Abstract
Yeast rad3-102, a mutant of the TFIIH complex involved in nucleotide excision repair (NER) and transcription, can perform NER initial steps but not late steps of postincision gap filing. Because removal of early-acting NER proteins prevents rad3-102 deleterious action, we used this feature to explore if chaperones act in early NER. We found that the cochaperone Ydj1 is required for NER and that Ydj1 guarantees TFIIH stoichiometry. Importantly, in the absence of Ydj1, the roles of TFIIH in transcription and transactivation, the ability to activate transcription by nuclear receptors in response to hormones, are strongly impaired. We propose that TFIIH constitutes a multitarget complex for Ydj1, as six of the seven TFIIH core components contain biologically relevant Ydj1- binding motives. Our results provide evidence for a role of chaperones in NER and transcription, with implications in cancer and TFIIH-associated syndromes.
Keywords: Rad3/XPD, Xeroderma pigmentosum, Cockayne syndrome, genome instability, HDJ-2
Genetic instability is prevented by the cooperation of multiple mechanisms, including replication fidelity, DNA damage checkpoint activation, and repair. Different pathways, whose intervention depends on lesions affecting the DNA helix, accomplish DNA repair. Double strand breaks are mainly repaired by nonhomologous end joining, a mechanism that seals the two ends of a break, or homologous recombination, which uses a homologous sequence to fill the break (1). Bulky adducts that distort the helix are repaired by nucleotide excision repair (NER). After detection of the lesion, NER proteins open the DNA, recognize the damaged strand, incise it at both sides of the damage, and fill in and seal the remaining gap (2).
TFIIH is a multifunctional complex composed of 10 subunits, with an essential role in transcription, in which it functions in promoter opening, RNA polymerase II (RNAPII) phosphorylation, and promoter escape; this along with a central role in NER by promoting damaged strand discrimination, DNA opening, and endonuclease recruitment. We recently unmasked an intimate connection between NER and homologous recombination repair pathways by characterizing the molecular defect present in rad3-102 yeast cells, which are mutated in the 5′>3′ TFIIH helicase needed for NER (3, 4). In these cells, the early steps of NER leading to incision can be performed, but recruitment of the gap-filling factors required for late steps fail, leaving an unrepaired ssDNA gap. After arrival of the replication fork, this gap is converted into a double strand break, requiring the recombination machinery for its repair. When the early steps of NER are prevented, the gap is not formed, explaining why mutations that inactivate early NER steps suppress rad3-102 main phenotypes (3, 4).
Chaperones are proteins in charge of modulating lifespan, activity, and cellular localization of proteins. The essential chaperone Hsp90 is central to a network in which cochaperones provide substrate specificity (5). Apart from its well defined roles in stress tolerance, protein folding, and their targeting to the proteasome, Hsp90 may function in the assembly and disassembly of multiprotein complexes such as the proteasome itself (6), the kinetochore complex (7), or transcriptional complexes (8). Hsp90 also promotes replication through UV-damaged templates in human cells (9). Another link between chaperones and replication was previously reported in Escherichia coli for the Hsp70 cochaperone homologue when template switching was needed to accomplish repair (10). Interestingly, chaperones have also been related to NER, as genetic and physical interactions were described between Hsp90-Sti1 and the other TFIIH helicase, Rad25 (11), and synthetic lethal interactions were found between the TFIIH kinase, Kin28, and the cochaperones Sti1 and Cdc37 (12).
Ydj1 is a cochaperone that belongs to the Hsp40 family, which in Saccharomyces cerevisiae is made up of 22 members (13). This family of proteins possesses a highly conserved N-terminal J domain, necessary to stimulate Hsp70 ATPase activity (14). Additionally, type I Hsp40 members, such as Ydj1, possess a zinc finger-like region that allows substrates to be transferred to Hsp70 (15). Interestingly, Ydj1 physically interacts with the TFIIH helicase Rad3 (16) and Tfb2 (17).
We investigated whether the role of chaperones in genetic stability could be extended to NER. Suppression of the phenotypes of rad3-102, defective in late NER steps, was used as a way to identify defects in early NER. Ydj1 was found to be necessary to guarantee genetic stability via NER, concretely by means of TFIIH. This has implications for the rest of the known TFIIH functions, such as transcription initiation and transactivation. Biochemical analysis of the core TFIIH in ydj1Δ reveals that stoichiometry of the complex is imbalanced. We uncover TFIIH as a multitarget substrate for Ydj1, as six of the seven components of the core complex bear biologically relevant Ydj1 binding motives. A nuclear role for chaperones was therefore revealed in TFIIH function and in preserving genomic integrity.
Results
NER Efficiency Is Reduced in ydj1Δ Cells.
Given the reported interactions between the cochaperone Ydj1 and the TFIIH complex (16, 17), we decided to ascertain whether Ydj1 had any importance in NER. First of all, UV sensitivity was assayed in ydj1Δ cells. They displayed loss of viability after UV irradiation with doses at which WT cells were unaffected (Fig. 1A and Fig. S1A), but grew in the presence of DNA-damaging agents such as methyl methanesulfonate (MMS), camptothecin (CPT), and hydroxyurea (HU; Fig. 1A). UV sensitivity at the assayed doses was specific to Ydj1 absence, as removal of other Hsp40s with similar subcellular localization (17) did not cause sensitivity to UV irradiation (Fig. S1B). These data indicate that the lack of this particular cochaperone specifically incapacitates cells to tolerate UV damage, but leaves other repair pathways unaltered.
