows a respirasome with dimensions of 27 × 21 nm in the membrane plane (Fig. 2 B and C) and a maximal height of 23 nm (Fig. 2B), made by the giant hydrophilic domain of complex I. By comparison with known 3D structures of the individual complexes, many relevant features are recognizable, such as protruding densities at the center, related by a local twofold symmetry axis (Fig. 2, arrows), that point to the position of the core I and II subunits of dimeric complex III. Complex III2 sits in the arc of the membrane arm of complex I and complex IV is attached to the tip of NADH dehydrogenase and complex III2 (Fig. 2, arrowheads). To closely interpret the cryo-EM map, we superimposed the high-resolution X-ray structures of bovine complexes III2 (12) and IV (13). For modeling of the complex I moiety, the medium-resolution X-ray data from the yeast Yarrowia lipolytica were used (14). Fitting was accomplished with correlation coefficients between 0.83–0.92 for the bovine structures, indicating a unique, close fit with high confidence of components in the final hybrid structure (Fig. 3 A, B, and D, and Fig. S3). No substantial parts of the single complexes extend the 3D cryo-EM reconstruction, indicating that the respirasome is basically the sum of its components. Based on the fitting, our 3D structure presents a complete respirasome, including the peripheral arm domain of complex I (Fig. 3 A and B, arrowheads), which sometimes is partly lacking in negatively stained samples (16). Some minor differences exist between the EM reconstruction and the X-ray maps (Fig. 3 A, B, and D, orange arrowheads), which are most likely detergent molecules surrounding the respirasome. Iterative multireference alignment and classification (17) showed the presence of the peripheral arm in all five classes with a slight amount of variation (Fig. S4). Subtraction of two reconstructions representing two different classes revealed a density (Fig. S4, red asterisk), which can be accounted for a possible conformational change in the hydrophilic domain of complex I. To prove it, additional experiments are required. The full complex I moiety of bovine and Yarrowia are equal; the peripheral matrix arms make the same angle (Fig. 3A, yellow) and the bend of the membrane arm is closely the same (Fig. 3D, double arrowhead). The superposition of the smaller prokaryotic complex I from Thermus thermophilus indicates a similar overall shape (Fig. S5) and, in addition, clearly shows the locations of the accessory subunits of the eukaryotic enzymes are located. These accessory proteins appear to be arranged mostly in clusters.
Fig. 2.
Cryo-EM map of I + III2 + IV supercomplex: (A and B) seen from aside, (C) seen from the matrix side; arrow points to complex III2, arrowhead to complex IV, and double arrowhead to the curved membrane arm of complex I, respectively. (D) Seen from aside from the tip. Contour level is 0.1. Horizontal lines on A and D indicate the position of the membrane. (Scale bar: 10 nm.)
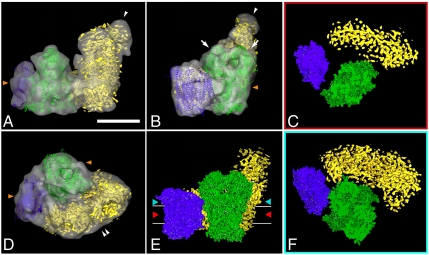
Fig. 3.
Fitting of the high- and medium-resolution structures of complexes I, III2, and IV to the 3D cryo-EM map of I + III2 + IV supercomplex: (A) side view, arrowhead points to flavoproteins; (B) side view from the membrane, arrows point to core I and II subunits of complex III2, arrowhead to flavoproteins; (C) section through the space-filling model of respirasome on the level of membrane, demonstrating gaps between complexes within the supercomplex; (D) top view from the intermembrane space, double arrowhead points to the bend of complex I in membrane; (E) space-filling model of respirasome seen from the membrane, red and light-blue arrowheads show the level of sections in C and F; (F) section through the space-filling model of respirasome on the level of matrix. In green, X-ray structure of the bovine dimeric complex III; in purple, X-ray structure of bovine monomeric complex IV; in yellow, the density map of complex I from Yarrowia lipolytica. Horizontal lines on E indicate the position of the membrane. Orange arrowheads on A, B, and D point to the position of detergent micelles. (Scale bar: 10 nm.)
