Abstract
The PI3K pathway is frequently activated in cancer; therefore, considerable effort is focused on identifying compounds that can inhibit specific pathway components, particularly the hallmark oncogene PIK3CA. Although targeted inhibition of a cancer survival gene holds significant promise, there are concerns that drug resistance may emerge within the cancerous cells, thus limiting clinical efficacy. Using genetically defined human mammary epithelial cells, we evolved resistance to the PI3K/mammalian target of rapamycin (mTOR) inhibitor BEZ235, and by genome-wide copy number analyses, we identified MYC and eIF4E amplification within the resistant cells. Importantly, either MYC or eukaryotic translation initiation factor 4E (eIF4E) was required to bypass pharmacological PI3K/mTOR inhibition in resistant cells. Furthermore, these cells displayed elevated 5′ cap-dependent protein translation. Collectively, these findings suggest that analysis of drivers of protein translation could facilitate the identification of cancer lesions that confer resistance to PI3K pathway-targeted drugs.
Various components of the PI3K pathway are frequently deregulated in human cancer through genetic alterations (point mutations and gene amplifications or deletions) or alternatively, epigenetic mechanisms [silencing of phosphatase and tensin homolog (PTEN) expression], resulting in constitutive pathway activation. Notably, PI3Kα is the only member of the PI3K pathway that bears frequent activating mutations in multiple major tumor types: breast, endometrial, ovarian, prostate, colorectal, pancreatic, liver, and lung cancers (1). The major oncogenic missense mutations in PI3K cluster in two separate regions of its p110α catalytic subunit—the helical (E542K and E545K) and the kinase (H1047R) domains—and both types yield constitutive lipid kinase activity (2, 3). In addition, the PIK3CA gene is amplified in a subset of head and neck, squamous cell lung, cervical, and gastric cancers (4).
Because of the high frequency of oncogenic activation of the PI3K pathway (1, 5–8), there is considerable interest in developing effective pharmacological inhibitors for cancer therapy. The expectation is that tumors bearing lesions along the PI3K pathway have acquired dependence on its activity and therefore, would exhibit augmented sensitivity to its inhibition, leading to growth arrest and/or induction of apoptosis. Currently, several PI3K inhibitors, including GDC0941 (Genentech) and BEZ235 (Novartis Pharmaceuticals), have entered phase I clinical trials, and in addition, isoform-specific compounds are being developed (9–11).
Despite the promise of targeted therapies, an emerging clinical obstacle is the acquisition of drug resistance within the tumor cells. To date, at least three distinct genetic mechanisms of resistance to kinase inhibitors have been described. First, the paradigm example for acquired drug resistance is the seminal observation that, during imatinib treatment of chronic myelogenous leukemia (CML), drug-resistant mutations arise in the drug target itself [breakpoint cluster region-Abelson leukemia homolog 1 (BCR-ABL)] (12, 13). Second, another documented mechanism of drug resistance derives from lesions in parallel pathways, which has been described for MET amplification leading to resistance to epidermal growth factor receptor (EGFR) inhibition by gefitinib and erlotinib in non-small cell lung carcinoma (NSCLC) (14, 15). Third, downstream lesions may occur in the same pathway, which has been described for trastuzumab resistance arising from PI3K pathway mutations (16, 17). Here, we explore acquired resistance to PI3K inhibitors and propose the prevailing mechanism of such resistance. Using genetically defined human mammary epithelial cells (HMECs), a model system that has previously been used for PI3K pathway-driven transformation because of its dependence on oncogenic PI3K signaling (18), we screened for emergence of BEZ235 resistance and identified genetic lesions involved.
Results
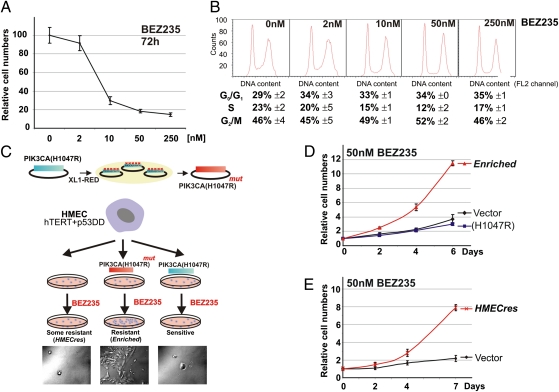
To develop a sensitive cell-based assay system to explore resistance to PI3K-targeted drugs, we chose to use immortalized HMECs, which had been modified to express both telomerase (hTERT) (19) and a synthetic dominant-negative p53 allele (p53DD) (20). Our overall goal was to explore whether mutations in the drug target itself or alternatively, acquisition of genetic alterations in downstream or parallel acting pathways, would yield drug resistance (12, 15, 21). Notably, these immortal HMECs are sensitive to PI3K inhibitors, as demonstrated by their response to treatment with the dual PI3K/mTOR inhibitor, BEZ235 (9), which significantly slows their growth and induces G1 cell cycle accumulation in a dose-dependent manner (Fig. 1 A and B).
Fig. 1.
Generation of BEZ235-resistant cells. (A) Growth curve of HMECs in the presence of BEZ235 (0–250 nM). (B) Cell cycle analysis by FACS counts of HMECs grown in indicated concentrations of BEZ235. (C) Schematic of the selection procedure used to generate BEZ235-resistant cells. (D) Growth curves of HMECs expressing vector control, PIK3CA(H1047R), and the Enriched cells in the presence of 50 nM BEZ235. (E) Growth curves of HMECs expressing vector control and the HMECres cells in the presence of 50 nM BEZ235. (Data are represented as mean ± SEM.)
Generation of BEZ235-Resistant Cells.
To investigate whether point mutations within the coding region of PIK3CA may drive resistance to BEZ235, we chose to bias our approach by generating a randomly mutagenized plasmid library of second site mutants in the background of oncogenic PIK3CA(H1047R). We accomplished this by passaging a PIK3CA retroviral expression vector [pBABE-puro-PIK3CA(H1047R)] through the mutator bacteria strain, XL1-Red (Stratagene) (22), and then introducing this library of mutant PIK3CA constructs into immortalized HMECs by retroviral transduction (Fig. 1C). Cultures of immortalized HMECs expressing mutagenized PIK3CA(H1047R), nonmutagenized PIK3CA(H1047R), and parental immortalized HMECs (HMEC/hTERT/p53DD) were exposed to increasing concentrations of BEZ235 in a stepwise fashion (Methods). Cells were maintained in the presence of BEZ235 for 2 mo until populations of proliferating cells emerged. In agreement with our expectations, this biased selection of HMECs expressing randomly mutagenized PIK3CA(H1047R) (Fig. 1C) produced a drug-resistant population of cells, which we will subsequently refer to as the Enriched population (BEZ235-resistant). Although PIK3CA(H1047R)-transformed cells failed to generate any cells resistant to BEZ235, rare cells originating from the parental HMECs emerged and could be propagated (referred to as HMECres). When these cultures were subsequently challenged with BEZ235, both the Enriched (Fig. 1D) and the HMECres (Fig. 1E) lines proliferated significantly faster than the parental (control) cells.
PIK3CA Point Mutations.