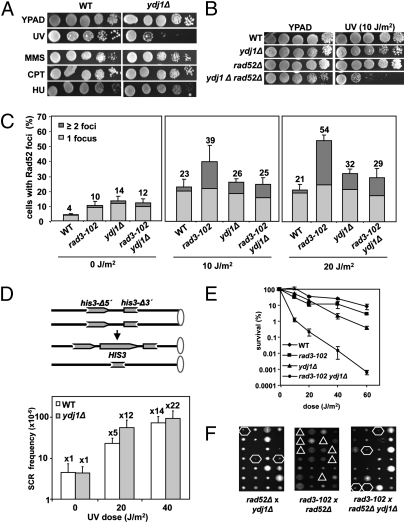
Fig. 1.
ydj1Δ cells are defective in NER. (A) Tenfold serial dilution assays to establish sensitivity to various DNA damaging agents. Doses used are 60 J/m2 UV, 0.01% MMS, 10 μg/mL CPT, and 50 mM HU. At the indicated doses of MMS, CPT, and HU, the positive control rad51Δ is dead (Fig. S1D). (B) Tenfold serial dilution assays to measure UV sensitivity at a 10-J/m2 dose in ydj1Δ cells in the absence of Rad52. (C) Spontaneous and UV-induced accumulation of Rad52 foci in WT, ydj1Δ, rad3-102, and their corresponding double mutant cells in S and G2 carrying the RAD52-YFP construct with or without UV irradiation. (D) Recombination frequency in response to UV in the direct-repeat system his3- Δ5′- his3-Δ3′, which measures unequal sister chromatid exchange in WT and ydj1Δ cells. Mean and SD of three independent fluctuation tests were calculated for each genotype. (E) Survival curves after UV irradiation of ydj1Δ, rad3-102, and their corresponding double mutant. (F) Tetrad dissection of crosses to obtain double mutants ydj1Δ rad3-102, rad52Δ rad3-102, and the triple mutant ydj1Δ rad52Δ rad3-102. Hexagons indicate viable combinations and triangles lethal combinations.
In WT cells, UV damage is mainly repaired by NER, although UV irradiation also moderately increases recombination and mutation levels. This is why, in NER-deficient backgrounds, such levels are exacerbated (18, 19). Recombination was chosen as a marker of the putative NER defect in ydj1Δ cells to prove that alternative pathways processed unrepaired UV damage. First of all, ydj1Δ cells lacking the recombination protein Rad52 are synergistically sensitive to UV (Fig. 1B). Consistently, levels of Rad52 foci, a general measure of recombination events (20), were 3.5-fold higher in ydj1Δ cells than in WT cells for spontaneous and UV-induced events, especially at 20 J/m2 (Fig. 1C). Sister chromatid recombination was also measured. A 12-fold increase in the mutant versus fivefold in the WT after 20 J/m2 irradiation, and 22-fold versus 14-fold after 40 J/m2, was found, meaning that there is more recombinogenic UV damage in the mutant (Fig. 1D). Altogether, the results suggest that Ydj1 ablation may cause a NER defect and that this confers genetic instability.
To prove that Ydj1 is important for NER, we took advantage of the fact that removal of early-acting NER proteins suppresses rad3-102 phenotypes. First, whereas rad3-102 cells are not UV-sensitive because they channel UV damage into recombination (4), they became highly sensitive to UV upon Ydj1 ablation (Fig. 1E). The rad3-102 ydj1Δ double mutant was also highly sensitive to 4-nitroquinolineoxide (Fig. S1C). Second, although Rad52 foci levels are strongly increased in rad3-102 cells upon UV (4), ydj1Δ suppressed that increase (10 and 20 J/m2; Fig. 1C). Third, rad3-102 cells are dependent on recombination for survival, with rad3-102 rad52Δ cells not being viable. ydj1Δ suppresses this lethality, as the triple mutant ydj1Δ rad52Δ rad3-102 is alive (Fig. 1F). Altogether, the results reveal that ydj1Δ suppresses rad3-102 phenotypes, indicating that Ydj1 functions in early NER.
Transcription Initiation Is Impaired in ydj1Δ Cells.
We considered TFIIH as a candidate to be altered in ydj1Δ cells. To test this, we took advantage of the transcription initiation role of TFIIH. First, we tested the effect of ydj1Δ in general transcription. The plasmid-born pLAUR construct, in which a lacZ-URA3 translational fusion is under control of the tet promoter (21) was used. ydj1Δ cells could not grow without uracil (Fig. 2A), indicating that they were not able to express the lacZ-URA3 fusion as a consequence of a defect in transcription. To prove a defect in transcription initiation, we analyzed transcription at the chromosomal GAL1 gene. Northern analysis after induction with galactose shows that ydj1Δ strongly delayed transcript apparition, suggesting a defect in transcription initiation (Fig. 2B).