Discussion
Single particle reconstruction can be performed in several ways. The most straightforward way is to reconstruct 3D structures from randomly oriented particles in a nontilted thin layer of vitreous ice (7). The respirasome, however, showed a strong preferential orientation on the carbon film. Therefore electron tomography reconstruction and subvolume averaging was applied. This method was previously used to visualize a number of membrane complexes in situ, like the nuclear pore complex in its native membrane (10) or protein–protein interactions in intact organelles (18, 19). Recent advances in automation of electron tomography data collection allowed more efficient subtomogram averaging, as applied for the study of capsids of herpes virions (20). Lately, the effectiveness of the maximum-likelihood processing approach of subtomograms was demonstrated on chaperonin GroEL/GroES complexes, showing a more reliable convergence at lower signal-to-noise levels (11). We performed an ML analysis to computationally extracted single respirasome subvolumes, which minimizes the reference bias for the reconstruction (21). Analysis of 2,466 subtomograms resulted in a 3D structure of the respirasome at an anisotropic resolution of 1.7–2.55 nm. However, the first zero in the contrast transfer function (CTF) curve at 2.2 nm-1. Hence, the details with spatial frequencies of 2.2 to 1.7 nm-1 have a reversed phase sign and are removed from the final reconstruction. This finding indicates that, in principle, the applied electron tomography and extensive 3D averaging could even be pushed to higher resolution, if reliable methods to perform CTF correction on the highly noisy images of the original tilt series would become available. From the 3D reconstructions, the position toward the support film of the single particles could be retrieved. About 70% of the respirasomes were found in one specific orientation, resulting in a triangular projection view (Fig. 1), and another 12% of the particles were in a side position with respect to the carbon support film (Fig. S2B). There was angular variation around both preferential orientations, and 10% of the particles were found in different, random orientations. This additional variation plus the tomographic tilting of the grids lead to a good sampling of angles over the Fourier space, demonstrating the advantages of subtomographic averaging for membrane proteins, when particles preferentially attach to a carbon film.
Although the 2.2-nm resolution cryo-EM model does not provide direct information about the secondary structure, the fitting allowed an interpretation at the level of α-helices, reflecting that the fitting procedure corresponds to a resolution gain by a factor of approximately 5 (22). From the obtained model, we can conclude that the interaction sites of complexes III2 and IV are different from the negatively stained data of the bovine respirasome (16) and the III2 + IV2 supercomplex from Saccharomyces cerevisiae (23). Complex IV contacts to complex III2 with the side opposite to the complex IV dimer interface in the X-ray structure (13) (Fig. 3 A, B, D, and E). Cytochrome b, the Rieske protein, and subunit 11 of one particular monomer of complex III are in close proximity to subunits III, VIa, and VIIa of complex IV. The cytochrome c1 subunit is far away from the complex III2–IV interface (Fig. 3 C, and F, blue helices), in contrast to what was concluded from the negative stain map of the bovine respirasome. The previously available respirasome EM map had resulted from negatively stained samples and did not show any distinct complex IV features that could have been of help in the proper positional assignment of complex IV (16). In contrast, in the current cryo-EM map, we can clearly distinguish subunits protruding outside the membrane. Subunit II of complex IV does not seem to be involved in the interaction between complex III and IV as it was proposed before (16). Subunits II, VIa, and VIb form a concave surface on top of subunit I, which could form an interaction site for a cytochrome c molecule (2.5 nm in diameter; ref. 13).
Interestingly, the single complexes I, III2, and IV appear after fitting to be at some distance in the model, suggesting that there is little close contact (Fig. 3 C, E, and F). The section through the model on the level of the membrane demonstrates the gap between complexes III2 and IV within the membrane (Fig. 3C). The complexes appear to contact each other in the membrane close to the matrix (Fig. 3F). It is possible that interactions are enhanced by lipid molecules such as cardiolipids, which were proposed to be essential in supercomplex formation (24, 25). In the structure of bovine complex IV, one phosphatidylethanolamine was found on subunit VIa and two phosphatidylethanolamines plus one phosphatidylglycerol molecule were revealed in the larger cleft of subunit III, which may serve as a lipid pool (13). The involvement of lipids may be special for maintaining the structure of membrane supercomplexes because in another large membrane protein complex, dimeric Photosystem II, 12 lipid molecules were at the interface between the monomers, of which four sulfoquinovosyldiacylglycerols might promote dimerization (26).
