Abstract
Inhibitor of apoptosis (IAP) and Heat shock proteins (HSPs) provide assistance in protecting cells from stresses of hypoxia, imbalanced pH, and altered metabolic and redox states commonly found in the microenvironmental mixture of tumor and nontumor cells. HSPs are upregulated, cell-surface displayed and released extracellularly in some types of tumors, a finding that until now was not shared by members of the IAP family. The IAP Survivin has been implicated in apoptosis inhibition and the regulation of mitosis in cancer cells. Survivin exists in a number of subcellular locations such as the mitochondria, cytoplasm, nucleus, and most recently, the extracellular space. Our previous work showing that extracellular survivin was able to enhance cellular proliferation, survival and tumor cell invasion provides evidence that Survivin might be secreted via an unidentified exocytotic pathway. In the present study, we describe for the first time the exosome-release of Survivin to the extracellular space both basally and after proton irradiation-induced stress. To examine whether exosomes contributed to Survivin release from cancer cells, exosomes were purified from HeLa cervical carcinoma cells and exosome quantity and Survivin content were determined. We demonstrate that although proton irradiation does not influence the exosomal secretory rate, the Survivin content of exosomes isolated from HeLa cells treated with a sublethal dose of proton irradiation (3 Gy) is significantly higher than control. These data identify a novel secretory pathway by which Survivin can be actively released from cells in both the basal and stress-induced state.
Keywords: Survivin, Exosomes, Apoptosis, Cancer, Microenvironment
Introduction
The heterogeneous and structurally complex tumor microenvironment is composed of cancer cells and stromal cells embedded in an extracellular matrix that is nourished by networks of vascular and fibroblast cells. In comparison to normal tissue, tumor stroma exhibits alterations in the amount and composition of extracellular matrix and has an increased number of fibroblasts, which secrete growth factors and chemokines [1]. It has been documented that tumor stroma is involved in malignant transformation, tumor invasion and metastasis, and can affect the sensitivity of tumor cells to drug treatment [2]. Furthermore, cancer treatments as well as microenvironmental stresses such as hypoxia, pH imbalance, and altered metabolic and redox states accompany the dynamic tumor pathobiology [3]. Intracellularly and compartmentally, these stresses are responded to by the increased production of chaperones or HSPs as well as a milieu of other stress activated proteins such as the IAPs. HSPs are necessary for the stability of protein folding, multiprotein complex formation, intraorganellar protein shuttling and protein degradation, and now there is evidence for a role in cancer development and cytoprotection [3]. Additionally, recent research has shown that tumor cells surface-display and even secrete HSPs, which has not been documented in normal cells [3–5].
Survivin, a member of the IAP gene family, has been implicated in both suppression of cell death, the regulation of mitosis, surveillance checkpoints and adaptation to unfavorable environments [6–8]. Its aberrant, high protein expression in cancer cells and concomitantly low expression in most normal tissues makes Survivin an important anticancer target. Unlike other IAP proteins, the position and role of Survivin in the apoptotic cascade has not been conclusively elucidated. While most IAP proteins are predominantly cytosolic and have roles in caspase binding and inhibition, Survivin has been found in the nucleus, cytosol, mitochondria [9] and most recently extracellularly [10]. Its mitochondrial role involves tumor growth in immunocompromised animals, and abolished tumor cell apoptosis in vivo [11], while its extracellular form has the ability to reenter cancer cells inducing increased proliferation, apoptosis resistance and invasion [10].
Survivin has also been shown to interact directly with intracellular Hsp60 [12] and Hsp90 [13], a relationship that has led to the engineering of binding interface mimetics [14] for anticancer therapy. The novelty of finding Survivin in yet another essential compartment speaks directly to the multifaceted functionality of this protein. Its interaction both intra- and extracellularly with HSPs emphasizes the link between cellular stress responses, apoptosis, and cell proliferation occurring in the tumor microenvironment. Recent research has identified many proteins associated with exosomes, including molecules crucial for initiating immune responses and/or apoptosis [15, 16]. In the present study, we identify exosomes as mediators of basal and stress-induced Survivin secretion from HeLa cells. These data identify a novel secretory pathway by which cancer cells are able to actively release Survivin.
Materials and methods
Cell lines and cultures
The cervical carcinoma (HeLaS) cell line, pancreatic carcinoma (Panc 1), prostate carcinoma (PC3), and human embryonic kidney epithelial (A293) cell lines were obtained from the American Type Culture Collection (ATCC) as were the human normal prostate stromal cells (PrSC). The normal human bone marrow stromal cells were a generous gift of Kimberly Payne (Loma Linda University). Stromal cells were isolated and propagated as previously described [17] from adult human bone marrow that was purchased from Poietics Cell Systems (Lonza Walkersville, Inc., Gaithersburg, MD). Human peripheral blood mononuclear cells were isolated from a peripheral blood draw using ficollhypaque (GE Healthcare Bio-Sciences. Corp., Piscataway, NJ, USA) as has been previously described [18]. The use of all human tissues was reviewed and approved by the Institutional Review Board at Loma Linda University with informed written consent obtained from each subject. Cells were maintained as defined in DMEM, McCoy’s, or IMEM (ATCC) supplemented with 100 units of penicillin, 100 μg/ml streptomycin, 300 μg of L-glutamine and 10–20% heat inactivated FBS (ATCC). Cells were grown at 37°C in a humidified atmosphere of 95% air and 5% CO2 until 80% confluent and supernatants were collected, centrifuged for 10 min at 2,500 rpm, and stored at −20°C until use. Because fetal bovine serum contains endogenous exosomes, cell culture medium is removed 24 h prior to exosome collection and replaced with serum deficient medium. HeLaS/POZnSurvivin cells were also grown as previously described [10].
Expression plasmid and generation of stable HeLaS/POZnSurvivin cell lines
The detailed cloning and propagation procedure has been described previously [19, 20]. In brief, recombinant retro-viruses expressing a bicistronic messenger RNA containing open reading frames of Flag-HA-tagged human Survivin and interleukin-2 receptor (IL-2R)-α were constructed and transduced into HeLaS cells. The infected HeLaS cells were sorted by anti-IL-2R monoclonal antibody (mAb) conjugated with magnetic beads, and the resulting Flag-HA-Survivin stable cell lines propagated as suspension cells. The expression level of Survivin was evaluated by Western analysis and immunohistochemistry with anti-Flag and HA antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
In-gel trypsin digestion and MS
Protein bands were excised manually and washed with 50% (v/v) methanol and 5% (v/v) acetic acid. The gel pieces were then dehydrated in acetonitrile and dried in a SpeedVac concentrator (Savant, Farmingdale, NY). Proteins were reduced using 10 mM dithiothreitol (DTT) in 100 mM ammonium bicarbonate for 30 min at room temperature. The DTT solution was removed and the proteins were alkylated for 30 min at room temperature using 100 mM iodoacetamide after which the gel pieces were dehydrated as before. Gel pieces were rehydrated in 100 mM ammonium bicarbonate and then dehydrated and dried as previously described. Proteins were tryptically digested using MS grade trypsin (Promega, Madison, WI), added at a final concentration of 20 ng/μl to fully cover the gel pieces. Digestion was performed at 37°C overnight. Peptides were recovered with 30 μl, 50% (v/v) acetonitrile and 5% (v/v) formic acid twice. All supernatants were pooled and dried in a SpeedVac concentrator for 1 h.