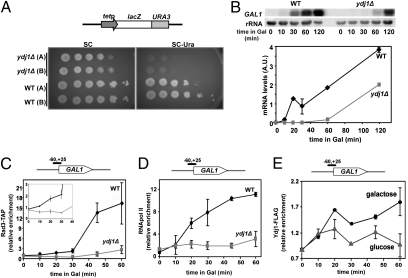
Fig. 2.
ydj1Δ cells are defective in transcription initiation. (A) Gene expression ability is measured as growth in SC-trp-ura using the pLAUR system. Serial dilutions assays show transformants of WT and ydj1Δ cells. (B) Northern analysis of GAL1 mRNA after galactose induction in WT and ydj1Δ. rRNA is shown as a loading control. Quantification is shown below in arbitrary units. Mean and SD of two independent experiments are depicted. (C) Chromatin immunoprecipitation of Rad3-TAP at the GAL1 promoter upon galactose addition in WT and ydj1Δ cells. The mean and SD of triplicate assays of two independent experiments are depicted at each point. A detail of early time points is shown. (D) ChIP with anti-RNAPII (details as in C). (E) ChIP of Ydj1-FLAG at the GAL1 promoter after induction of transcription (galactose) or under repression conditions (glucose). Mean and SD of three independent experiments are depicted at each point.
At the molecular level, ChIP of a TAP-tagged version of Rad3 was performed within the first hours of GAL1p activation, when the transcription defect was more evident. Recruitment of Rad3 to GAL1p upon activation was impaired in the mutant from early time points (Fig. 2C). The same approach was used to assess RNAPII loading. Again, RNAPII was barely recruited to GAL1p in the ydj1Δ mutant (Fig. 2D). The data demonstrate that transcription initiation does not take place properly without Ydj1. Altogether our results suggest that YDJ1 deletion leads to functional defects within the TFIIH complex.
To further prove that Ydj1 was involved in TFIIH-mediated DNA transactions, a competent C-terminal FLAG-tagged Ydj1 strain was created (Fig. S2) and assessed as to whether the Ydj1-FLAG was recruited to chromatin upon transcription activation. ChIP experiments revealed that Ydj1 was present at GAL1p upon induction with galactose, and not when transcription was repressed with glucose (Fig. 2E), displaying a mild recruitment that peaked at 60 min, which correlates with the higher levels detected for Rad3 and the RNAPII (Fig. 2 C and D). This suggests that the cochaperone may function in situ at the transcription initiation site.
Direct Action of Ydj1 on TFIIH Function.
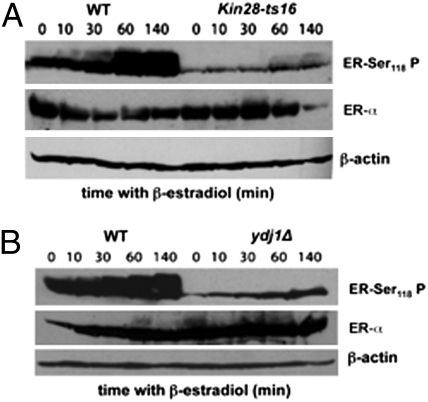
As Ydj1 ablation causes NER and transcription initiation deficiencies and Ydj1 may be recruited to chromatin in a transcription-dependent manner, it is possible that Ydj1 played a role in TFIIH assembly or function. However, the Hsp90 pathway has been proposed as helping to remove nucleosomes at sites of transcription (22), which could explain the phenotypes reported here. To assay that other DNA modifications could be responsible for the repair and transcription phenotypes of ydj1Δ cells, we assessed a well documented TFIIH function in human cells that is chromatin-independent: transactivation. This consists of the phosphorylation of nuclear receptors in response to hormones, which in turn are able to bind to responsive elements in the DNA and activate transcription. In the case of the human steroid and retinoid receptors, this phosphorylation is accomplished by the kinase of the CAK subcomplex of TFIIH (23, 24). To this end, the estrogen-responsive system from human cells was transplanted into yeast with a plasmid bearing the human estrogen receptor gene (ER) under control of the GPD1 constitutive promoter (25), as well as the reporter lacZ gene under control of an estrogen-responsive element (ERE) (26). Upon β-estradiol addition to liquid cultures, the ER becomes quickly phosphorylated at Ser118 and binds to the ERE, thereby activating lacZ expression (26). Nevertheless, the interest lay in measuring only ER phosphorylation (Fig. S3), as lacZ expression also implies transcription, a feature altered in ydj1Δ cells (Fig. 2). Although this heterologous system works in yeast (27), whether the kinase of the CAK complex from yeast was, as in humans, responsible for ER phosphorylation, was not known. To test this, ER phosphorylation was assayed in a TFIIH kinase-defective mutant, kin28-ts16 (28). ER phosphorylation kinetics in WT and kin28-ts16 cells were determined. Because induced ER phosphorylation at serine 118 was impaired in the mutant (Fig. 3A), this confirms that the yeast TFIIH kinase is also responsible for ER phosphorylation in response to estrogen. Finally, it was reasonable that, if Ydj1 acted directly on TFIIH to preserve its integrity, transactivation would also be altered in ydj1Δ cells. Therefore, the experiment described earlier was repeated in ydj1Δ cells in comparison with the WT strain. Fig. 3B clearly shows that, in the absence of Ydj1, the ER, which is expressed at the same levels in both WT and ydj1Δ cells, barely becomes phosphorylated, even for a long time after induction (140 min). Altogether, these results demonstrate that Ydj1 directly acts on the TFIIH complex to preserve its functionality.