Because the Enriched BEZ235-resistant population arose from cells expressing the randomly mutagenized PIK3CA(H1047R) library, it would be consistent with previous observations (22) that the resistant phenotype of these cells could be explained by mutations within the ectopic PIK3CA(H1047R) alleles. To this end, we used genomic DNA from the Enriched cells to amplify the integrated proviral DNA using proofreading PCR. In parallel, we amplified the ectopic PIK3CA allele from the naïve cells (i.e., cells expressing the randomly mutagenized PIK3CA library before selection with BEZ235). These two samples were analyzed by SOLiD sequencing (Applied Biosystems) and differential sequence analysis (Fig. S1A). Approximately 40% of the PIK3CA cDNA molecules from the drug-resistant cells contained a point mutation (G959A) resulting in a G320E amino acid substitution. Another 20% contained a point mutation (T1457C) resulting in an F486S amino acid substitution. Importantly, the mutagenized preselection PIK3CA alleles did not show any mutational prevalence. SOLiD sequencing is based on parallel sequencing of short DNA fragments that are 50 nt long; thus, it was unclear whether the mutations detected were positioned in cis or in trans. To determine this, we cloned the 3.2-kb proviral PCR fragments (containing the ectopic PIK3CA) and sequenced a cohort of them by conventional Sanger sequencing. We detected G320E in one-half (14 of 28) of the sequenced PIK3CA molecules, whereas 10 of 28 harbored the F486S mutation juxtaposed with a 10-bp deletion (nucleotides 1,030–1,039) (Fig. S1B). One clone harbored both G320E and F486S in cis. All sequenced clones retained the original H1047R mutation.
Owing to the fact that no point mutations were found within the catalytic domain, which would be indicative of altered drug binding, it was not immediately clear by what mechanism the identified mutations might contribute to the observed BEZ235 resistance. The G320E mutation is in the linker preceding the C2 domain, whereas the F486S mutation is in the linker after the C2 domain and co-occurs with an additional 10-nt frame-shift deletion inside the C2 domain (Fig. S1 B and C). To assess these alleles’ inherent capacity to mediate BEZ235 resistance, we introduced these variants into the PIK3CA(H1047R) cDNA, transduced the parental HMECs with each allele individually (G320E or del;F486S), and measured their growth relative to controls in the presence of the drug (Fig. S1D). Using this approach, neither of the two variant alleles could recapitulate the drug resistance.
Although unable to yield BEZ235 resistance, we also examined these allele variants with regard to downstream signaling (Fig. S1E) and their inherent lipid kinase activity (Fig. S1F). Although overexpression of PIK3CA(G320E;H1047R) resulted in constitutive downstream signaling, measured as phospho-S473-AKT and heightened lipid kinase activity [similar to the signaling of the oncogenic PIK3CA(H1047R)], the PIK3CA(del;F486S;H1047R) allele did not induce significant AKT phosphorylation, and there was no lipid kinase activity detectable (Fig. S1 E and F). Because we could not detect the ectopic PIK3CA alleles within the Enriched cells by Western analysis and the PIK3CA(del;F486S;H1047R) allele is expected to yield a truncated product, we measured the RNA expression of the ectopic virally encoded PIK3CA and the antibiotic marker puromycin. Notably, we could not detect even nominal RNA levels of the ectopic alleles within the Enriched cells, although expression of puromycin could be verified, indicating that these allele variants had lost expression during selection for drug resistance (Fig. S1 G and H).
Integrated Copy Number and Expression Analysis Indicates MYC Amplification in BEZ235-Resistant Cells.
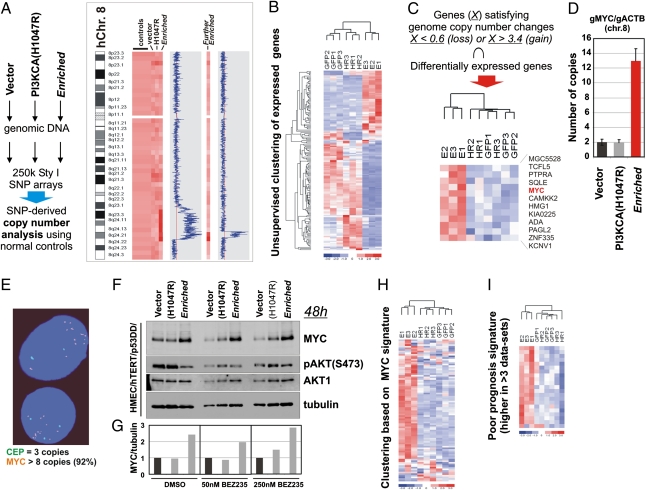
Because the identified PIK3CA point mutant alleles could not recapitulate the drug resistance phenotype (Fig. S1D), we hypothesized that alternative genetic changes might have occurred. Therefore, we used 250,000 genome-wide SNP-based copy number analysis to determine if any chromosomal gains or losses were discernable. Comparison of genomic DNA from the Enriched cells with the genomic DNA of the cells expressing vector or oncogenic PIK3CA(H1047R) revealed a discrete region of genomic amplification on chromosome 8q24, which became more focal upon additional selection in BEZ235 (Fig. 2A). We chose to use gene expression analysis to limit the number of amplified candidate genes mediating the observed drug resistance (Table S1). Using total RNA extracted from immortalized HMECs expressing vector (control), PIK3CA(H1047R), and the Enriched cells (in biological triplicates), which had been grown in the presence of 50 nM BEZ235 for 72 h, we performed a comparative gene expression microarray analysis. Cluster analysis revealed that triplicates of vector control and PIK3CA(H1047R) cells cluster closer together than Enriched cells (Fig. 2B), indicating that the latter population has a gene expression signature that is significantly different from its preselection counterparts. By reducing the analysis to genes contained within regions (X) of chromosomal gain (>3.4) or loss (<0.6) in the Enriched genome (Fig. 2A), we generated a narrow list of 12 genes, which are differentially expressed and could potentially be responsible for the observed BEZ235 resistance (Fig. 2C). This integrated analysis identified MYC, a well-known human oncogene present in the 8q24 amplicon. Notably, MYC is implicated as a downstream target of the PI3K/mTOR pathway (23) and frequently deregulated in breast cancer (24); thus, we considered it a likely candidate for mediating BEZ235 resistance. We verified that MYC was amplified in the Enriched cells by measuring MYC genomic copy numbers by quantitative real-time PCR (QPCR) relative to controls (Fig. 2D). In addition, FISH confirmed MYC amplification in the Enriched cells (Fig. 2E). Furthermore, we could detect a twofold increase in MYC protein levels in the Enriched cells relative to the controls, independent of BEZ235 treatment (Fig. 2 F and G).
Fig. 2.