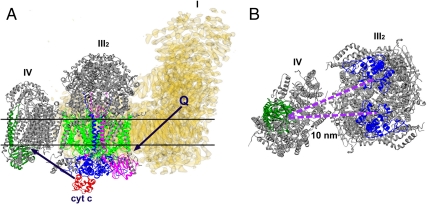
Complexes I and III2 are physiologically linked by the movement of quinones. The ubiquinone binding site at complex I in the matrix arm at the interface with the membrane arm (27) and cytochrome b of complex III binding hydroquinone (12) are facing each other in our cryo-EM map and are at a distance of 13 nm (Fig. 4A), allowing efficient ubiquinone transport between binding sites of complexes I and III. The distance that cytochrome c has to diffuse between the sites of complexes III2 and IV is 10 nm in the respirasome (Fig. 4B), but less than 4 nm in the III2 + IV2 supercomplex (23). Nevertheless, the formation of the respirasome brings these sites closer to each other than in a situation of randomly distributed complexes III and IV molecules in the membrane, which is probably beneficial, because in free random diffusion the time is proportional to the square of the distance. But although diffusion coefficients have been determined for cytochrome c (28), the rate at which cytochrome c diffuses is not directly measurable, because it depends on the local free water volume. Hence, we cannot quantify the effect of the (variable) distance on cytochrome c diffusion time. However, it is questionable if distance between electron transport components is the main reason for the supercomplex formation. Moreover, flux control experiments have not demonstrated substrate channeling between complexes III and IV via cytochrome c, although substrate channeling was found between bovine complexes I and III via ubiquinone (29). Likely, there are other reasons for the respirasome formation, such as overall regulation of oxidative phosphorylation. Another one is the assembly of the components. It was shown earlier that complex III can stabilize complex I in mammalian mitochondria (30). Another may be on an even higher level of organization, which is the respiratory string, a linear association of respirasomes. Our model is not in conflict with previously proposed models of respiratory strings (31, 32). According to Wittig et al. (31), the ratio between complex I, III, and IV in bovine mitochondria is equal to 1∶3∶6. Based on this ratio, they proposed the string in which every respirasome is connected via complex IV to another respirasome and III2 + IV2 supercomplex. Although the interfaces of complexes III2–IV are different than in yeast supercomplex, the site of complex IV, which is involved in dimerization, as it is revealed by X-ray crystallography, is available for the attachment of the next respiratory unit via the complex IV–complex IV interface. Finally, in mammalian mitochondria, the respirasome may prevent superoxide formation, a very harmful, naturally occurring process in mitochondria. Most of the mitochondrial superoxide originates from complex I and complex III (33).
Fig. 4.
Scheme of electron flow within the respirasome mediated by cytochrome c and ubiquinol. (A) Side view of the I + III2 + IV supercomplex. Black arrows point how the ubiquinol and the cytochrome c move between complexes I, III2, and IV. (B) Top view of the respirasome fragment consisting of complexes III2 and IV, seen from the intermembrane space. The distance between cytochrome c binding sites of complexes III2 and IV is 10 nm. In yellow, the density map of complex I from Yarrowia lipolytica, marked as I; in gray, X-ray structures of bovine complexes III2 and IV, marked as III2 and IV correspondingly. Q, ubiquinol; cyt c, cytochrome c. Colors: red, cytochrome c; dark green, subunit II of complex IV; blue, cytochrome c1 subunit of complex III2; green, cytochrome b of complex III2; magenta, “Rieske” protein of complex III2. Horizontal lines on A indicate the position of the membrane.
Materials and Methods
Beef heart mitochondria were solubilized in 30 mM Hepes buffer with 10% digitonin, 0.15 M potassium acetate, 10% glycerol, pH 7.4 at 4 °C at a ratio of 25 g detergent per 1 g of protein. The solubilized fraction was separated with BN-PAGE as described (2). The band with respirasomes was excised from an unstained gel, supplemented with 0.01% digitonin and electroeluted in a Bio-Rad electroelutor (34). For tomography, the sample was concentrated by centrifugation in Microcon tubes; aliquots of 2.5 μL were mixed with 10-nm gold particles as fiducial markers and applied on glow-discharged 200 mesh Quantifoil support grids (Quantifoil Micro Tools, GmbH) with an additional carbon support film. Grids were blotted at 100% humidity for 4–6 s at a blot offset (the longitudinal grid positioning) setting of -3.5, using a Vitrobot Mk3 (FEI). Data collection was performed with a 300-kV G2 Polara electron microscope (FEI) equipped with a Gatan energy filter. Images were recorded at 200 kV accelerating voltage for tomography or 300 kV for 2D projections processing with a 2,000 × 2,000 CCD camera (Gatan) at 2-μm underfocus (first CTF zero at 2.2 and 2 nm, respectively) and 78,000× magnification, resulting in a sampling with 0.38-nm pixels. Tilt series were acquired at 2° tilt angle increments covering a range of ± 65°, with a total dose of 8,000 electrons/nm2. Two-dimensional image processing of projections was done with the Groningen Image Processing software package (2).