Fig. 3.
ydj1Δ cells are defective in transactivation. (A) Immunodetection of phosphorylation at Ser118 of ER at basal levels (0 min) and after β-estradiol addition in WT and the TFIIH kinase-defective mutant kin28-ts16. Total ER protein (ER-α) and β-actin loading controls are shown. (B) Immunodetection of phosphorylation at Ser118 of ER at basal levels (0 min) and after β-estradiol addition in WT and in ydj1Δ. Total ER protein (ER-α) and β-actin loading controls are shown.
TFIIH Stoichiometry Is Altered in ydj1Δ Cells.
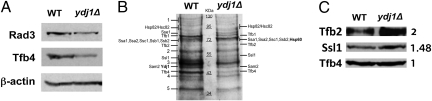
One possibility to explain the NER phenotypes of ydj1Δ cells was via an alteration of TFIIH composition. Thus, we analyzed the total levels of the TFIIH structural component Tfb4 and its catalytic subunit Rad3 with (Fig. 4A) or without (Fig. S4A) a TAP tag. The levels of both proteins were diminished in ydj1Δ cells, as seen by Western analysis (Fig. 4A and Fig. S4A), whereas the control Rad14 NER protein, which does not form part of the TFIIH complex, did not show such a decrease (Fig. S4A). This is not a result of a different distribution of cells throughout the cell cycle, as both WT and ydj1Δ display identical FACS patterns (Fig. S4B). Thus, this may indicate insufficient concentration of TFIIH or lower proportion of functional TFIIH caused by incorrect assembly in ydj1Δ.
Fig. 4.
TFIIH stoichiometry is altered in ydj1Δ cells. (A) Immunodetection of Tfb4-TAP and Rad3-TAP by using anti-TAP antibody in WT and in ydj1Δ crude extracts from cells grown to exponential phase in SC. β-Actin loading control is shown. (B) Coomassie gel comparing the purified TFIIH complex obtained by Tfb4-TAP immunoprecipitation in WT and in ydj1Δ strains. Proteins identified by MALDI-TOF are indicated. Additional proteins, such as Pse1 and Eft2 (“1”), Cdc19 and Vma2 (“2”), Tef2 (“3”), Psa1 (“4”), or Thd2/3 (“5”) were identified in both strains. (C) Immunodetection of Tfb2, Ssl1, and Tfb4-TAP in purified extracts from two independent purifications to determine their relative abundance. The ydj1Δ: WT ratios of the level of each protein relative to Tfb4 are shown on the right.
The TFIIH complex was purified in isogenic WT and ydj1Δ strains carrying a TAP-tagged Tfb4, a valid structural component to this end (29, 30). The expected core components for this kind of immunoprecipitation (31) were found in both strains (Fig. 4B). Notably, Ydj1 was purified in the WT strain, whereas in ydj1Δ, a chaperone was purified that was not present in the purifications of the WT strain, Hsp60 (Fig. 4B). This may suggest the existence of a compensatory effect between different chaperones. Nevertheless, in agreement with the absence of UV sensitivity when other Hsp40s are removed, no other Hsp40 was found in our purification (Fig. 4B). Of note, the Hsp70 family members Ssa1 and Ssa2, well known partners of Ydj1, were also present in both purifications (Fig. 4B). To determine if these chaperones were also involved in the TFIIH function, the ssa1Δ ssa2Δ double mutant was analyzed to see if it recapitulated some of the ydj1Δ phenotypes. The ssa1Δ ssa2Δ double mutant was UV-sensitive (Fig. S4C) and showed a transcription defect compared with WT and simple mutants (Fig. S4D), although these effects were milder than in ydj1Δ cells. This confirms a functional link between Ydj1 and Hsp70 proteins regarding TFIIH integrity. Indeed, these partial phenotypes and the presence in the purification of other Hsp70s, such as Ssb1 and Sse1, suggest that Ydj1 works in concert with several other Hsp70s.
Next, the relative amount of some TFIIH proteins was determined in the purified extracts by Western analysis. We found that the relative amount of Ssl1 and Tfb2 with respect to the immunoprecipitated component Tfb4-TAP could be detected in the ydj1Δ purification at higher levels than in the WT (Fig. 4C). A Western analysis and the relative increase from one representative purification using Tfb4-TAP are shown to illustrate this finding (Fig. 4C). Altogether, these data indicate that, in the absence of Ydj1, the TFIIH composition is suboptimal or, alternatively but not exclusively, only a subpopulation of correctly assembled TFIIH is present. This alteration in TFIIH stoichiometry, although mild, could contribute to the ydj1Δ defects in repair, transcription, and transactivation.
TFIIH Displays Biologically Relevant Ydj1 Binding Motives.
Ydj1 has been crystallized in a complex with its peptide substrate, which consists of seven amino acids, GWLYEIS (32). The amino acid motif to which Ydj1 binds has been later refined to a more flexible consensus sequence by means of computational analysis, and some of the targets experimentally verified to prove the biological relevance of the motif (33). The consensus sequence is GX[LMQ](P)X(P)(CIMPVW), where [XY] denotes either X or Y, and (XY) denotes neither X nor Y.