Genome-wide copy number and expression analyses of BEZ235-resistant cells. (A) SNP array analysis of HMECs expressing vector control (GFP), PIK3CA(H1047R) (HR), or the Enriched (E) cells indicating a region of amplification at chromosome 8 present in the BEZ235-resistant Enriched cells that becomes narrowed on additional drug selection (Further Enriched). (B, C, H, and I) Expression array analysis of triplicate samples of HMECs expressing vector control (GFP), PIK3CA(H1047R) (HR), or the Enriched (E) cells. (B) Unsupervised clustering of expressed genes. (C) Differential expression of genes within the amplified regions in A. (D) Genomic QPCR analysis of the lines in A confirming MYC amplification. (E) FISH analysis of the Enriched cells using the genomic probe for MYC (orange) and centromeric 8 control probe (green). (F) Western analysis in the absence or presence of BEZ235 (50 and 250 nM) of the cell lines in A. (G) Quantification of MYC protein levels normalized to tubulin from the Western assay in F. (H) Clustering based on published MYC signature (25). (I) Clustering based on the published poor prognosis signature (26).
Enrichment of a MYC Signature in the BEZ235-Resistant Cells.
If MYC amplification were indeed the genetic event that mediated the observed BEZ235 resistance, it would be expected to be traceable by changes in expression of MYC-regulated genes. To test the hypothesis that MYC may be the driver of resistance to BEZ235 in our system, we applied a previously derived MYC signature (25), which we curated for higher stringency to contain only those probe sets (genes) that are up-regulated. When this signature was applied to our dataset, the Enriched cells exhibited a close relationship with the MYC signature, whereas the controls [vector and PIK3CA(H1047R)] did not (Fig. 2H). In addition, it was recently reported that induction of MYC-regulated genes is associated with poor outcome in human cancers (26). Comparison of the Enriched profile with the most common poor prognosis genes (curated to contain probe sets that were associated with higher expression in at least four datasets) revealed that the Enriched cells display this signature (Fig. 2I). Collectively, the expression array analysis suggested that the Enriched cells exhibit expression patterns resembling those patterns of human tumors with deregulated MYC.
MYC Overexpression Is Sufficient to Confer Resistance to PI3K and mTOR Inhibition.
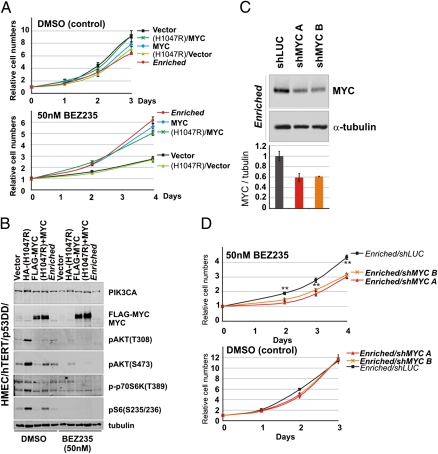
We next wanted to test whether overexpression of MYC could functionally recapitulate BEZ235 resistance. Immortalized HMECs transduced to express MYC were exposed to BEZ235 in growth assays (Fig. 3A) and found to generate a BEZ235 resistance phenotype similar to the resistance of the Enriched cells. MYC overexpression on its own seems to be dominant in rendering resistance to BEZ235, because we did not observe a synergistic effect of MYC overexpression in combination with PIK3CA(H1047R). Importantly, overexpression of Cyclin D1, a documented downstream effector of MYC (27), or overexpression of a suggested gatekeeper mutant PIK3CA allele (I848V) (28) did not parallel the resistance conferred by MYC (Fig. S2). Notably, introduction of ectopic MYC in HMECs expressing oncogenic PIK3CA(H1047R) suppressed the expression of the ectopic PIK3CA allele and resulted in reduced downstream PI3K signaling measured as pS473-AKT (Fig. 3B), and thus, it may explain why the Enriched cells evolved to attenuate expression of ectopic PIK3CA.
Fig. 3.
MYC is sufficient and necessary for BEZ235 resistance. (A) Growth curves in the absence (DMSO) or presence of 50 nM BEZ235 for HMECs expressing vector control, PIK3CA(H1047R), MYC, PIK3CA(H1047R)+MYC, or the Enriched cells. (B) Western analysis of the cell lines in A grown in the absence or presence of 50 nM BEZ235. (C) Western assay of Enriched cells with shLUC, shMYC A, or shMYC B accompanied by quantification of MYC levels normalized to tubulin. (D) Growth curves of the cell lines from C in the absence or presence of 50 nM BEZ235 (**P < 0.005; Student two-tailed t test for paired samples). (Data are represented as mean ± SEM.)
Having established that MYC overexpression is sufficient to recreate the BEZ235 resistance observed in the Enriched cells, we wanted to examine if MYC was functionally necessary. Therefore, we used lentiviral shRNA knockdown of MYC in the drug-resistant cells (Fig. 3C). Notably, we achieved ∼40% MYC protein depletion in the Enriched cells, which taking into account the twofold increase caused by MYC amplification, effectively reverted MYC expression to nominal endogenous levels. When grown in the presence of BEZ235, Enriched cells expressing shRNA against MYC showed a decreased drug resistance phenotype relative to the shRNA against luciferase (shLUC) -infected control (Fig. 3D).
MYC Confers Resistance to Pan-PI3K and TORC1/2 Inhibition.
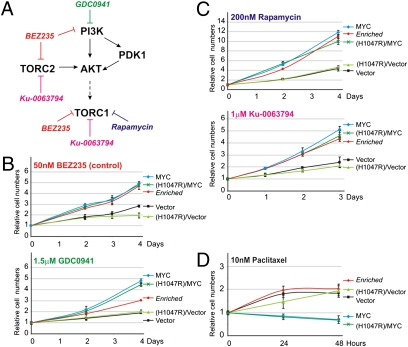
To determine if the drug resistance phenotype resulting from MYC overexpression and amplification was specific for BEZ235, we tested response of these cells to other kinase inhibitors that share targets with BEZ235–PI3K and/or mTOR (Fig. 4A). First, we examined if MYC could confer resistance to GDC0941 (29), a pan-PI3K inhibitor that does not inhibit mTOR (Fig. 4B), and found that, indeed, MYC overexpression results in resistance to GDC0941. Because BEZ235 is a dual PI3K/mTOR inhibitor, we also tested if MYC renders cells insensitive to TORC1 inhibition by Rapamycin as well as the catalytic inhibitor of mTOR (TORC1 and TORC2) and Ku-0063795 (30) (Fig. 4C). As was seen for dual PI3K/mTOR inhibition by BEZ235, we found that MYC overexpression was sufficient to render resistance to both Rapamycin and Ku-0063795. In addition to targeted inhibitors, we analyzed whether MYC overexpression may affect general drug resistance. MYC overexpression had no effect on cellular sensitivity to Doxorubicin (Fig. S3) but rendered cells even more sensitive to Paclitaxel (Fig. 4D), which are two classical chemotherapeutic agents.
Fig. 4.
MYC confers resistance to pharmacological PI3K or mTOR inhibitors. (A) Schematic representation of the PI3K-mTOR pathway with the inclusion of kinase inhibitors used in this study. (B–D) Growth curves of HMECs expressing empty vector, PIK3CA(H1047R), MYC, PIK3CA(H1047R)+MYC, or the Enriched cells grown in (B) 50 nM BEZ235 (control) or 1.5 μM GDC0941, (C) 200 nM rapamycin or 1 μM Ku-0063794, or (D) 10 nM Paclitaxel. (Data are represented as mean ± SEM.)