Tryptic peptides were analyzed on our ThermoFinnigan LCQ Deca XP system that includes a surveyor HPLC and a PicoView 500 (New Objective, Woburn, MA) for performing nanoflow electrospray ionization. The flow of the surveyor HPLC pump was split to achieve a 200–300 nanoliter/min flow exiting a PicoFrit column (New Objective) packed with BioBasic C18 beads (10 cm, 5 μm, 300 Å). Samples were loaded onto a Michrom Bioresources (Auburn, CA) cap-trap at 5 μl/min and washed with mobile phase A (aqueous 2% acetonitrile with 0.1% formic acid). Peptides were then eluted onto the column and into the mass spectrometer using a gradient of 0–75% mobile phase B (aqueous 90% acetonitrile with 0.1% formic acid). The mass spectra acquisition was operated in the data dependent mode with one MS scan (300–1,500 m/z) and three MS/MS scans of the most intense ions in the MS scan.
We used the Sequest algorithm implemented on the TurboSequest software package to identify proteins based on the MS/MS spectra. The resulting Sequest hits were filtered based on the charge state and Xcorr value to require Xcorr ≥ 1.5, 2.0, and 2.5 for single, double, and triple charged ions, respectively.
The MS/MS fragmentation spectra were searched against a current human protein database (March 2009) containing 37,391 reference sequences. The search algorithms Sequest [21], Mascot [22], and X! TANDEM [23], were used to identify peptides and proteins. The significance of identified peptides and proteins were determined using the Peptide-Prophet [24] and ProteinProphet [25], respectively, algorithms as implemented in Scaffold 2 (Proteome Software, Portland, OR). We included only peptides with a Scaffold score of ≥95% (5% false discovery rate) in the results.
Immunoprecipitation and western blot analysis
For Western blots of purified exosomes, exosomes were solubilized with lysates loaded onto gels according to either protein concentration or acetylcholinesterase (AChE) activity (described below). For protein concentrations the BCA assay (Pierce, Rockford, USA) was used. For Western blots of whole cell lysates, cells were lysed with lysates centrifuged at 13,000 rpm at 4°C for 20 min to remove cell debris. The cleared supernatants were transferred to fresh tubes and protein concentrations were determined by BCA assay (Pierce, Rockford, USA). Proteins (20–40 mg) were separated using 12% Bis–Tris polyacrylamide gels, transferred onto polyvinylidene difluoride membranes (Millipore) and probed using the following antibodies: mouse monoclonal anti-LAMP1 (Abcam, Cambridge, MA), mouse monoclonal anti-c-Src (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and rabbit polyclonal anti-Survivin (Novus, Littleton, CO) and anti-Hsp70, anti-H-Ras, anti-Calnexin (Santa Cruz Biotechnology) and rabbit polyclonal antibodies to β-actin (AbCam, Cambridge, MA). Secondary antibodies (IR-Dye-conjugated) were goat anti-rabbit and goat anti-mouse immunoglobulin (LICOR, Lincoln, Nebraska). Immunoreactive bands were detected using the Odyssey imaging system (LICOR) and quantified using ImageQuant software. For extracellular protein immunoprecipitation, conditioned medium was precleared using protein A/G beads (Sigma Chemical Co., St. Louis, MO) followed by an overnight incubation using rabbit polyclonal anti-Survivin (Novus, Littleton, CO) or control polyclonal anti-IgG (Santa Cruz Biotechnology, Santa Cruz, CA). Protein A/G immunobeads were then used for a 1 h immunoprecipitation followed by washing and resolving of proteins on 4–12% gradient Bis–Tris polyacrylamide gels and overnight Coomassie staining for further mass spectrometry analysis.
Exosome isolation
Exosomes were isolated according to the protocols of Savina et al. [26] and supplement 30 from Current Protocols in Cell Biology [27]. Because fetal bovine serum contains endogenous exosomes, cell culture medium was removed 24 h prior to exosome collection and replaced with serum deficient medium. Briefly, exosomes were collected from 10 ml of serum free medium removed from HeLaS, HeLaS/POZnSurvivin, HeLaS/POZnSurvivin + 10 μg/ml cytochalasin D and HeLaS/POZnSurvivin + 10 μM monensin medium (6 × 106 cells) cultured over 24–48 h. The culture mediums were collected on ice and centrifuged at 800g for 10 min to pellet the cells and then centrifuged at 12,000g for 30 min to pellet any cellular debris. Exosomes were separated from the supernatant by centrifugation at 110,000g for 15 h. The exosomal pellet was washed once in PBS and then resuspended in 200 μl of PBS (exosome fraction). Exosome protein amounts were measured using the BCA protein assay kit (Pierce, Rockford, IL).
Exosome quantification
To quantify the amount of exosomes released we assessed the activity of acetylcholinesterase, an enzyme that is specific to these vesicles [28]. Acetylcholinesterase activity was assessed as described by Savina et al. [26]. Briefly, 40 μl of the exosome fraction was suspended in 110 μl of PBS. 37.5 μl of this PBS-diluted exosome fraction was then added to individual wells on a 96-well flat-bottomed microplate. 1.25 mM acetylthiocholine and 0.1 mM 5,5′-dithiobis(2-nitrobenzoic acid) were then added to exosome fractions in a final volume of 300 μl, and the change in absorbance at 412 nm was monitored every 5 min for 30 min.
Exosome preparation for transmission electron microscopy
Transmission Electron Microscopy was performed at Real-view Analytical Laboratories, Boston, MA according to standard protocols. Briefly, exosome pellets were fixed in 3% formaldehyde and 0.1% glutaraldehyde (EM grade) for 10 min at 37°C. Samples were incubated in 50 mM NH4Cl in PBS for 60 min at 22°C to aminidate free aldehydes, dehydrated through a gradual series of ethanol to 100%, and transferred into a mixture of 50:50 (vol/vol) Lowicryl K4 M resin/100% ethanol overnight at 22°C.
Exosome isolation and fixation (Lowicryl embedding) for transmission electron microscopy
Exosomes were pelleted as described above and fixed as a suspension in 2% paraformaldehyde and 50 mM KH2PO4, pH 7.4, at room temperature for 1 h. Afterward, the exosomes were again centrifuged, and the pellets washed three times in 50 mM KH2PO4, pH 7.4, to remove the fixative. Final exosome pellets were resuspended in 200 μl of 50 mM KH2PO4, pH 7.4, and diluted 1:1 (v/v) with 3% agarose. The mixture was allowed to solidify and was stored at 4°C. Samples were dehydrated in a graded series of methanol, infiltrated and embedded in Lowicryl, a low temperature embedding resin, and polymerized overnight with a UV lamp.