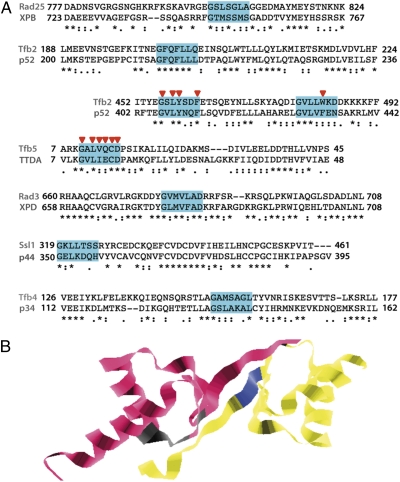
The amino acid sequences of the 10 components of TFIIH were searched for putative Ydj1 targets. Because it is a short peptide, the probability of finding the motif throughout a sequence without a biological relevance may exist. Therefore, we tried to diminish this probability by studying only the motives present simultaneously in the yeast and the human protein upon sequence alignment (Fig. 5A). Interestingly, the Ydj1-binding consensus sequence is present in six of the seven components of the TFIIH core, except for Tfb1.
Fig. 5.
The Ydj1 binding consensus motif is present in six TFIIH subunits. (A) Representative fragments of the alignments of yeast and human TFIIH components showing the regions where Ydj1 binding motif was conserved are annotated by residue positions. Motives are highlighted in blue. An asterisk denotes full residue conservation. A colon denotes equivalent residue change. Arrowheads indicate residues required to establish contact with another TFIIH component (as detailed in the text). (B) Three-dimensional view of the Tfb2 C-terminal residues 432–507 (yellow) interacting with Tfb5 residues 2–66 (pink). The key residues of the Ydj1 binding motif (arrowheads in A) are displayed in blue for Tfb2 and in gray for Tfb5. The Protein Data Bank ID code is 3DGP.
Table S1 collects the information about the point mutations and deletions present in or covering the motives highlighted in Fig. 5A. As can be seen, motives are located at sites that prove to be a key to allow correct assembly and subsequent functioning of the complex. Along this line, phosphorylation of Ser751 in XPB needs to be carefully regulated to permit the incision during NER (34). XPB retention and ATPase stimulation for appropriate NER and transcription are dependent on the integrity of Ydj1-binding motif present in Tfb2/p52 to avoid strong defects in Drosophila (35). The other two motives present in Tfb2/p52, located in the C-terminal part, contain key residues to establish contact with Tfb5 (36) (Fig. 5A, arrowheads). Reciprocally, the seven residues that compose the motif in Tfb5/TTDA are essential to interact with Tfb2/p52 (37, 36) (Fig. 5A, arrowheads). Rad3/XPD loses its interaction with Ssl1/p44 after mutation of the Gly675 residue of the motif (38), whereas Ssl1/p44 and Tfb4/p34 lose their mutual interactions when the fragments containing the motives are removed (39, 40).
Finally, we performed a statistical analysis to clarify whether this was significant (Fig. S5 and SI Materials and Methods). We randomly created groups of six proteins whose amino acid alignments between yeast and human were similar to those of the six TFIIH proteins of interest (Fig. S5A). We grouped the proteins in classes by using the aligned amino acid length as a criterion and analyzed the distribution of the motives in such classes. The control groups of proteins displayed a frequency of motives that was in agreement with a higher number of motives as the protein length increases (Fig. S5B). In contrast, in the TFIIH group, the frequency of motives was significantly differently distributed to that of the controls, as the motif was more abundant in proteins sharing less than 600 aligned amino acids (P = 0.99; Fig. S5B). This overrepresentation of Ydj1-binding motives is observed in proteins that perform regulatory roles in TFIIH (Tfb5, Tfb4, Ssl1, Tfb2) but not in those with helicase catalytic functions (Rad3 and Rad25). Altogether, the in silico and statistical analyses suggest that Ydj1 has binding consensus sites in different components of the TFIIH core, which are fundamental for TFIIH integrity.
Discussion
Here we show that the cochaperone Ydj1 has a central role in TFIIH function. Ydj1 is needed for full TFIIH performance in NER, transcription, and transactivation, and its absence leads to genetic instability. This action is exerted directly on TFIIH and does not rely on DNA, even though Ydj1 may be present at the site of chromatin by means of its action on TFIIH. In the absence of Ydj1, TFIIH composition is imbalanced and in silico analysis reveals Ydj1- binding domains in six of the seven TFIIH core proteins. Statistical analysis refines this putative binding to the regulatory core subunits Tfb5, Tfb4, Ssl1, and Tfb2. We propose that TFIIH is a multitarget substrate for Ydj1, on which it exerts a regulatory or an assembly action. This finding opens a perspective to understand TFIIH function, as mutations in the Ydj1-binding residues are known to provoke TFIIH dysfunction and disease. Our study reveals the importance of Ydj1 in the early steps of NER, as deduced from the UV sensitivity phenotype of ydj1Δ cells and the capability of ydj1Δ to suppress the rad3-102 phenotypes affecting late postincision steps in NER. The additional impairment of transcription initiation (Fig. 2) suggests that indeed the target of Ydj1 is the TFIIH complex.