MYC Overexpression Renders Breast Cancer Cell Lines Resistant to BEZ235.
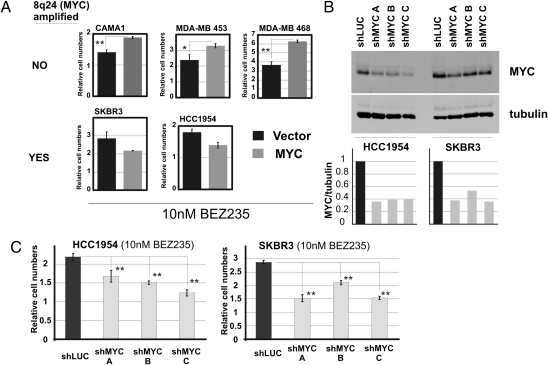
We next explored if manipulation of MYC levels could affect the response to BEZ235 in established human breast cancer cell lines. To this end, we used five breast cancer cell lines, two of which (SKBR3 and HCC1954) bear amplification of the 8q24 genomic region (Cosmic; Sanger Wellcome Trust). The remaining three lines, CAMA-1, MDA-MB-453, and, MDA-MB-468, are diploid over the 8q24 region (Cosmic; Sanger Wellcome Trust) (Fig. 5A). Notably, overexpression of MYC in the lines without 8q24 amplification resulted in increased resistance to BEZ235. However, overexpression of MYC in lines where MYC was already amplified did not result in additional drug resistance. We next tested if MYC is necessary for growth in the presence of BEZ235 in the MYC genomic region-amplified cell lines SKBR3 and HCC1954 (Fig. 5B). Knockdown of MYC in both of these cell lines resulted in the impaired growth in the presence of BEZ235 relative to the controls (shLUC) (Fig. 5C).
Fig. 5.
MYC confers resistance to BEZ235 in breast cancer cell lines without MYC amplification. (A) Expression of vector control vs. MYC in breast cancer cell lines without (CAMA1, MDA-MB-453, and MDA-MB-468) or with (SKBR3 and HCC1954) 8q24 genomic amplification grown in 10 nM BEZ235. (B) Knockdown of MYC in HCC1954 and SKBR3. Western blot assay showing protein levels and quantification of MYC protein levels normalized to tubulin. (C) Growth in the presence of 10 nM BEZ235 of the cell lines in B (*P < 0.02, **P < 0.005; Student two-tailed t test for paired samples). (Data are represented as mean ± SEM.)
Amplification of eIF4E Can Mediate Resistance to BEZ235.
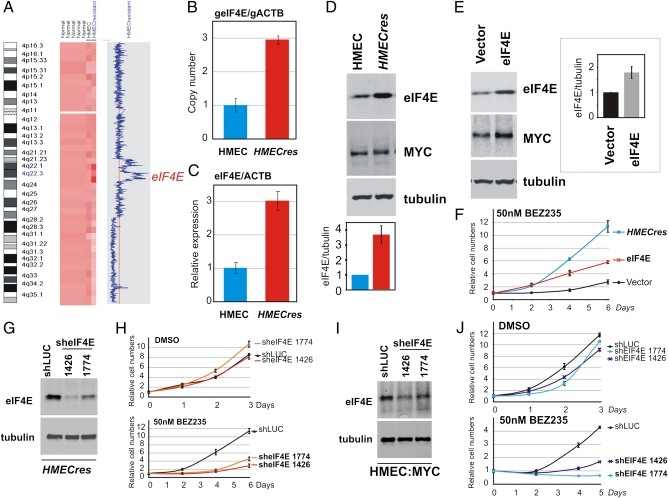
Upon extended selection in BEZ235, we propagated an additional seemingly drug-resistant population of cells, referred to as HMECres, that originated from the original immortal HMEC cells. Compared with the Enriched population, HMECres emerged 1 mo later during the course of the selection procedure. Notably, when assayed, these cells proliferate significantly faster in the presence of BEZ235 relative to the parental cells in a manner paralleling the Enriched cells (Fig. 1E). Importantly, these cells retain their resistance phenotype upon drug recovery (i.e., prolonged propagation in the absence of inhibitor). Hence, the nature of their acquired BEZ235 resistance would likely be genetically mediated. Thus, we used genome-wide copy number analysis to investigate if any detectable chromosomal changes were identifiable. A comparison of the genomic DNA from the HMECres BEZ235-resistant cells to the genomic DNA of the parental HMEC line revealed a region of genomic amplification centered around chromosome position 4q22 (Fig. 6A). Among the genes contained within the chromosome 4 amplicon (Table S2), the eukaryotic translation initiation factor 4E, eIF4E, seemed to be a likely candidate mediating resistance to PI3K/mTOR inhibition because of its downstream position in translation control relative to the PI3K-mTOR axis (31). Interestingly, it also has a well-documented role as a MYC-regulated gene (32, 33). Genomic QPCR analysis of HMECres cells confirmed eIF4E amplification in these cells, with an indicated four-copy gain of this gene (threefold increase) (Fig. 6B). Concordantly, real-time mRNA quantification revealed a threefold increase in eIF4E mRNA levels in HMECres relative to HMEC control, hence paralleling the copy number increase (Fig. 6C). Similarly, quantitative Western blot analysis also confirmed these findings, showing a 3- to 4.5-fold increase in eIF4E protein levels in the HMECres cells relative to control (Fig. 6D).
Fig. 6.
eIF4E is amplified and overexpressed in HMECres and confers resistance to BEZ235. (A) SNP array analysis of HMECres vs. parental HMECs indicating a region of amplification at chromosome 4 present in the BEZ235-resistant HMECres. (B) Genomic QPCR confirming eIF4E amplification in HMECres. (C) QPCR analysis comparing eIF4E mRNA expression levels normalized to ACTB in HMECres with parental HMEC. (D) Western blot assay confirming eIF4E overexpression in HMECres. (E) Western blot analysis of HMECs transduced with vector control or eIF4E. (F) Growth assay in the presence of 50 nM BEZ235 comparing HMECres with HMECs transduced with vector control or eIF4E. (G) Western analysis of HMECs transduced with shLUC (control) or two sheIF4E constructs (1426 and 1774). (H) Growth curves of the cell lines in G in the absence or presence of 50 nM BEZ235. (I) Western analysis of HMECs overexpressing MYC transduced with shLUC (control) or two sheIF4E constructs (1426 and 1774). (J) Growth curves of the cell lines in I in the absence or presence of 50 nM BEZ235. (Data are represented as mean ± SEM.)