Immunoelectron microscopy to identify antigen-existence in the exosome by immunogold labeling
Samples were transferred to aliquots of fresh resin (3 changes, 1 h each) and applied to filling embedding capsules for 24 h at 60°C. Thin sections were cut using a diamond knife on a Reichert-Jung Ultracut E ultramicrotome and placed on gold support grids for immunostaining. For labeling, sections were incubated in blocking solution (Zymed Laboratories Inc.) for 30 min at 22°C, rinsed, incubated with primary antibodies to Survivin, Hsp70 or control IgG, rinsed again, and incubated with gold-conjugated secondary reagents (Hsp70, 5 nm diameter; Survivin, 10 nm diameter) of appropriate specificities (1:20; Jackson ImmunoResearch Laboratories Inc.). After washes, samples were exposed to OsO4 vapor for 1 h at 22°C, post-stained with uranyl acetate and lead citrate, and analyzed on a Philips CM10 electron microscope (Philips Electronics) at 80 kV. For quantification of exosomal colocalization of Survivin and Hsp70, 19 individual electron microscopy fields were individually scored for accumulation of Survivin- and Hsp70-associated gold particles.
Flow cytometry of exosomes
Flow cytometry of exosomes was accomplished as previously described [29] with some modifications. Anti-MHC II or anti-CD9 antibodies (Becton–Dickinson) were coupled to aldehyde-sulfate latex beads (4 μm; Invitrogen) by rotation overnight at 4°C. The reaction was quenched with 100 mM glycine and the beads washed with 1 ml PBS. Isolated exosomes (30 μg) were incubated with 10 μl antibody-conjugated beads at room temperature for 15 min, 1 ml PBS added and incubated for an additional 1 h at room temperature with rotation. Exosome conjugated beads were washed twice with 1 ml PBS and resuspended in 20 μl PBS. Exosomes were stained with 15 μl of PE or FITC-conjugated anti-CD9, anti-CD54 (Becton–Dickinson), anti-LAMP1 (AbCam), anti-Hsp70 (Novus) or anti-Survivin Alexa 647 (AbCam) for 30 min at 4°C, then washed with PBS. PE or FITC-conjugated mouse IgG antibodies were added for 30 min at 4°C to the stained samples. All samples were washed twice with PBS and fixed with 200 μl 1% paraformaldehyde. Samples were collected using the MACSQuant Analyzer (Miltenyi Biotech) and data analyzed using FlowJo software (Tree Star Inc.).
Quantification of Survivin from conditioned media
Conditioned Media (CM) was collected from 90% confluent cell cultures with cell viability assessed by Trypan-Blue exclusion (Sigma–Aldrich, St. Louis, MO). Samples were assayed for the presence of Survivin using the Quantikine Human Survivin Immunoassay kit (R&D Systems, Inc., Minneapolis, MN), according to the manufacturer’s instructions.
Immunofluorescence localization of protein
Cells were plated in 6 well-plates on 22 × 22 mm cover-slips and cultured with 2 ml of control or cytochalasin D media (WT-CM). Wells were washed with PBS and fixed in 4% paraformaldehyde for 30 min at room temperature. Cells were permeabilized with 0.5% NP-40 in PBS for 15 min, blocked with 5% fetal bovine serum in 0.01% NP40/PBS for 30 min, incubated for 1 h with 1:1,000 dilution of anti-beta actin polyclonal antibodies (AbCam, Cambridge, MA). Cells were incubated with FITC-conjugated rabbit anti-mouse IgG secondary antibody (Invitrogen/Molecular Probes, Eugene, OR), mounted with Vectashield mounting media containing DAPI (Vector Laboratories, Burlingame, CA) and observed under an Olympus BX50 fluorescent microscope (Olympus America Inc., Center Valley, PA).
Proton irradiation
All radiation procedures were accomplished in the Loma Linda University Radiobiology Proton Treatment Facility, now the James M. Slater, MD, Proton Treatment and Research Center. Cells were exposed in vitro to 250 MeV protons with a single dose of 3 Gy. Three Gy has been determined in our lab to be a sublethal dose of proton radiation in these HeLa cells. Subconfluent cultures of HeLaS/POZnSurvivin cells were exposed to proton irradiation (0 or 3 Gy/h) and then incubated for 24 h at 37°C after which conditioned medium was collected and processed for exosomes as described above. Cells were harvested, prepared, and analyzed for cell death using trypan blue exclusion and DNA content.
Apoptosis and cell cycle analysis
Cells were harvested, prepared, and analyzed for DNA content. DNA content was analyzed using a Becton–Dickinson FACScan flow cytometer (Becton–Dickinson, San Jose, CA). The distribution of cells in the different phases of the cell cycle was analyzed from DNA histograms using BD CellQuest software (Becton–Dickinson and Company, San Jose, CA).
Statistical analysis
Data are presented as means ± standard deviation. Comparisons between groups were performed using the Student’s t test. A P value < 0.05 was considered statistically significant.
Results and discussion
Survivin is found associating with HSPs in extracellular media
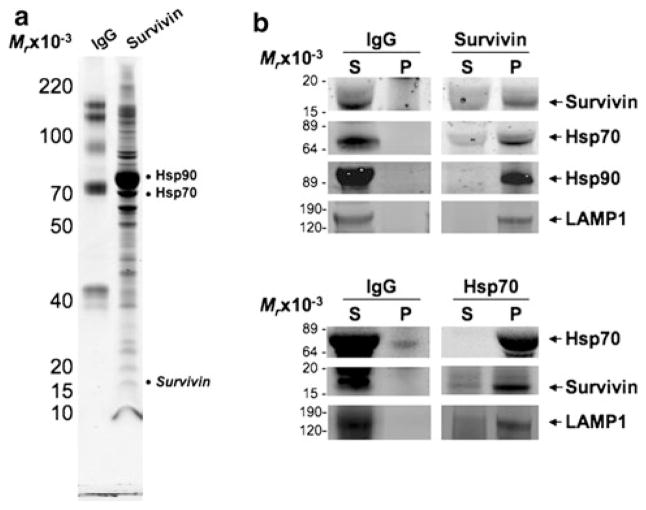
Previous work accomplished by our laboratory [10] described for the first time an extracellular pool of Survivin that when re-incubated with cancer cell lines enhanced functional characteristics associated with more aggressive cancer phenotypes such as proliferation, anti-apoptosis and invasion. We therefore undertook this work in order to evaluate more deliberately this pool of extracellular Survivin and determine specifically whether it was secreted or released as a result of cellular demise. HeLaS/POZnSurvivin cells were incubated in serum-free medium. After 48 h, the conditioned medium was collected and subjected to purification using anti-Survivin antibodies. Immunoprecipitates were separated by 4–12% gradient SDS–PAGE and detected by Coomassie blue staining (Fig. 1a). To identify the proteins that were precipitated in association with Survivin, protein bands were excised, tryptically digested, and analyzed using sequential chromatography, electrospray ionization and tandem mass spectrometry. A database search identified several proteins specifically associated in protein union with Survivin (Fig. 1a and Supplemental Table 1). These proteins include HSPs such as Hsp90, Hsp70, and many unrelated or unidentified proteins (data not shown). While exosome immunoprecipitation followed by Western blots with a Survivin antibody detected the expected band at 16.5 kDa (Fig. 1b), Survivin was not easily observed by mass spectrometry. However, when the beads containing the Survivin antibody were directly digested with trypsin, tryptic peptides from Survivin were readily observed (Supplemental Table 1). The MS/MS fragmentation spectra of these Survivin peptides from the beads were similar to the spectra obtained in control experiments with authentic recombinant Survivin protein (data not shown). To confirm these findings, we performed reciprocal immunoprecipitations in which purified exosomes were sonicated after which Survivin was immunoprecipitated from them directly, ran on SDS–PAGE and blotted for Survivin, Hsp70, Hsp90 and LAMP1 (Fig. 1b). Again, the presence of Survivin precipitated directly from purified exosomes and associating with Hsp70 and Hsp90 provides further evidence that the observed Survivin release is exosome-dependant. The reverse was also evaluated where Hsp70, immunoprecipitated from exosome sonicates, was resolved on SDS–PAGE and blotted for Hsp70, Survivin and LAMP1. As before, Hsp70, Survivin and LAMP1 are found precipitating together in exosomes (Fig. 1b).