Evidence for the conformational changes of protein complexes at their site of action has been provided for nuclear proteins (9, 10), and our study proposes TFIIH as a nuclear substrate of chaperones. Physical interactions between Ydj1 and TFIIH components had been previously reported for Rad3 (16) and Tfb2 (17). We found that Ydj1 copurifies with TFIIH in WT yeast strains, revealing a physical interaction of Ydj1 with the whole complex. Interestingly, the Hsp70 cochaperones Ssa1 and Ssa2, previously purified in association with TFIIH but considered as contaminants (31), were consistently copurified in our assays, in agreement with the notion that Hsp40 proteins work in concert with Hsp70s. Our study shows that the Ydj1-Hsp70s association has a functional role regarding TFIIH, as the simultaneous absence of the Ssa1 and Ssa2 cochaperones partially mimics the ydj1Δ phenotypes (compare Figs. 1A and 2A and Fig. S4). Ydj1 may work in association with several Hsp70s, explaining why ssa1Δ ssa2Δ cells display milder phenotypes than ydj1Δ cells (compare Fig. 2A and Fig. S4D). Indeed, functional modules containing Ydj1 as the main Hsp40 also include the Hsp70s Ssa1, Ssa2, Ssb1, and Sse1 (17). Interestingly, these four Hsp70s are the ones that copurify with TFIIH in the WT strain (Fig. 4B). Instead, in ydj1Δ, TFIIH copurifies with Hsp60 (Fig. 4B). Hsp60 is able to form functional chaperone modules in concert with the natural Ydj1 partners Ssa1, Ssa2, and either Ssb1 or Sse1 (17). This interchange may reveal a cell response to partially overcome Ydj1 absence. Indeed, Ydj1 and the Hsp60 human homologues HDJ-2 and Hsp60 have been reported to be simultaneously overexpressed in kidney biopsy specimens of patients undergoing acute and chronic rejection (41). The relevance of chaperones in TFIIH function is also supported by our purification of the chaperone Hsp90 (Hsc82-Hsp82) in WT and ydj1Δ cells (Fig. 4B).
It has been proposed that chaperones of the Hsp90 pathway are involved in nucleosome positioning at sites of transcription (22), which could, in principle, explain some of the phenotypes of genetic instability, NER, and transcription reported for ydj1Δ cells (Fig. 1 and 2). However, we ruled out this possibility by assaying transactivation, a TFIIH function that is DNA- and chromatin-independent. With a humanized yeast system, we have shown that ER phosphorylation is dependent on the yeast TFIIH kinase, Kin28, as it is on the Cdk7 homologue in humans. This is noteworthy, considering that S. cerevisiae lacks an ER system, and implies a strong conservation of TFIIH functions. Importantly, a low ER phosphorylation takes place in the absence of Ydj1 (Fig. 3B), indicating that Ydj1 directly acts on TFIIH independently of DNA or chromatin.
Our search for the consensus Ydj1 binding motif in the 10 subunits of the TFIIH complex revealed that six of the seven subunits composing the core bear at least one motif conserved between yeast and humans (Fig. 5A). Interestingly, all these motives lead to a molecular defect when mutated or removed (Table S1), consistent with the biological relevance of the Ydj1–TFIIH interaction. However, this does not necessarily mean that Ydj1 simultaneously interacts with all these proteins. Statistical analysis pointed to those performing a regulatory role, namely Tfb5, Tfb4, Ssl1, and Tfb2. It may happen that, depending on whether TFIIH may be acting on NER, transcription, or transactivation, one subunit or a subset of them may become the relevant target. A good example is given by a 3D view in Fig. 5B in which the residues previously published as essential for Tfb2–Tfb5 interaction (36, 37) are highlighted. Such residues fall within Ydj1-binding consensus motives, facing each other in a mirror-like distribution to allow contact between molecules (Fig. 5B). Altogether, alterations in the scheduled interactions among the TFIIH subunits may explain the relevance of the Ydj1 chaperone in TFIIH function.
Ydj1 is an abundant cytosolic protein with roles in the endoplasmic reticulum (42, 43), but no evidence was provided for nuclear functions. The biological impact of Ydj1 ablation in the nucleus shown in this work may help our understanding of the molecular basis of particular cancer treatments. This is the case of the inhibition of farnesylation of the Ydj1 homologue HDJ-2 by the drug R115777 that sensitizes human glioblastoma to radiation (44). HDJ-2 migrates from the cytosol to the nucleus upon irradiation, with concomitant resistance to irradiation. R115777 prevents HDJ-2 from entering the nucleus, abrogating irradiation resistance. Our findings open the possibility that HDJ-2 may have a role in DNA repair by acting directly on a DNA repair complex or on transcription of some repair factors.
The observation of a unique pathway of TFIIH regulation suggests that NER, transcription, and transactivation depend on Ydj1. There are situations in which the same TFIIH mutation leads to different clinical manifestations. This is the case of XPB mutation F99C, which caused mild Xeroderma pigmentosum (XP) in two sisters but XP combined with the Cockayne syndrome (XPCS) in two brothers (45). This difference could be explained by genetic background, which might affect their chaperone system. Also, the changes G675R and D681N in XPD/Rad3 cause XPCS and XP, respectively. Both residues are comprised in the Ydj1-binding motif but, because of the motif versatility, although the D681N change would still allow Ydj1 binding, the G675R destroys the Ydj1-binding site. An effect on Ydj1 binding could impair p44/Ssl1 recruitment (38), leading to an additional defect in transcription (46) compatible with XPCS.
In conclusion, the present work demonstrates that Ydj1 is a direct player in TFIIH function, providing a new perspective to understand TFIIH functions and their clinical manifestations.