To determine whether eIF4E amplification could be a causal event mediating BEZ235 resistance in HMECres cells, we overexpressed eIF4E in the parental HMECs by retroviral transduction (Fig. 6E). We were able to obtain only a twofold increase in eIF4E protein levels by ectopic expression, an amount lower than yielded by amplification in HMECres (3- to 4.5-fold) (Fig. 6D). Nevertheless, we proceeded to analyze whether eIF4E overexpression could yield resistance to BEZ235 by measuring growth in the presence of the drug. Indeed, a twofold increase in protein conferred resistance to BEZ235, although notably less pronounced compared with HMECres cells that bear higher eIF4E protein levels (Fig. 6F). Furthermore, eIF4E overexpression also improves growth in the presence of the PI3K inhibitor GDC0941 and mTOR inhibitor Ku-0063794 (Fig. S4A). To underscore the capacity of eIF4E and MYC to drive resistance to PI3K/mTOR inhibitors, we also increased the dosage of these inhibitors and could determine maintained ability to evade drug inhibition of growth (Fig. S4B). As expected, increased BEZ235 dosage resulted in potentiated suppression of downstream pathway signaling measured as phosphorylation of AKT and p70S6K (Fig. S4C).
In addition, using a loss of function approach, we investigated if eIF4E is necessary for BEZ235 resistance in HMECres cells. Two short-hairpin eIF4E-targeting constructs (1426 and 1774) resulted in significant reduction of eIF4E protein levels relative to control (shLUC) (Fig. 6G). Although neither of the shRNA constructs affected normal cellular growth in the absence of the drug, the two that conferred significant protein-level reduction also revealed an increased sensitivity to BEZ235 relative to control (Fig. 6H). Collectively, these findings suggest that eIF4E amplification, resulting in increased eIF4E protein levels, may be an important factor mediating resistance to BEZ235.
We also investigated whether eIF4E expression is required for MYC-mediated BEZ235 resistance, because it has been suggested to be a downstream target of MYC function (32, 33). Using the two verified shRNA constructs against eIF4E (Fig. 6G), we could show reduced eIF4E protein levels in MYC-transduced cells (Fig. 6I). When grown in the presence of BEZ235, MYC-expressing cells with reduced eIF4E expression displayed increased sensitivity to BEZ235 relative to the shLUC control (Fig. 6J). Notably, this finding suggests that eIF4E expression is a required downstream factor enabling MYC-mediated resistance to PI3K/mTOR inhibition. Furthermore, we examined whether expression of canonical PI3K-mTOR pathway components was affected in drug-resistant cell lines (Fig. S4D). We could not detect any significant variations in the expressed amounts of these proteins, indicating that the effects that we document are likely driven by downstream lesions.
MYC- and eIF4E-Driven Resistance Is Indicative of Up-Regulated Cap-Dependent Translation.
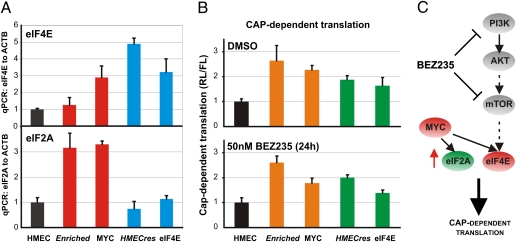
Because eIF4E is a known canonical effector of the PI3K/mTOR pathway and a critical translation initiation factor that regulates 5′ cap-dependent translation (32–34), we postulated that both eIF4E and MYC may up-regulate cap-dependent translation, which would otherwise be down-regulated by the dual inhibition of PI3K and mTOR. To test this hypothesis, we investigated if MYC affects eIF4E expression in our system. As shown in Fig. 7A, overexpression of MYC increases cellular levels of eIF4E. In addition, when overexpressed, MYC up-regulates translation initiation factor eIF2A RNA levels (34, 35), which is also observed in the Enriched cells (Fig. 7A). Given the increased expression of translation initiation factors in the BEZ235-resistant cells, we examined whether cap-dependent translation may be generally altered. Using a bicistronic luciferase reporter construct pRL-HCV-FL (36), we assessed the relative ratios of cap-dependent vs. -independent [internal ribosome entry site (IRES)-dependent] translation in the BEZ235-resistant cells. Both in the absence and presence of BEZ235, our derived resistant cell lines, as well as the MYC- and eIF4E-overexpressing cells, exhibit elevated levels of cap-dependent translation relative to control cells (Fig. 7B). Consequently, this finding suggests that both MYC- and eIF4E-mediated BEZ235 resistance display common effects leading to an increase in 5′ cap-dependent translation, a convergence point with the PI3K-mTOR pathway.
Fig. 7.
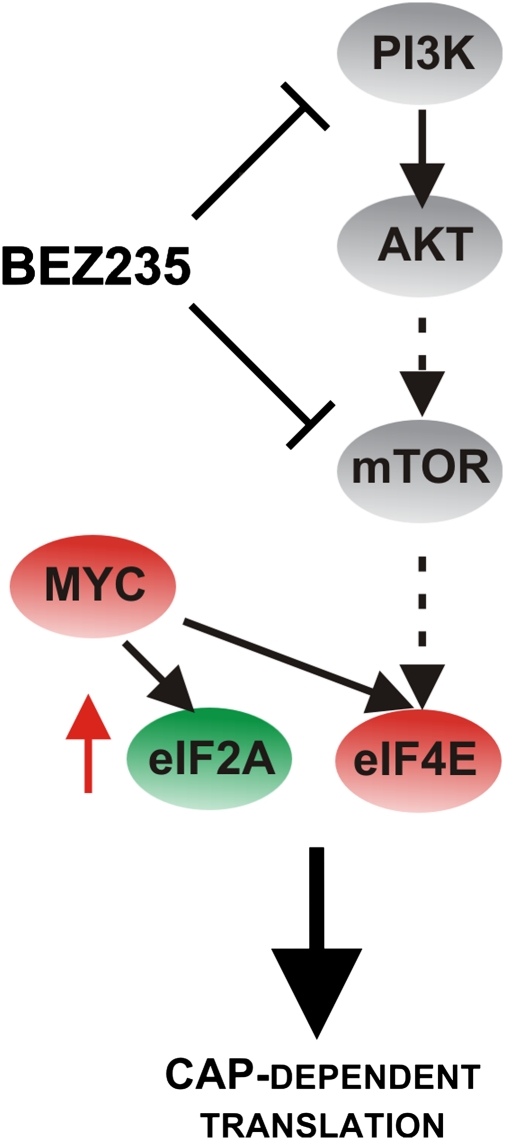
MYC and eIF4E overexpression up-regulate cap-dependent translation. (A) QPCR analysis comparing eIF4E and eIF2A mRNA expression levels in parental HMECs or HMECs expressing MYC or eIF4E as well as the Enriched and HMECres cells. (B) Relative levels of cap-dependent translation. Cell lines from A transfected with the bicistronic luciferase reporter pRL-5′-IRES-FL are grown in the absence or presence of 50 nM BEZ235 and assayed 24 h posttransfection; ratios of RL to FL are represented and normalized to HMEC control, indicating changes in cap-dependent translation. (C) Schematic model of our results indicating that both MYC and eIF4E mediate resistance to BEZ235 through up-regulation of cap-dependent translation. (Data are represented as mean ± SEM.)