Fig. 1.
Survivin interacts with HSPs from conditioned medium. a Survivin and control IgG immunoprecipitates from HeLaS/POZn-Survivin conditioned medium were separated on a 4–12% gradient SDS gel and stained with Coomassie brilliant blue. The major bands were analyzed by trypsin digestion and LC–MS/MS mass spectrometry (see Supplemental Table 1). b Exosomes purified from the conditioned medium of HeLaS/POZnSurvivin cells were lysed and immunoprecipitated with pAb Survivin or pAb Hsp70 and pAb IgG. Aliquots of pellet (P) or supernatant (S) were sequentially immuno-blotted with pAb Survivin or pAb Hsp70 or mAb Hsp90 or mAb LAMP1. Molecular-weight markers in kilodaltons (KDa) are shown on the left
Secretion of intracellular proteins in response to heat shock and inflammation has been widely reported [30] and similar to Survivin, oxidative stress-induced factors such as cyclophilins and HSPs have been shown to be secreted and present in biological fluids, which include plasma and synovial fluid [30–32], possibly with exosome association. Proteomic analysis of exosomes has provided evidence of exosomes enriched with several members of the HSP family [33] and thus we next chose to investigate if Survivin also occupied exosomal space.
Survivin is found, like Hsp70, both on and in exosomes
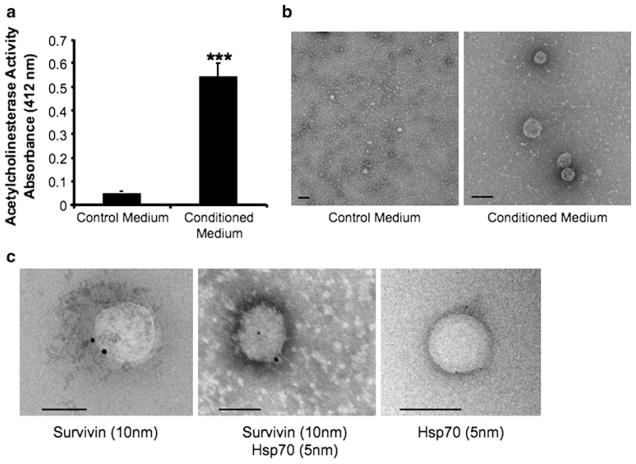
Signal sequences mediate classical protein secretion, and software programs [34] are now being employed to more easily and accurately determine signal sequence existence. Neither Survivin nor Hsp70 proteins can be predicted by these criteria to be secreted proteins. In addition, it has been previously shown that small vesicles called exosomes provide a vehicle by which cells actively release Hsp70 [35]. We therefore hypothesized that as a result of Survivin’s association in conditioned medium with Hsp70, its extracellular localization may also be dependent upon exosomes. Exosomes were purified and validated by acetylcholinesterase (AChE) activity according to the methods of Savina et al. [26]. AChE, an enzyme specific to exosomes [36], assays showed significantly more activity in conditioned medium taken from Survivin-releasing HeLaS/POZnSurvivin cells [10] compared to non-conditioned medium (Fig. 2a). The vesicular nature of these isolates was further confirmed by electron microscopy as shown in Fig. 2b in which the exosomes of the characteristic size and shape (50–150 nm) were easily seen in the conditioned medium isolates (right panel) and not from the control medium isolates (left panel). The presence of Survivin (10 nm) (left panel) and Hsp70 (5 nm) (right panel) and then their colocalization (middle panel), was also confirmed using immunoelectron microscopy (Fig. 2c and Fig. S1).
Fig. 2.
HeLaS/POZnSurvivin cells secrete Survivin and Hsp70-containing exosomes. a Exosome presence, measured as AChE activity from either 24 h control media or conditioned medium fractions. The 110,000g HeLaS/POZnSurvivin pellets were defined for AChE activity as described in the “Methods and materials”. Each fraction was evaluated by colorimetry. Values represent the means of three media control samples and nine conditioned medium samples. *** P <0.001. b Exosomes were characterized from 24 h HeLaS/POZnSurvivin conditioned medium using electron microscopy from both nonconditioned (left) and conditioned medium (right) respectively. c Immunoelectron microscopy on anti-Survivin- and anti-Hsp70-stained cryosections of HeLaS/POZnSurvivin cells shows that Survivin and Hsp70 localization in the exosome. Immunogold nanoparticles of 10 nm were conjugated to anti-Survivin pAb while 5 nm nanoparticles were conjugated to anti-Hsp70 mAb. Exosomes are cup-shaped and 50–150 nm in size. Bar 100 nm
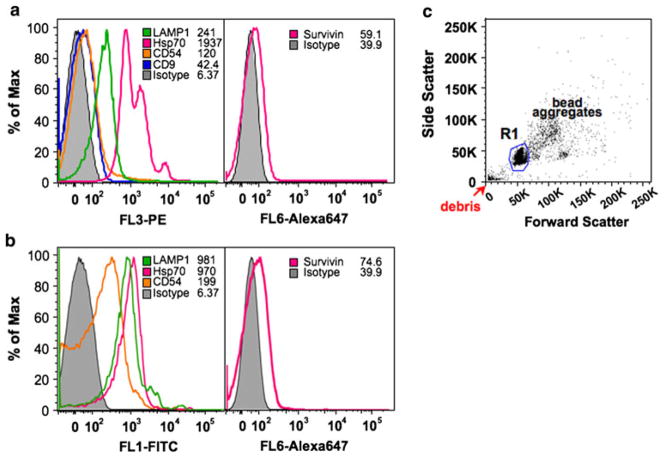
A more simple and rapid method [29, 37], compared to the serial centrifugation and immunogold labeling combined with electron microscopy, suitable for the routine isolation and analysis of exosomes was next accomplished. Exosomes have been shown to exhibit a phenotype with high expression of MHC class I and II, the lysosomal-associated membrane protein 1 (LAMP1), as well as expression of several tetraspanin proteins like CD9, CD63, CD81 and CD82 [38]. The presence of adhesion proteins CD11b, CD11c, CD54 (ICAM-1) and CD58 have also been detected on the purified exosomes [29]. Based upon immuno-magnetic extraction of exosomes bearing human MHC Class II (HLA-DR) (Fig. 3a, c) or bearing CD9 (Fig. 3b, c), this rapid semi-quantitative method takes advantage of flow cytometry. HeLa cell-secreted exosomes, coupled to MHC class II or CD9 aldehyde-sulfate latex beads, exhibited a phenotype with high expression of LAMP1 and Hsp70, as well as expression of CD54, CD9 and Survivin (Fig. 3a, b). HeLa cell-derived exosomes exhibited a strong surface expression of Hsp70 which resembles that reported from exosomes released from pancreas and colon carcinoma cells [39] and not that recorded from B cell exosomes [37] where HSPs were restricted from the exosome surface to its lumen. The presence of HSPs in and on these tumor-derived exosomes has been proposed to be an indicator of cellular injury, with important roles in inflammation and immunity [37]. Hsp70 has been proposed to play a role in exosome formation and/or release [40]. Survivin’s exosomal surface expression may be low in order to reduce the likelihood of it taking part in presentation and priming of the immune system. In determining if cellular stress would change the localization of Survivin from the lumen preference to the exosome surface, exosomes were collected from 3 Gy irradiated cells. Surface levels of Survivin, measured using flow cytometry as described above did not increase (data not shown).