Materials and Methods
All experimental procedures are detailed in SI Materials and Methods and Table S2. Recombination frequencies were obtained as the mean value of three median recombination frequencies obtained each from six independent colonies. Spontaneous and UV-induced Rad52 foci were counted in DAPI-stained nuclei from S-G2 midlog cells bearing plasmid pWJ1344. For ChIP, strains were grown in SGL (synthetic complete medium with 2% glycerol-2% lactate) overnight and glucose or galactose added to a 2% final concentration. Samples (50 mL) were processed as described previously (4). IgG Sepharose (GE Healthcare), anti-FLAG (F3165; Sigma), or anti-RNAPII 8WG16 antibodies (Covance) were used to precipitate TAP-tagged Rad3, Ydj1-FLAG, or total RNAPII, respectively. Quantitative PCR was performed at coordinates 278969 to 279048 on chromosome II. Normalization was done with values of amplification at ARS504 (coordinates 9754–9837 on chromosome V). For the estrogen receptor phosphorylation kinetics, WT, kin28-ts16, and ydj1Δ strains were grown until the exponential phase and β-estradiol was added to a final concentration of 0.1 μM. For Western blots, TCA-extracted proteins were separated in 10% PAGE. Antibodies anti-ERα (ab2746; Abcam), anti- ERSer118-P (ab32396; Abcam), anti-TAP (Thermo Scientific), anti-actin (ab8224; Abcam), anti-Ssl1, anti-Tfb2, and anti-Tfb4 (gift from Yuichiro Takagi, Indiana University, Indianapolis, IN), anti-Rad3 (sc-11963; Santa Cruz Biotechnology), and anti-Rad14 (ab22092; Abcam) were used. TAP-tagged purification of TFIIH from strains CB010-TFB4 and MMIII-60 (ydj1Δ) was performed followed by affinity purification using an IgG-Sepharose column (GE Healthcare). Proteins were identified by MALDI-TOF. For bioinformatics, proteins sequences were obtained from the PubMed database and aligned by using the ClustalW and LAlign tools (EBI). Molecules were processed by using RasMol.
Supplementary Material
Acknowledgments
We thank S. Buratowski, A. G. Hinnebusch, M. Fasullo, R. Rothstein, D. Picard, R. Kornberg, and Y. Takagi for providing reagents; J. C. Reyes for critical reading of the manuscript; and D. Haun for style supervision. This work was supported by Spanish Ministry of Science and Innovation Grants BFU2006-05260 and CSD2007-015 and Junta de Andalucía Grants BIO102 and CVI2549, and by the European Union (Regional Development European Funds, FEDER). M.M.-C. was the recipient of a predoctoral Formación de Profesorado Universitario training grant from the Spanish Ministry of Science and Innovation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107425108/-/DCSupplemental.
References
- 1.Pardo B, Gómez-González B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci. 2009;66:1039–1056. doi: 10.1007/s00018-009-8740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nouspikel T. DNA repair in mammalian cells: Nucleotide excision repair: Variations on versatility. Cell Mol Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montelone BA, Hoekstra MF, Malone RE. Spontaneous mitotic recombination in yeast: The hyper-recombinational rem1 mutations are alleles of the RAD3 gene. Genetics. 1988;119:289–301. doi: 10.1093/genetics/119.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moriel-Carretero M, Aguilera A. A postincision-deficient TFIIH causes replication fork breakage and uncovers alternative Rad51- or Pol32-mediated restart mechanisms. Mol Cell. 2010;37:690–701. doi: 10.1016/j.molcel.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 6.Imai J, Maruya M, Yashiroda H, Yahara I, Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003;22:3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bansal PK, Abdulle R, Kitagawa K. Sgt1 associates with Hsp90: An initial step of assembly of the core kinetochore complex. Mol Cell Biol. 2004;24:8069–8079. doi: 10.1128/MCB.24.18.8069-8079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- 9.Sekimoto T, et al. The molecular chaperone Hsp90 regulates accumulation of DNA polymerase eta at replication stalling sites in UV-irradiated cells. Mol Cell. 2010;37:79–89. doi: 10.1016/j.molcel.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Goldfless SJ, Morag AS, Belisle KA, Sutera VA, Jr, Lovett ST. DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol Cell. 2006;21:595–604. doi: 10.1016/j.molcel.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 11.Flom G, Weekes J, Johnson JL. Novel interaction of the Hsp90 chaperone machine with Ssl2, an essential DNA helicase in Saccharomyces cerevisiae. Curr Genet. 2005;47:368–380. doi: 10.1007/s00294-005-0580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valay JG, et al. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- 13.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cyr DM, Douglas MG. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- 15.Fan CY, Ren HY, Lee P, Caplan AJ, Cyr DM. The type I Hsp40 zinc finger-like region is required for Hsp70 to capture non-native polypeptides from Ydj1. J Biol Chem. 2005;280:695–702. doi: 10.1074/jbc.M410645200. [DOI] [PubMed] [Google Scholar]