Discussion
Because many tumor types exhibit survival and/or growth dependence on a particular mutationally activated gene, commonly a kinase, through a process termed oncogene addiction, the principle of targeted kinase inhibition has provided clinical success in treating diverse cancer types (37–41). However, a major concern hampering the sustained clinical benefits of targeted therapies is the observed emergence of acquired drug resistance. As a result of inhibitor specificity as well as the inherent capacity of cancer cells to select for growth-promoting genetic variants, several resistance mechanisms have been documented involving either secondary mutations in the drug target gene (12, 13, 21) or alternatively, activation of parallel-acting or downstream pathway components (14). Here, we report that genomic amplification of either of two protooncogenes, MYC and eIF4E, is able to provide resistance to PI3K/mTOR pathway inhibitors, apparently bypassing the inhibitors by acting downstream of the pharmacologically inhibited targets.
Notably, our initial expectation was that mutations in the oncogenic PI3K, possibly in the kinase domain (inhibitor binding), would yield resistance to catalytic PI3K inhibition. It is possible that we could not achieve this mode of resistance with our approach, because the inhibitor that we chose for analysis (BEZ235) has since been found to exhibit dual specificity to both PI3K and mTOR. Although we biased our selection process to second site mutations within the PIK3CA gene, the emerging variant alleles in our Enriched population did not recapitulate the resistance to BEZ235 (Fig. S1). One intriguing possibility is that MYC seems to suppress the expression of ectopic PIK3CA alleles in our system (Fig. 3B), which may have resulted in selection of rare cells with preexisting MYC amplification during growth in the presence of the drug. This result would also consolidate the presence of two different acting variant alleles identified, G320E;H1047R and F486S:del;H1047R, exhibiting oncogenic and null lipid kinase activity, respectively (Fig. S1F).
After we eliminated the possibility that the selected PIK3CA alleles were contributing to the observed resistance (Fig. S1 G and H), we opted to search for genomic alterations in the host cells, which might be responsible for the observed acquired resistance. Our data firmly suggest that two independent selection processes have identified two oncogenes already closely tied to a common pathway. Using an integrated copy number and expression-based approach within the Enriched cells, we determined that MYC, a commonly deregulated breast cancer oncogene, was responsible for the acquired BEZ235 resistance that we had evolved (Fig. 3D). It was striking to us that the Enriched cells exhibit a gene expression signature like the one reported by Bild et al. (25) that was obtained by overexpression of MYC in HMECs (Fig. 2H). It was, therefore, not surprising that overexpression of MYC in immortal HMECs could confer BEZ235 resistance (Fig. 3A), indicating that MYC is not only necessary but also sufficient to drive this drug-resistant state. In addition, an independent selection of immortalized HMECs using BEZ235 resulted in the emergence of a second drug-resistant cell line, HMECres, containing a discrete region of genomic amplification on chromosome 4 (Fig. 6A). From among the genes in this region, we selected eIF4E as a likely BEZ235 resistance candidate gene, because eIF4E is already a known downstream effector of the PI3K-mTOR pathway (31). Moreover, eIF4E lies downstream of and is regulated by MYC (32–34). In addition, eIF4E expression is indeed elevated in MYC overexpressing HMECs (Fig. 7A), and this expression seems to be required for resistance to BEZ235 (Fig. 6J). Notably, MYC and eIF4E have been shown to collaborate to transform cells in vitro (42) and in vivo (43). However, we could not detect elevated expression of eIF4E in the Enriched cells, which may be because of lower MYC levels than in the ectopically expressing MYC cells (Fig. S4C).
It is intriguing how amplification of MYC and eIF4E might contribute to the observed PI3K/mTOR inhibitor resistance. By far, the simplest explanation of the MYC-eIF4E resistance axis is that, under the growth conditions used for selection, the critical element downstream of PI3K/mTOR signaling is initiation of 5′ cap-dependent translation (Fig. 7). Certainly, a great deal of literature shows the conserved role of MYC in driving cell growth and the requirement for eIF4E in controlling growth (44, 45). However, the PI3K pathway promotes the activity of a large number of proteins involved in driving key cellular processes. It is, therefore, somewhat surprising that acquired resistance to PI3K/mTOR pathway inhibitors provoked selection and elevation of the 5′ cap-dependent translation machinery. By allowing for cap-dependent translation to proceed despite upstream pathway blockage by BEZ235 (for example, by overexpressing eIF4E), mitogenic proteins with structure-rich 5′ UTRs like Cyclin D1 (46) and ornithine decarboxylase (47) can be produced. Forced expression of Cyclin D1 alone could not recapitulate resistance in our system (Fig. S2), arguing again for the global effects on the translation machinery that MYC and eIF4E exert downstream of the PI3K-mTOR pathway. If our selection had been done in a cell line that undergoes apoptosis in response to pharmacological PI3K inhibition, it is quite possible that the resistance genes identified would have been different, because it would not strictly involve growth promoting functions but rather, cell survival.
Because PI3K inhibitors have only recently entered phase I/II clinical trials, resistance to this class of targeted therapies has not yet been studied in clinical materials. However, it is of crucial importance to predict possible mechanisms of resistance to these inhibitors, especially if such mechanisms may preexist within the tumors, because this information will help guide clinicians in selecting patients who may benefit maximally from this class of therapy. In addition, identification of rare drug-resistant clones from a pretreatment specimen may help in targeting resistance before it is selected for by combining PI3K inhibition with inhibition of the lesion that leads to resistance. Since both MYC and eIF4E amplification and overexpression are found in human tumor cells (24, 48) and may coexist with mutations along the PI3K pathway, it may be advantageous to assess MYC and eIF4E levels before considering PI3K inhibitors as a therapeutic strategy and also, monitor changes during therapy, as our data implicate these changes as drivers of resistance. Furthermore, considering that both of these lesions seem to converge on up-regulation of cap-dependent translation, it is plausible that alternate genetic events that positively regulate the same functional cascade may also mediate resistance to PI3K/mTOR inhibitors.
Methods
Mammalian Cell Culture.
Immortal HMECs expressing hTERT and p53DD (previously described in refs. 18 and 49) were cultured in DMEM/F-12 (1:1) with 0.5% FBS, hEGF (10 ng/mL), insulin (10 μg/mL), hydrocortisone (0.5 μg/mL), and cholera toxin (1 μg/mL). PIK3CA(H1047R) transductions were performed as described earlier (50). HMECs were treated with the drugs NVP-BEZ235 (Novartis), GDC0941 (Selleck Chemicals), Rapamycin (Cell Signaling), Ku-0063794 (Chemdea), Paclitaxel (Sigma), and Doxorubicin (Sigma) at the indicated doses and indicated time intervals. At various time points, cells grown on tissue culture dishes in the presence of drugs were fixed with 10% ethanol/10% acetic acid, subsequently stained with 0.4% crystal violet in 20% ethanol, and washed; after drying, the dye was extracted with 10% acetic acid. The absorbance, corresponding to cell numbers attached on the dishes, was measured at 595 nm on a multiwell plate reader (Bio-Rad).
Expression and shRNA Vectors.