Fig. 3.
Histogram profile of surface CD antigens on purified exosomes bound to anti-MHC class II (a) or CD9 (b) -coupled aldehyde beads. Presence of LAMP1, Hsp70, CD9, CD54 and Survivin antigens were analyzed using exosomes bound to anti-MHC class II- or CD9-coupled aldehyde beads. c Beads are identified on the forward and side scatter plot that represents single beads (R1), bead aggregates, and debris. The region R1 denotes the region selected for gating in all the experiments. Typically, single beads represent 50–80% of total beads
To this point, the data that we present comes entirely from the HeLaS/POZnSurvivin cervical cancer cell line which was engineered to overexpress a FLAG/HA-tagged Survivin protein. For these findings to be relevant to tumor biology and not be simply an artifact of Survivin-protein overexpression, conditioned medium from cell lines derived from pancreatic (Panc1) and prostate (PC3) cancers as well as from the non tumor-derived cells, Peripheral Blood Mononuclear Cells (PBMC), Human Bone Marrow Stroma (HBMS), Prostate Stromal Cells (PrSC) and human embryonic kidney epithelial (A293) cells have been processed and their exosomes analyzed by Western blot analysis (Fig. S2A) and AChE activity (Fig. S2B). Our findings indicate that Survivin’s localization in cancer cell line exosomes is a general finding for tumor-cell derived exosomes. It is not specific to this HeLa overexpression cell line as the non tumor-derived cells, though expressing exosomes as analyzed by Western blot using LAMP1, do not express Survivin (Fig. S2A). Preparation purity was evaluated using antibody against Calnexin, a protein reported not to be found in exosomal isolations [27].
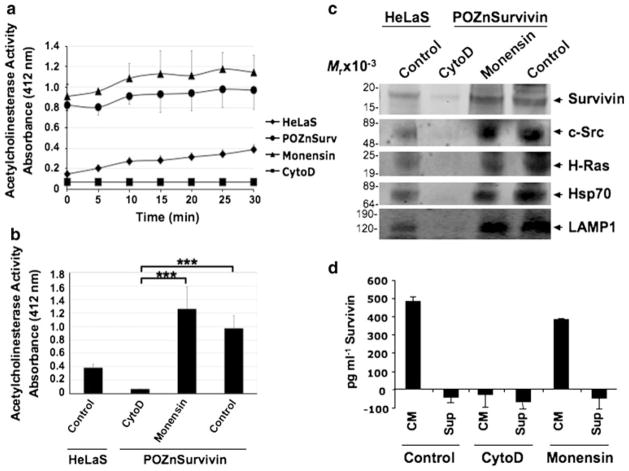
To evaluate further that Survivin’s extracellular location is being modulated solely by exosomes, two compounds, cytochalasin D (cytoD) and monensin were acquired. CytoD, though not previously reported to directly prohibit exosome release, has been shown to disrupt actin cytoskeletal filaments [41, 42]. Because direct modulation of actin polymerization affects membrane trafficking [42], we hypothesized that cytoD may block exosome release as well. Monensin blocks protein release by the classical secretory-pathway [39]. HeLa cells treated with vehicle alone or with monensin expressed the same AChE activity, while those treated with cytoD exhibited a marked reduction in AChE activity across time (Fig. 4a). This prohibition of exosome release by cytoD when evaluated at the 30 min time period within this activity assay is significant compared to the control or the monensin treatment (Fig. 4b). Furthermore, exosomes were tested by Western blotting for the presence of proteins known to be specific for (LAMP1) or enriched (c-Src, H-Ras, Hsp70) in exosomes [43, 44]. In addition to expressing Survivin, the purified exosomes were also enriched with c-Src tyrosine kinase, H-Ras, Hsp70 and the exosome specific LAMP1 (Fig. 4c). We next investigated whether disrupting the actin cytoskeleton or inhibiting the common secretory pathway would prohibit the release of Survivin-containing exosomes. Cells treated with cytoD were prohibited from releasing exosome-containing proteins, Survivin, c-Src, H-Ras, Hsp70 or LAMP1 while treatment with monensin had no effect (Fig. 4c). One further experiment using a commercially available ELISA for Survivin quantification was utilized in examining Survivin’s exosomal localization. Conditioned mediums underwent ultracentrifugation to remove the exosomes and comparison was made between the supernatants from these centrifugations and the conditioned medium prior to centrifugation. As would be predicted from an exosomally packaged, non-classically secreted protein, removal of exosomal Survivin all but depleted the conditioned medium of Survivin (Fig. 4d). ELISA quantitation of conditioned medium taken from CytoD treated cells revealed complete prohibition of Survivin in the medium even prior to exosome removal while Monensin treatment did not prohibit the measuring of Survivin in the conditioned medium (Fig. 4d). In our previous work [10] we reported the extracellular compartmentalization of Survivin and its ability to participate in the cytoprotection, tumorigenesis and enhanced invasion of cancer cells. It remained unclear whether Survivin was released as a result of an excretion, secretion, or cellular necrosis. These findings provide further evidence of Survivin’s localization in the extracellular space being primarily through exosomal rather than soluble mechanisms, a finding we believe will prove useful in the designing of therapeutic interventions in cancer patients.
Fig. 4.