A total of 21 tomograms were reconstructed by weighted back-projection using IMOD software (35). No CTF correction was performed. In total, 2,466 subvolumes of respirasomes with a box size of 128 × 128 × 128 pixels were selected from the tomograms by cross-correlation to an artificial globular reference followed by manual selection of particle volumes. An average structure was generated using the XMIPP ML for tomography software (11) on volumes downsampled to a pixel size of 0.76 nm. Averaging was initiated with a sum of randomly oriented particles and after 20 iterations with 20° global angular search the structure reliably converged. Further decreasing the angular search to 10° and 5° yielded an average at resolution of 3.2 nm. XMIPP ML calculations were run on 16 or 32 cores on the Maia cluster of the University of Basel. The alignment transforms were imported to the “AV3” routine for Matlab (15) and local 6D refinements on the volumes with initial voxel sizes of 0.38 nm were performed reaching the final average structure at resolution 2.2 nm. About one-half of the particles (1,174) with the best correlation coefficients were used to produce the final average. Resolution estimation was performed by the 0.5 information threshold criterion (36). Resolution criterion yielded a value that is below the first zero of the CTF, thus the resolution was assumed to be 2.2 nm. Maximum likelihood classification (17) into five classes was performed starting with the average structure at 3.2 nm with eight iterations allowing local angular search of 10°.
For the anisotropy test, we measured Fourier shell correlation in cones of Fourier space for different orientation of the cone according to unit sphere [φ-θ]. The cone had a semiaperture angle of 22.5°, angles were spaced every 18°. The resolution in the direction of each cone was determined by the 0.5 threshold. This routine is a feature of a yet unreleased Dynamo software package for subtomogram alignment.
X-ray structures of the bovine dimeric cytochrome bc1 complex [Protein Data Bank (PDB) ID 1BGY; ref. 12], monomeric cytochrome c oxidase (PDB ID 1OCC; ref. 13), Yarrowia lipolytica NADH: ubiquinone oxidoreductase (14), and NADH∶ubiquinone oxidoreductase from Thermus thermophilus (PDB ID 3M9S; ref. 27) were used for modeling of the electron density map of the respirasome at a voxel size 3.8 Å. The monomeric complex I was generated from the density map by manually erasing of the densities assigned to the neighboring molecule in the crystal unit by using the Chimera program from University of California, San Francisco (UCSF) (37). The maps were first prealigned manually and then automatically by cross-correlation using the Fit in Map tool in the UCSF Chimera program (37).
Supplementary Material
Acknowledgments.
We thank Dr. U. Brandt (Germany) for kindly providing the complex I density map from Yarrowia lipolytica, Dr. G. Oostergetel (Netherlands) for help with electron microscopy, Dr. Y. Chaban (United Kingdom), Dr. Daniel Castano-Diaz (Switzerland) for discussions and for determining resolution anisotropy, and Martin Jacquot (Switzerland) for support with computation. N.V.D. is supported by a Veni grant from the Netherlands Organization for Scientific Research. H.S. acknowledges support from the SystemsX.ch initiative and the Swiss National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The EM map of the respirasome has been deposited in the EMDataBank, www.emdatabank.org (accession no. EMD-5319).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107819108/-/DCSupplemental.