The pWZL-blast-FLAG-MYC retroviral construct was generated by cloning an FLAG-MYC cDNA from pCMV5-FLAG-MYC (Michael Cole, Dartmouth University, Hanover, NH) into the pWZL-blast backbone by cohesive EcoRI digests. The pBABE-puro-eIF4E retroviral construct was generated by excising an eIF4E cDNA from the pHA-eIF4E construct (Addgene plasmid 17343 from Dong-Er Zhang, University of California at San Diego, La Jolla, CA), blunting the ends with Klenow polymerase, and ligating into the pBABE-puro backbone cut with SnaBI. The retroviral construct pBABE-puro-HA-PIK3CA(H1047R) has been previously described (18). The shRNA constructs targeting MYC were obtained from the Broad Institute RNAi Consortium as pLKO.puro-shMYC vectors. Three reproducible and efficient shMYC target sequences (shMYC A: CAGTTGAAACACAAACTTGAA, shMYC B: CAGGAACTATGACCTCGACTA, and shMYC C: GAACTATGACCTCGACTACGA) were selected, and the pLKO.puro-shMYC constructs were modified by replacing puromycin with blasticidin. The shRNA lentiviral constructs targeting eIF4E were purchased from Sigma-Aldrich, and two constructs were validated for efficient and reproducible knockdown (sheIF4E 1426, clone ID: NM_001968.2–1426s1c1; sheIF4E 1774, clone ID: NM_001968.2–1774s1c1).
Random Mutagenesis and Generation of BEZ235-Resistant Cells.
XL1-RED bacteria (Stratagene), which induce random mutations into DNA, were transformed with the pBABE puro-HA-PIK3CA(H1047R) retroviral construct. Plasmid DNA was purified, and the resulting library of mutated PIK3CA constructs was retrovirally transduced into HMECs. HMECs expressing the mutagenized PIK3CA library were exposed to increasing concentrations of BEZ235. BEZ235 concentrations were increased stepwise from 10 to 250 nM for 2 mo total, at which point the cells acquired growth rates similar to the untreated control cells. The cells were allowed to grow in the absence of the drug for 2 wk after the drug selection before being tested for resistance. To confirm the phenotype of resistant cells (Enriched cells), crystal violet growth assays were performed at different drug concentrations. The Enriched cells were passaged in the absence of drug for 6 mo and still maintained BEZ235 resistance, which was confirmed by crystal violet growth assays.
Sequencing.
Retrovirally integrated mutagenized ectopic PIK3CA cDNA was PCR-amplified from the naive cells (before BEZ235 selection) and the Enriched BEZ235-resistant cells. The primers used for PCR amplification spanned the vector backbone–cDNA boundary and were FWD-TCCAAGCTTCACCATGGGGTACCCAT and REV-ATGCATTGAACTGAGTCGACCAGCTGTGG. Each PCR product was processed to generate a fragment library according the manufacturer's protocol (Applied Biosystems). The library was then sequenced on an SOLiD version 3 Plus instrument to generate sequences 50 bases long from one end of each fragment. Resulting sequence data were aligned with the PIK3CA cDNA sequence using Corona Lite software (Applied Biosystems). In addition, PCR fragments were cloned by pCR2.1-TOPO-TA (Invitrogen), and clones with inserts of expected size were analyzed by standard automated Sanger sequencing.
Cell Cycle Analysis.
Cells were harvested by trypsin digestion at 48 h postdrug addition and fixed/permeabilized in 35% ethanol; after RNaseH treatment and propidium iodide staining, the samples were subsequently analyzed on a BD FACS Canto flow cytometer for DNA content. Experiments were conducted in triplicates.
Western Assays.
Immunoblotting was performed using standard procedures. The commercial antibodies used were HA (clone 6E2), p110α, pAKT(S473), pAKT(T308), p-p70S6K (T389), pS6(S234/236), c-MYC (clone D84C12), AKT1, and pMAPK(Y42/44) from Cell Signaling and α-tubulin (clone DM1A) from Sigma.
Lipid Kinase Assay.
Lipid kinase assays were performed as previously described (51). In brief, PI3K was purified from cells by nondenaturing immunoprecipitation and preincubated at room temperature with a lipid mix containing sonicated lipids phosphatidylserine (2 μg/μL; Avanti) and phosphatidylinositol (4 μg/μL; Avanti). Subsequently, ATP mix was added to the reaction [10 μCi 32P-γ-ATP, ATP (0.5 mM), MgCl2 (100 mM), 67 mM Hepes, pH 7.4]. After 10 min, reactions were stopped with 4 N HCl. After extraction, lipids from the chloroform (organic) layer were spotted onto the TLC plate and separated overnight by TLC in n-propanol:2 M acetic acid (65:35). Results were visualized using a Typhoon phosphoimager, and image acquisition and analysis were performed with the accompanying software.
Microarray Analyses.
Genomic DNA (DNeasy; Qiagen) and total RNA (RNeasy Plus; Qiagen) were extracted from logarithmically growing cultures of the indicated genotypes of HMECs. Genomic DNA was processed for interrogation on StyI 250k SNP arrays (Affymetrix) according to the manufacturer's recommendations. SNP-derived copy number analysis was performed using dChip (52) using normal controls set to diploid for standardization of calls and signal intensities. Qualifying genomic alterations were determined as regions X (containing more than 4 SNPs) outside of 0.6 > X or X > 3.5. Expression array analysis was conducted using Affymetrix HG-U133AV2.0 arrays (in triplicate) using RNA extracted from samples grown in the presence of BEZ235 for 72 h before harvesting. Resulting Affymetrix Cell Intensity (.CEL) files were normalized in intensity, and signal/calls were computed using dChip. Expression analysis was conducted on filtered variant probe sets (genes) that satisfy variation among samples: 0.5 < SD, mean < 1,000, and present calls in 33% of samples. Clustering was done by calculating average Euclidean distance as a metric. The data have been deposited to the Gene Expression Omnibus under accession number GSE25173.
QPCR.
Expression levels of eIF4E and eIF2A mRNA were determined by QPCR using the amplicons eIF4E (FWD: CAATCCGGTTTGAATCTCAT and REV: AGTCCACTCTGCTTTTTGAAGA) and eIF2A (FWD: CTGGACCTCATGCAGCTTTAGC and REV: CTCCATAGTAGGAAGCTCCTGTC) and normalized to ACTB (FWD: ATTGCCGACAGGATGCAGAA and REV: GCTGATCCACATCTGCTGGAA) using SYBR chemistry on an ABI StepONE plus instrument (94 °C, 2 min; 40 cycles: 94 °C, 20 s; 51 °C, 20 s).
Genomic QPCR.
MYC amplification levels were confirmed by real-time genomic PCR using the primers qgMYC_FWD: CACCAGGCTTAGATGTGGCTCTTT and qgMYC_REV: CTTCCTCATCTTCTTGTTCCTCCTC and qgeIF4E_FWD: CTAGGAAACCACCCCTACTCCTA and qgeIF4E_REV: TCCAAGTGAAAATAGAACCCTCA compared with qgACTB_FWD: CTCCATCATGAAGTGTGACGTGGA and qgACTB_REV: CAGGAAAGACACCCACCTTGATCT in the sample measured relative to control normal DNA set to diploid (gene copy number ≡ 2) using an Applied Biosystems 7300 with SYBR green chemistry and ROX as a passive reference.
Luciferase Reporter Assays.