Survivin’s exosomal release from cancer cells is blocked by cytoD but not monensin. 24 h exosome release is quantitated from HeLaS cells as control or HeLaS/POZnSurvivin cells treated with vehicle, monensin or cytoD. a Exosome presence was measured as AChE activity. Direct modulation of actin polymerization with cytoD reduced exosome release measured for 30 min while inhibition of the common secretory pathway using monensin had no effect. b Measured at 30 min, this reduction in exosome presence after cytoD treatment was significant when compared to both monensin and control samples. AChE activity values represent the means of three samples. Exosomes were collected after 24 h of treatment. c Western blots of Survivin, c-Src, H-Ras, Hsp70 and the exosome specific marker protein LAMP1 in the conditioned medium of exosome isolations. C-Src, H-Ras and Hsp70 were found and are known exosome-associated proteins while LAMP1 is exosome specific. As in A, cytoD prohibited exosome proteins from accumulating in the exosomes while monensin did not. Protein concentrations for Western blot loading were determined using the AChE activity. Molecular-weight markers in kilodaltons (kDa) are shown on the left. *** P <0.001. d ELISA quantitation was accomplished according to the manufacturer’s instructions. Survivin was quantitated from conditioned medium (CM) and from CM having exosome removed using ultracentrifugation (Sup-supernatant) in the presence and absence of cytoD and monensin
Previous experiments have shown using cytoD that the actin cytoskeletal network is required for secretion by providing contractile forces that expulse secretory proteins [45]. Visualization of these cytoD-treated cells, probing with antibodies to actin, shows the result of cytoD treatment on the actin cytoskeleton and cell morphology (Fig. S3A). Upon evaluating this cytoD-induced prohibition of exosome release, we record that there is an increase in the number of cells undergoing apoptosis (Fig. S3B) with a concomitant reduction in exosome release (Fig. S3C). When cytoD or monensin is combined with proton irradiation, the previously sublethal 3 Gy radiation dose becomes lethal (Fig. S3B) and Survivin, rather than being released in the exosome is retained in the cytosol of the treated cell (Fig. S3D). This finding is in line with our unpublished observations that cells induced to undergo apoptosis using chemotherapy or radiation therapy have a reduced exosome number by AChE activity and no measurable Survivin in exosome protein Western blots. Whether Survivin’s exosomal trafficking is in part controlled by the type and level of stress is unknown. It does not appear that this cytoD inhibition of Survivin-containing exosomes is cell type specific as we have made similar observations in cell lines derived from prostate cancer patients (data not shown). Disruption of microtubules has been reported to inhibit exocytosis in some instances [46] but to increase exocytosis in others [47].
Exosomal Survivin release is affected by stress
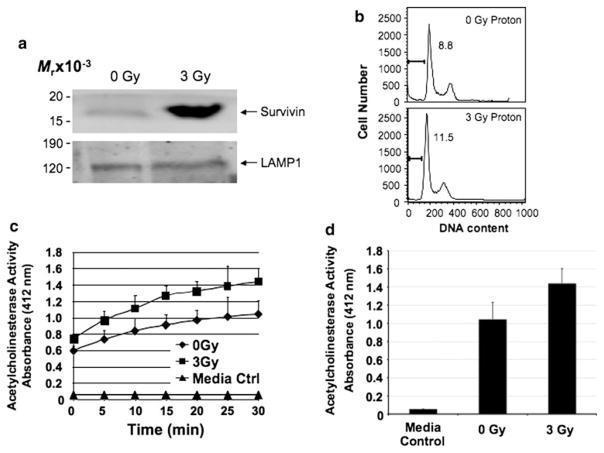
Survivin overexpression in cancer has been described as a predictive factor in determining response to chemotherapy and radiotherapy. Thus, its role in cancer biology exceeds by far its simple description as an inhibitor of apoptosis. Repeatedly, Survivin is described as a mitotic spindle checkpoint-associated protein and shown to play a role in tumor progression and chemoresistance. Survivin also reduces cell death induced by several anticancer agents including paclitaxel, etoposide, and tumor necrosis factor alpha. Conversely, inhibition of Survivin reduces tumor growth potential and sensitizes tumor cells to chemotherapeutic agents, such as paclitaxel, cisplatin, etoposide, gamma irradiation, and immunotherapy [48]. More recently, we have investigated Survivin’s role in resistance to proton irradiation of pancreatic cancer cells and found that stress proteins such as Survivin and the X-linked inhibitor of apoptosis (XIAP) protein play important roles in resistance to proton-based treatment options [49]. We therefore investigated whether stress, delivered by proton irradiation to HeLaS/POZnSurvivin cells, stimulated enhanced Survivin release to the extracellular conditioned medium compared with control conditions. As expected, sublethal proton irradiation (3 Gy) resulted in a marked accumulation of Survivin in the exosomal fraction taken from the conditioned medium of these cells 24 h post irradiation (Fig. 5a). We performed trypan blue exclusion analysis (data not shown) and propidium iodide (PI) flow cytometry (Fig. 5b) in order to confirm that 3 Gy was sublethal. Cell viability was always greater than 95% and we did not observe any significant viability differences between proton irradiation (3 Gy) and controls (0 Gy). Additionally, in order to determine whether proton irradiation stress-induced increases in exosomal Survivin was the result of enhanced exosome release or enriched exosome packaging of Survivin, we again performed exosomal AChE activity assays on the exosome purifications from conditioned medium taken from 3 Gy and 0 Gy proton-irradiated cells. Figure 5c shows, similar to Figs. 2a and 4b, conditioned medium contained significantly more exosomes than control media. However, there was no appreciable difference in exosome number in conditioned medium taken from 3 Gy or 0 Gy proton-irradiated cells. AChE activity was followed every 5 min for 30 min without significant change (Fig. 5d). Our findings, that proton-induced stress did not result in the enhanced secretion of exosomes, matches reports by others investigating heat stress and exosomes [37]. Given the intracellular roles performed by Survivin, it seems counterintuitive that a secretory pathway exists that allows cells to release Survivin both basally and in response to cellular stress. However, it may be plausible that Survivin’s both basal and stress-enhanced release is to communicate with adjacent cells, which may not themselves have been initially affected by the stress, and thus produce a more coordinated response to stress by the surrounding cells or tissues, a term or phenomenon called the bystander effect.
Fig. 5.
Stress-activated Survivin protein content increases in exosomes after proton irradiation of HeLaS/POZnSurvivin cells. a Western blotting shows an increase in Survivin in exosome vesicles after 3 Gy of proton irradiation. Protein concentrations for Western blot loading were determined using the AChE activity. Loading is controlled for by the Lysosome Associated Membrane Protein (LAMP1). Molecular-weight markers in kilodaltons (KDa) are shown on the left. Exosomes were collected after 24 h of treatment. b Cervical carcinoma HeLaS/POZnSurvivin cells were irradiated using proton radiation (0 or 3 Gy) and after 24 h were harvested and analyzed for DNA content by propidium iodide staining and flow cytometry. Percentages of apoptotic cells with hypodiploid (sub-G1) DNA content or 4 N DNA representative of cells in G2/M are indicated per each condition tested. Data are representative of one of two independent experiments with comparable results. c Exosomes were purified from control media, 0 Gy and 3 Gy proton-irradiated HeLaS/POZnSurvivin conditioned medium after which AChE activity was determined as described under “Methods and materials”. Values represent the means of three samples. d Measured at 30 min, no significant difference was recorded
While these studies do not identify the functional consequence of Survivin release via exosomes, our recent publication [10] provides evidence for extracellular Survivin affecting cellular proliferation, apoptosis-control and invasion. Although it has been suggested that cellular release of Survivin may be the result of non-specific processes, such as cell lysis, we believe that the work presented here provides a reasonable argument against this impression. It has also been suggested that exosomes released from B- and T-cells, macrophages, platelets and dendritic cells modulate immune responses [33]. Furthermore, Survivin has been found in the circulation and synovial fluid of certain patients with rheumatoid arthritis [50, 51] where its presence is correlated with destructive (erosive) disease. In contrast, high levels of antibodies against Survivin were found in patients with non-erosive rheumatoid arthritis. It is therefore plausible that Survivin is secreted by normal and cancer cells alike. The confirmation of this possibility is the focus of an ongoing study in our laboratory.