- 16.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 17.Gong Y, et al. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: Implications to protein folding pathways in the cell. Mol Syst Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguilera A, Klein HL. Yeast intrachromosomal recombination: Long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics. 1989;123:683–694. doi: 10.1093/genetics/123.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang ME, Rio AG, Galibert MD, Galibert F. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase delta, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics. 2002;160:1409–1422. doi: 10.1093/genetics/160.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jimeno S, Rondón AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Floer M, Bryant GO, Ptashne M. HSP90/70 chaperones are required for rapid nucleosome removal upon induction of the GAL genes of yeast. Proc Natl Acad Sci USA. 2008;105:2975–2980. doi: 10.1073/pnas.0800053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rochette-Egly C, Adam S, Rossignol M, Egly JM, Chambon P. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 1997;90:97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 24.Keriel A, Stary A, Sarasin A, Rochette-Egly C, Egly JM. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell. 2002;109:125–135. doi: 10.1016/s0092-8674(02)00692-x. [DOI] [PubMed] [Google Scholar]
- 25.Liu JW, Picard D. Bioactive steroids as contaminants of the common carbon source galactose. FEMS Microbiol Lett. 1998;159:167–171. doi: 10.1111/j.1574-6968.1998.tb12856.x. [DOI] [PubMed] [Google Scholar]
- 26.Picard D, et al. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- 27.Johnson JL, Craig EA. A role for the Hsp40 Ydj1 in repression of basal steroid receptor activity in yeast. Mol Cell Biol. 2000;20:3027–3036. doi: 10.1128/mcb.20.9.3027-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cismowski MJ, Laff GM, Solomon MJ, Reed SI. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takagi Y, et al. Revised subunit structure of yeast transcription factor IIH (TFIIH) and reconciliation with human TFIIH. J Biol Chem. 2003;278:43897–43900. doi: 10.1074/jbc.C300417200. [DOI] [PubMed] [Google Scholar]
- 30.Yang C, Khaperskyy DA, Hou M, Ponticelli AS. Improved methods for expression and purification of Saccharomyces cerevisiae TFIIF and TFIIH; identification of a functional Escherichia coli promoter and internal translation initiation within the N-terminal coding region of the TFIIF TFG1 subunit. Protein Expr Purif. 2010;70:172–178. doi: 10.1016/j.pep.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranish JA, et al. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat Genet. 2004;36:707–713. doi: 10.1038/ng1385. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Qian X, Sha B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure. 2003;11:1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 33.Kota P, Summers DW, Ren HY, Cyr DM, Dokholyan NV. Identification of a consensus motif in substrates bound by a Type I Hsp40. Proc Natl Acad Sci USA. 2009;106:11073–11078. doi: 10.1073/pnas.0900746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coin F, et al. Phosphorylation of XPB helicase regulates TFIIH nucleotide excision repair activity. EMBO J. 2004;23:4835–4846. doi: 10.1038/sj.emboj.7600480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fregoso M, et al. DNA repair and transcriptional deficiencies caused by mutations in the Drosophila p52 subunit of TFIIH generate developmental defects and chromosome fragility. Mol Cell Biol. 2007;27:3640–3650. doi: 10.1128/MCB.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kainov DE, Vitorino M, Cavarelli J, Poterszman A, Egly JM. Structural basis for group A trichothiodystrophy. Nat Struct Mol Biol. 2008;15:980–984. doi: 10.1038/nsmb.1478. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y, Kou H, Wang Z. Tfb5 interacts with Tfb2 and facilitates nucleotide excision repair in yeast. Nucleic Acids Res. 2007;35:861–871. doi: 10.1093/nar/gkl1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coin F, et al. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet. 1998;20:184–188. doi: 10.1038/2491. [DOI] [PubMed] [Google Scholar]
- 39.Fribourg S, et al. Structural characterization of the cysteine-rich domain of TFIIH p44 subunit. J Biol Chem. 2000;275:31963–31971. doi: 10.1074/jbc.M004960200. [DOI] [PubMed] [Google Scholar]
- 40.Fribourg S, et al. Dissecting the interaction network of multiprotein complexes by pairwise coexpression of subunits in E. coli. J Mol Biol. 2001;306:363–373. doi: 10.1006/jmbi.2000.4376. [DOI] [PubMed] [Google Scholar]
- 41.Alevy YG, Brennan D, Durriya S, Howard T, Mohanakumar T. Increased expression of the HDJ-2 heat shock protein in biopsies of human rejected kidney. Transplantation. 1996;61:963–967. doi: 10.1097/00007890-199603270-00020. [DOI] [PubMed] [Google Scholar]
- 42.Caplan AJ, Douglas MG. Characterization of YDJ1: A yeast homologue of the bacterial dnaJ protein. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vergés E, Colomina N, Garí E, Gallego C, Aldea M. Cyclin Cln3 is retained at the ER and released by the J chaperone Ydj1 in late G1 to trigger cell cycle entry. Mol Cell. 2007;26:649–662. doi: 10.1016/j.molcel.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 44.Wang CC, et al. HDJ-2 as a target for radiosensitization of glioblastoma multiforme cells by the farnesyltransferase inhibitor R115777 and the role of the p53/p21 pathway. Cancer Res. 2006;66:6756–6762. doi: 10.1158/0008-5472.CAN-06-0185. [DOI] [PubMed] [Google Scholar]
- 45.Oh KS, et al. Phenotypic heterogeneity in the XPB DNA helicase gene (ERCC3): xeroderma pigmentosum without and with Cockayne syndrome. Hum Mutat. 2006;27:1092–1103. doi: 10.1002/humu.20392. [DOI] [PubMed] [Google Scholar]
- 46.Seroz T, Perez C, Bergmann E, Bradsher J, Egly JM. p44/SSL1, the regulatory subunit of the XPD/RAD3 helicase, plays a crucial role in the transcriptional activity of TFIIH. J Biol Chem. 2000;275:33260–33266. doi: 10.1074/jbc.M004764200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.