References
- 1.Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc Natl Acad Sci USA. 2005;102:3225–3229. doi: 10.1073/pnas.0408870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minauro-Sanmiguel F, Wilkens S, Garcia JJ. Structure of dimeric mitochondrial ATP synthase: Novel F0 bridging features and the structural basis of mitochondrial cristae biogenesis. Proc Natl Acad Sci USA. 2005;102:12356–12358. doi: 10.1073/pnas.0503893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dudkina NV, Heinemeyer J, Keegstra W, Boekema EJ, Braun HP. Structure of dimeric ATP synthase from mitochondria: An angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett. 2005;579:5769–5772. doi: 10.1016/j.febslet.2005.09.065. [DOI] [PubMed] [Google Scholar]
- 5.Dudkina NV, Kouřil R, Peters K, Braun HP, Boekema EJ. Structure and function of mitochondrial supercomplexes. Biochim Biophys Acta. 2010;1797:664–670. doi: 10.1016/j.bbabio.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Jin L, Fang Q, Wong HH, Zhou ZH. 3.3 Å Cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Heel M. Angular reconstitution—a posteriori assignment of projection directions for 3-D reconstitution. Ultramicroscopy. 1987;21:111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 8.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random-conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 9.Leis A, Rockel B, Andrees L, Baumeister W. Visualizing cells at the nanoscale. Trends Biochem Sci. 2009;34:60–70. doi: 10.1016/j.tibs.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryoelectron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 11.Scheres SH, Melero R, Valle M, Carazo JM. Averaging of electron subtomograms and random conical tilt reconstructions through likelihood optimization. Structure. 2009;17:1563–1572. doi: 10.1016/j.str.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwata S, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 13.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 14.Hunte C, Zickermann V, Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2010;329:448–451. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- 15.Förster F, Hegerl R. Structure determination in situ by averaging of tomograms. Methods Cell Biol. 2007;79:741–767. doi: 10.1016/S0091-679X(06)79029-X. [DOI] [PubMed] [Google Scholar]
- 16.Schäfer E, Dencher NA, Vonck J, Parcej DN. Three-dimensional structure of the respiratory chain supercomplex I1III2IV1 from bovine heart mitochondria. Biochemistry. 2007;46:12579–12585. doi: 10.1021/bi700983h. [DOI] [PubMed] [Google Scholar]
- 17.Stölken M, et al. Maximum likelihood based classification of electron tomographic data. J Struct Biol. 2011;173:77–85. doi: 10.1016/j.jsb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Dudkina NV, Oostergetel GT, Lewejohann D, Braun HP, Boekema EJ. Row-like organization of ATP synthase in intact mitochondria determined by cryoelectron tomography. Biochim Biophys Acta. 2010;1797:272–277. doi: 10.1016/j.bbabio.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Kouřil R, Oostergetel GT, Boekema EJ. Fine structure of granal thylakoid membrane organization using cryoelectron tomography. Biochim Biophys Acta. 2011;1807:368–374. doi: 10.1016/j.bbabio.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Grünewald K, et al. Three-dimensional structure of herpes simplex virus from cryoelectron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 21.Zeng X, Stahlberg H, Grigorieff N. A maximum likelihood approach to two-dimensional crystals. J Struct Biol. 2007;160:362–374. doi: 10.1016/j.jsb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossmann MG. Fitting atomic models into electron-microscopy maps. Acta Crystallogr D Biol Crystallogr. 2000;56:1341–1349. doi: 10.1107/s0907444900009562. [DOI] [PubMed] [Google Scholar]
- 23.Heinemeyer J, Braun HP, Boekema EJ, Kouřil R. A structural model of the cytochrome C reductase/oxidase supercomplex from yeast mitochondria. J Biol Chem. 2007;282:12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 25.Wenz T, et al. Role of phospholipids in respiratory cytochrome bc1 complex catalysis and supercomplex formation. Biochim Biophys Acta. 2009;1787:609–616. doi: 10.1016/j.bbabio.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 26.Guskov A, et al. Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- 27.Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 28.Hochman J, Ferguson-Miller S, Schindler M. Mobility in the mitochondrial electron transfer chain. Biochemistry. 1985;24:2509–2516. doi: 10.1021/bi00331a017. [DOI] [PubMed] [Google Scholar]
- 29.Bianchi C, Genova ML, Parenti Castelli G, Lenaz G. The mitochondrial respiratory chain is partially organized in a supercomplex assembly: Kinetic evidence using flux control analysis. J Biol Chem. 2004;279:36562–36569. doi: 10.1074/jbc.M405135200. [DOI] [PubMed] [Google Scholar]
- 30.Acín-Pérez R, et al. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell. 2004;13:805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wittig I, Carrozzo R, Santorelli FM, Schägger H. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim Biophys Acta. 2006;1757:1066–1072. doi: 10.1016/j.bbabio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Bultema JB, Braun HP, Boekema EJ, Kouřil R. Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim Biophys Acta. 2009;1787:60–67. doi: 10.1016/j.bbabio.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 34.Werhahn W, Braun HP. Biochemical dissection of the mitochondrial proteome from Arabidopsis thaliana by three-dimensional gel electrophoresis. Electrophoresis. 2002;23:640–646. doi: 10.1002/1522-2683(200202)23:4<640::AID-ELPS640>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 35.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 36.van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.