Our finding, that Survivin has intercellular transport and signaling capabilities, is a significant, and potentially seminal discovery. Our data show that Survivin is expressed within exosomes and that exosomes contribute to the release of Survivin from HeLa cells in both the basal and proton radiation, stress-induced state. Although proton irradiation had no effect on the exosome secretory rate, exosomal Survivin content was increased in these irradiated cells. In addition, our data do not support a role for the common cellular secretory pathway in the etiology of Survivin’s export into the extracellular environment. Consistent with Survivin’s association with unfavorable clinicopathological parameters, trafficking Survivin throughout the tumor microenvironment could potentially be responsible for driving the aggressive status of the tumor, prohibiting or minimizing therapeutic results. Thus, treatment of a tumor, using antibody against Survivin, could neutralize Survivin extracellularly by the formation of immune complexes. Alternatively, if Survivin’s intercellular signaling and transport are dependent upon formation of a complex with HSPs, treating tumors with binding mimetics to disrupt their interaction should neutralize Survivin’s extracellular localization and functions. This work is currently under consideration in our and other laboratories.
Supplementary Material
Acknowledgments
NCMHD Project EXPORT Program 5P20MD001632/Project 3 (N.R. Wall). Funding was also obtained as part of a start-up package from Loma Linda University’s Center for Molecular Biology and Gene Therapy, now the Center for Health Disparities Research and Molecular Medicine (NRW) and a National Merit Test Bed (NMTB) award sponsored by the Department of the Army under Cooperative Agreement Number DAMD17-97-2-7016 (NRW). Proton irradiation was accomplished at the Loma Linda University Radiobiology Proton Treatment Facility, now the James M. Slater, MD, Proton Treatment and Research Center. The authors would like to personally thank Dr. James Slater, Dr. Daila Gridley, Steven Rightnar and Celso Perez for all their help and we dedicate this work to our dear friend and colleague Dr. Lora Green who passed away unexpectedly.
Abbreviations
- IAP
Inhibitor of apoptosis
- HSPs
Heat shock proteins
- AChE
Acetylcholinesterase
- LAMP1
Lysosomal-associated membrane protein 1
- cytoD
Cytochalasin D
- XIAP
X-linked inhibitor of apoptosis
- ATCC
American Type Culture Collection
- IL2R
Interleukin-2 receptor
- DTT
Dithiothreitol
- CM
Conditioned medium
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10495-010-0534-4) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflict of interest.
Contributor Information
Salma Khan, Email: salmakhan@llu.edu, Center for Health Disparities Research and Molecular Medicine, Loma Linda University, 11085 Campus Street, Mortensen Hall, Room 162, Loma Linda, CA 92350, USA. Department of Basic Sciences, Division of Biochemistry and Microbiology, Loma Linda University, Loma Linda, CA 92350, USA.
Jessica M. S. Jutzy, Email: jeslater@llu.edu, Center for Health Disparities Research and Molecular Medicine, Loma Linda University, 11085 Campus Street, Mortensen Hall, Room 162, Loma Linda, CA 92350, USA. Department of Basic Sciences, Division of Biochemistry and Microbiology, Loma Linda University, Loma Linda, CA 92350, USA
Jonathan R. Aspe, Email: jaspe@llu.edu, Center for Health Disparities Research and Molecular Medicine, Loma Linda University, 11085 Campus Street, Mortensen Hall, Room 162, Loma Linda, CA 92350, USA. Department of Basic Sciences, Division of Biochemistry and Microbiology, Loma Linda University, Loma Linda, CA 92350, USA
Dalmor W. McGregor, Email: dmcgregor@llu.edu, Center for Health Disparities Research and Molecular Medicine, Loma Linda University, 11085 Campus Street, Mortensen Hall, Room 162, Loma Linda, CA 92350, USA. Department of Basic Sciences, Division of Biochemistry and Microbiology, Loma Linda University, Loma Linda, CA 92350, USA
Jonathan W. Neidigh, Email: jneidigh@llu.edu, Department of Basic Sciences, Division of Biochemistry and Microbiology, Loma Linda University, Loma Linda, CA 92350, USA
Nathan R. Wall, Email: nwall@llu.edu, Center for Health Disparities Research and Molecular Medicine, Loma Linda University, 11085 Campus Street, Mortensen Hall, Room 162, Loma Linda, CA 92350, USA. Department of Basic Sciences, Division of Biochemistry and Microbiology, Loma Linda University, Loma Linda, CA 92350, USA
References
- 1.Aznavoorian S, Stracke ML, Krutzsch H, Schiffmann E, Liotta LA. Signal transduction for chemotaxis and haptotaxis by matrix molecules in tumor cells. J Cell Biol. 1990;110:1427–1438. doi: 10.1083/jcb.110.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohtani H. Stromal reaction in cancer tissue: pathophysiologic significance of the expression of matrix-degrading enzymes in relation to matrix turnover and immune/inflammatory reactions. Pathol Int. 1998;48:1–9. doi: 10.1111/j.1440-1827.1998.tb03820.x. [DOI] [PubMed] [Google Scholar]
- 3.Graner MW, Cumming RI, Bigner DD. The heat shock response and chaperones/heat shock proteins in brain tumors: surface expression, release, and possible immune consequences. J Neurosci. 2007;27:11214–11227. doi: 10.1523/JNEUROSCI.3588-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eustace BK, Jay DG. Extracellular roles for the molecular chaperone, hsp90. Cell Cycle. 2004;3:1098–1100. [PubMed] [Google Scholar]
- 5.Radons J, Multhoff G. Immunostimulatory functions of membrane-bound and exported heat shock protein 70. Exerc Immunol Rev. 2005;11:17–33. [PubMed] [Google Scholar]
- 6.Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 7.Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 8.Altieri DC. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr Opin Cell Biol. 2006;18:609–615. doi: 10.1016/j.ceb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Fortugno P, Wall NR, Giodini A, et al. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci. 2002;115:575–585. doi: 10.1242/jcs.115.3.575. [DOI] [PubMed] [Google Scholar]
- 10.Khan S, Aspe JR, Asumen MG, et al. Extracellular, cell-permeable survivin inhibits apoptosis while promoting proliferative and metastatic potential. Br J Cancer. 2009;100:1073–1086. doi: 10.1038/sj.bjc.6604978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dohi T, Beltrami E, Wall NR, Plescia J, Altieri DC. Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J Clin Invest. 2004;114:1117–1127. doi: 10.1172/JCI22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh JC, Dohi T, Kang BH, Altieri DC. Hsp60 regulation of tumor cell apoptosis. J Biol Chem. 2008;283:5188–5194. doi: 10.1074/jbc.M705904200. [DOI] [PubMed] [Google Scholar]