HMEC cells were transiently transfected with a bicistronic luciferase reporter plasmid, pRL-5′-IRES-FL (36), using Polyfect (Qiagen). The reporter plasmid directs cap-dependent translation of the Renilla luciferase and hepatitis C virus (HCV) IRES-dependent translation of the firefly luciferase gene. Twenty-four hours posttransfection, dual luminescence was measured according to the manufacturer's instructions (Promega) using a Veritas Microplate Luminometer. Assays were performed in quadruplicate from lysed cells grown in the presence of 50 nM BEZ235 or vehicle (DMSO), and results were expressed as average ± SEM and normalized to controls.
FISH.
Growing cultures of cells were treated with colcemid, harvested, and treated with hypotonic buffer before fixation onto slides. FISH analysis was performed using the CEP8 reference probe (green fluorescence) and the human c-Myc probe (orange fluorescence) to assess the c-Myc copy numbers. Fluorescent probes were generated by standard nick translation DNA labeling and hybridized to interphase cells followed by fluorescent microscopy analysis and statistical counting; 100 nuclei were scored for each cell line.
Supplementary Material
Acknowledgments
The authors thank Dr. Edward Fox and the Dana–Farber Cancer Institute Microarray Core for excellent assistance with microarray studies and SOLiD sequencing, Dr. Julien Senechal for FACS operation, and Dr. Malcolm Whitman for helpful advice regarding lipid kinase assays. We are grateful to Drs. Daniel Silver and Linda Clayton for critical comments on the manuscript. We thank the Cytogenetics Core of Dana–Farber Harvard Cancer Center (P30 CA006516) for performing FISH analysis. This work was supported by Department of Defense Breast Cancer Research Program Predoctoral Fellowship W81XWH-08-1-0747 (to N.I.); National Institutes of Health Grants CA089393 (to T.M.R.), CA030002 (to T.M.R.), and CA89021 (to T.M.R.); and Stand Up to Cancer (to T.M.R.).
Footnotes
Conflict of interest statement: In compliance with Harvard Medical School guidelines, we disclose the consulting relationships: Novartis Pharmaceuticals, Inc. (T.M.R.).
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE25173).
This article is a PNAS Direct Submission.
See Author Summary on page 15027.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108237108/-/DCSupplemental.
References
- 1.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: Variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samuels Y, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 4.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 5.Sauter G, Maeda T, Waldman FM, Davis RL, Feuerstein BG. Patterns of epidermal growth factor receptor amplification in malignant gliomas. Am J Pathol. 1996;148:1047–1053. [PMC free article] [PubMed] [Google Scholar]
- 6.Tornillo L, Terracciano LM. An update on molecular genetics of gastrointestinal stromal tumours. J Clin Pathol. 2006;59:557–563. doi: 10.1136/jcp.2005.031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moasser MM. The oncogene HER2: Its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26:6469–6487. doi: 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 9.Serra V, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 10.Junttila TT, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 11.Kong D, Yamori T. Advances in development of phosphatidylinositol 3-kinase inhibitors. Curr Med Chem. 2009;16:2839–2854. doi: 10.2174/092986709788803222. [DOI] [PubMed] [Google Scholar]
- 12.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 13.Shah NP, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 14.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 15.Turke AB, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagata Y, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 17.Berns K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 18.Zhao JJ, et al. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyerson M, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 20.Shaulian E, Zauberman A, Ginsberg D, Oren M. Identification of a minimal transforming domain of p53: Negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol. 1992;12:5581–5592. doi: 10.1128/mcb.12.12.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yun CH, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, Blenis J, Yuan J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc Natl Acad Sci USA. 2008;105:6584–6589. doi: 10.1073/pnas.0802785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Olopade OI. MYC in breast tumor progression. Expert Rev Anticancer Ther. 2008;8:1689–1698. doi: 10.1586/14737140.8.10.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 26.Wolfer A, et al. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc Natl Acad Sci USA. 2010;107:3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obaya AJ, Mateyak MK, Sedivy JM. Mysterious liaisons: The relationship between c-Myc and the cell cycle. Oncogene. 1999;18:2934–2941. doi: 10.1038/sj.onc.1202749. [DOI] [PubMed] [Google Scholar]
- 28.Zunder ER, Knight ZA, Houseman BT, Apsel B, Shokat KM. Discovery of drug-resistant and drug-sensitizing mutations in the oncogenic PI3K isoform p110 alpha. Cancer Cell. 2008;14:180–192. doi: 10.1016/j.ccr.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folkes AJ, et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J Med Chem. 2008;51:5522–5532. doi: 10.1021/jm800295d. [DOI] [PubMed] [Google Scholar]
- 30.García-Martínez JM, et al. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421:29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin KA, Blenis J. Coordinate regulation of translation by the PI 3-kinase and mTOR pathways. Adv Cancer Res. 2002;86:1–39. doi: 10.1016/s0065-230x(02)86001-8. [DOI] [PubMed] [Google Scholar]
- 32.Jones RM, et al. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23:3217–3221. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- 34.Rosenwald IB, Rhoads DB, Callanan LD, Isselbacher KJ, Schmidt EV. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2 alpha in response to growth induction by c-myc. Proc Natl Acad Sci USA. 1993;90:6175–6178. doi: 10.1073/pnas.90.13.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson JD, Oster SK, Shago M, Khosravi F, Penn LZ. Identifying genes regulated in a Myc-dependent manner. J Biol Chem. 2002;277:36921–36930. doi: 10.1074/jbc.M201493200. [DOI] [PubMed] [Google Scholar]
- 36.Krüger M, et al. Involvement of proteasome alpha-subunit PSMA7 in hepatitis C virus internal ribosome entry site-mediated translation. Mol Cell Biol. 2001;21:8357–8364. doi: 10.1128/MCB.21.24.8357-8364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Druker BJ, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 38.Demetri GD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 39.Inoue A, et al. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24:3340–3346. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- 40.Sequist LV, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 41.Mok TS, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 42.Lazaris-Karatzas A, Sonenberg N. The mRNA 5′ cap-binding protein, eIF-4E, cooperates with v-myc or E1A in the transformation of primary rodent fibroblasts. Mol Cell Biol. 1992;12:1234–1238. doi: 10.1128/mcb.12.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruggero D, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 44.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith MR, et al. Translation initiation factors induce DNA synthesis and transform NIH 3T3 cells. New Biol. 1990;2:648–654. [PubMed] [Google Scholar]
- 46.Rosenwald IB, Lazaris-Karatzas A, Sonenberg N, Schmidt EV. Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol Cell Biol. 1993;13:7358–7363. doi: 10.1128/mcb.13.12.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shantz LM, Pegg AE. Overproduction of ornithine decarboxylase caused by relief of translational repression is associated with neoplastic transformation. Cancer Res. 1994;54:2313–2316. [PubMed] [Google Scholar]
- 48.Sorrells DL, et al. Detection of eIF4E gene amplification in breast cancer by competitive PCR. Ann Surg Oncol. 1998;5:232–237. doi: 10.1007/BF02303778. [DOI] [PubMed] [Google Scholar]
- 49.Kiyono T, et al. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 50.Zhao JJ, et al. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell. 2003;3:483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 51.Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- 52.Li C, Wong WH. DNA-Chip Analyzer (dChip) New York: Springer; 2003. [Google Scholar]