- 13.Fortugno P, Beltrami E, Plescia J, et al. Regulation of survivin function by Hsp90. Proc Natl Acad Sci USA. 2003;100:13791–13796. doi: 10.1073/pnas.2434345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plescia J, Salz W, Xia F, et al. Rational design of shepherdin, a novel anticancer agent. Cancer Cell. 2005;7:457–468. doi: 10.1016/j.ccr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 15.Farsad K. Exosomes: novel organelles implicated in immunomodulation and apoptosis. Yale J Biol Med. 2002;75:95–101. [PMC free article] [PubMed] [Google Scholar]
- 16.Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 17.Hao QL, Shah AJ, Thiemann FT, Smogorzewska EM, Crooks GM. A functional comparison of CD34 + CD38− cells in cord blood and bone marrow. Blood. 1995;86:3745–3753. [PubMed] [Google Scholar]
- 18.Wang CY, Azzo W, Al-Katib A, Chiorazzi N, Knowles DM., 2nd Preparation and characterization of monoclonal antibodies recognizing three distinct differentiation antigens (BL1, BL2, BL3) on human B lymphocytes. J Immunol. 1984;133:684–691. [PubMed] [Google Scholar]
- 19.Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F-and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y, Sawada J, Sui G, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 21.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 22.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 24.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 25.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 26.Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 27.Thery C, Clayton A, Amigorena S, Raposo G. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;(Suppl 30):3.22.21–23.22.29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 28.Savina A, Vidal M, Colombo MI. The exosome pathway in K562 cells is regulated by Rab11. J Cell Sci. 2002;115:2505–2515. doi: 10.1242/jcs.115.12.2505. [DOI] [PubMed] [Google Scholar]
- 29.Clayton A, Court J, Navabi H, et al. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods. 2001;247:163–174. doi: 10.1016/s0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- 30.Liao DF, Jin ZG, Baas AS, et al. Purification and identification of secreted oxidative stress-induced factors from vascular smooth muscle cells. J Biol Chem. 2000;275:189–196. doi: 10.1074/jbc.275.1.189. [DOI] [PubMed] [Google Scholar]
- 31.Caroni P, Rothenfluh A, McGlynn E, Schneider C. S-cy-clophilin. New member of the cyclophilin family associated with the secretory pathway. J Biol Chem. 1991;266:10739–10742. [PubMed] [Google Scholar]
- 32.Sherry B, Yarlett N, Strupp A, Cerami A. Identification of cyclophilin as a proinflammatory secretory product of lipopolysaccharide-activated macrophages. Proc Natl Acad Sci USA. 1992;89:3511–3515. doi: 10.1073/pnas.89.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iero M, Valenti R, Huber V, et al. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 2008;15:80–88. doi: 10.1038/sj.cdd.4402237. [DOI] [PubMed] [Google Scholar]
- 34.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 35.Lancaster GI, Febbraio MA. Exosome-dependent trafficking of HSP70: a novel secretory pathway for cellular stress proteins. J Biol Chem. 2005;280:23349–23355. doi: 10.1074/jbc.M502017200. [DOI] [PubMed] [Google Scholar]
- 36.Johnstone RM. Exosomes biological significance: a concise review. Blood Cells Mol Dis. 2006;36:315–321. doi: 10.1016/j.bcmd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Clayton A, Turkes A, Navabi H, Mason MD, Tabi Z. Induction of heat shock proteins in B-cell exosomes. J Cell Sci. 2005;118:3631–3638. doi: 10.1242/jcs.02494. [DOI] [PubMed] [Google Scholar]
- 38.Lamparski HG, Metha-Damani A, Yao JY, et al. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270:211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 39.Gastpar R, Gehrmann M, Bausero MA, et al. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005;65:5238–5247. doi: 10.1158/0008-5472.CAN-04-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathew A, Bell A, Johnstone RM. Hsp-70 is closely associated with the transferrin receptor in exosomes from maturing reticulocytes. Biochem J. 1995;308(Pt 3):823–830. doi: 10.1042/bj3080823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tajika Y, Matsuzaki T, Suzuki T, et al. Differential regulation of AQP2 trafficking in endosomes by microtubules and actin filaments. Histochem Cell Biol. 2005;124:1–12. doi: 10.1007/s00418-005-0010-3. [DOI] [PubMed] [Google Scholar]
- 42.Takata K. Aquaporin-2 (AQP2): its intracellular compartment and trafficking. Cell Mol Biol (Noisy-le-grand) 2006;52:34–39. [PubMed] [Google Scholar]
- 43.Mignot G, Roux S, Thery C, Segura E, Zitvogel L. Prospects for exosomes in immunotherapy of cancer. J Cell Mol Med. 2006;10:376–388. doi: 10.1111/j.1582-4934.2006.tb00406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 45.Valentijn KM, Gumkowski FD, Jamieson JD. The sub-apical actin cytoskeleton regulates secretion and membrane retrieval in pancreatic acinar cells. J Cell Sci. 1999;112(Pt 1):81–96. doi: 10.1242/jcs.112.1.81. [DOI] [PubMed] [Google Scholar]
- 46.Matter K, Dreyer F, Aktories K. Actin involvement in exocytosis from PC12 cells: studies on the influence of botulinum C2 toxin on stimulated noradrenaline release. J Neurochem. 1989;52:370–376. doi: 10.1111/j.1471-4159.1989.tb09131.x. [DOI] [PubMed] [Google Scholar]
- 47.Sandvig K, van Deurs B. Selective modulation of the endocytic uptake of ricin and fluid phase markers without alteration in transferrin endocytosis. J Biol Chem. 1990;265:6382–6388. [PubMed] [Google Scholar]
- 48.Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000–5005. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 49.Galloway NR, Aspe JR, Sellers C, Wall NR. Enhanced antitumor effect of combined gemcitabine and proton radiation in the treatment of pancreatic cancer. Pancreas. 2009;38:782–790. doi: 10.1097/MPA.0b013e3181a85999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mera S, Magnusson M, Tarkowski A, Bokarewa M. Extracellular survivin up-regulates adhesion molecules on the surface of leukocytes changing their reactivity pattern. J Leukoc Biol. 2008;83:149–155. doi: 10.1189/jlb.0507287. [DOI] [PubMed] [Google Scholar]
- 51.Bokarewa M, Lindblad S, Bokarew D, Tarkowski A. Balance between survivin, a key member of the apoptosis inhibitor family, and its specific antibodies determines erosivity in rheumatoid arthritis. Arthritis Res Ther. 2005;7:R349–R358. doi: 10.1186/ar1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kapp EA, Schutz F, Connolly LM, et al. An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics. 2005;5:3475–3490. doi: 10.1002/pmic.200500126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.